Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
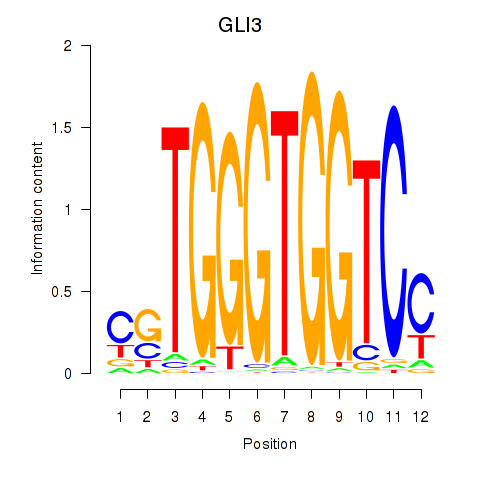
Results for GLI3
Z-value: 1.63
Transcription factors associated with GLI3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
GLI3
|
ENSG00000106571.8 | GLI3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| GLI3 | hg19_v2_chr7_-_42276612_42276782 | -0.58 | 1.3e-01 | Click! |
Activity profile of GLI3 motif
Sorted Z-values of GLI3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of GLI3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_21529811 | 5.37 |
ENST00000588004.1 |
LAMA3 |
laminin, alpha 3 |
| chr12_-_52887034 | 3.08 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr19_-_51456344 | 3.06 |
ENST00000336334.3 ENST00000593428.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr19_-_51456321 | 3.06 |
ENST00000391809.2 |
KLK5 |
kallikrein-related peptidase 5 |
| chr12_-_52845910 | 2.87 |
ENST00000252252.3 |
KRT6B |
keratin 6B |
| chr1_+_183155373 | 2.82 |
ENST00000493293.1 ENST00000264144.4 |
LAMC2 |
laminin, gamma 2 |
| chr6_+_125540951 | 2.70 |
ENST00000524679.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr1_-_160990886 | 2.57 |
ENST00000537746.1 |
F11R |
F11 receptor |
| chr19_+_35606692 | 2.46 |
ENST00000406242.3 ENST00000454903.2 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
| chr12_-_52867569 | 2.36 |
ENST00000252250.6 |
KRT6C |
keratin 6C |
| chr19_-_51456198 | 2.33 |
ENST00000594846.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr20_+_62327996 | 2.31 |
ENST00000369996.1 |
TNFRSF6B |
tumor necrosis factor receptor superfamily, member 6b, decoy |
| chr19_-_35992780 | 2.23 |
ENST00000593342.1 ENST00000601650.1 ENST00000408915.2 |
DMKN |
dermokine |
| chr16_+_68679193 | 2.18 |
ENST00000581171.1 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr13_-_20806440 | 2.04 |
ENST00000400066.3 ENST00000400065.3 ENST00000356192.6 |
GJB6 |
gap junction protein, beta 6, 30kDa |
| chr1_+_44399466 | 1.94 |
ENST00000498139.2 ENST00000491846.1 |
ARTN |
artemin |
| chr16_+_68771128 | 1.86 |
ENST00000261769.5 ENST00000422392.2 |
CDH1 |
cadherin 1, type 1, E-cadherin (epithelial) |
| chr20_+_44637526 | 1.80 |
ENST00000372330.3 |
MMP9 |
matrix metallopeptidase 9 (gelatinase B, 92kDa gelatinase, 92kDa type IV collagenase) |
| chr1_+_152956549 | 1.75 |
ENST00000307122.2 |
SPRR1A |
small proline-rich protein 1A |
| chr16_+_68678739 | 1.57 |
ENST00000264012.4 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr19_-_51522955 | 1.54 |
ENST00000358789.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr8_+_31497271 | 1.44 |
ENST00000520407.1 |
NRG1 |
neuregulin 1 |
| chr14_-_61748550 | 1.43 |
ENST00000555868.1 |
TMEM30B |
transmembrane protein 30B |
| chr1_+_233463507 | 1.39 |
ENST00000366623.3 ENST00000366624.3 |
MLK4 |
Mitogen-activated protein kinase kinase kinase MLK4 |
| chr11_+_32851487 | 1.39 |
ENST00000257836.3 |
PRRG4 |
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr12_-_8814669 | 1.35 |
ENST00000535411.1 ENST00000540087.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr16_-_68269971 | 1.34 |
ENST00000565858.1 |
ESRP2 |
epithelial splicing regulatory protein 2 |
| chr1_+_3388181 | 1.31 |
ENST00000418137.1 ENST00000413250.2 |
ARHGEF16 |
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr8_-_145642267 | 1.28 |
ENST00000301305.3 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
| chr7_-_41742697 | 1.27 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr15_+_40531621 | 1.27 |
ENST00000560346.1 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr11_+_69931519 | 1.25 |
ENST00000316296.5 ENST00000530676.1 |
ANO1 |
anoctamin 1, calcium activated chloride channel |
| chr12_-_54779511 | 1.24 |
ENST00000551109.1 ENST00000546970.1 |
ZNF385A |
zinc finger protein 385A |
| chr1_+_35220613 | 1.24 |
ENST00000338513.1 |
GJB5 |
gap junction protein, beta 5, 31.1kDa |
| chr15_+_90728145 | 1.21 |
ENST00000561085.1 ENST00000379122.3 ENST00000332496.6 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr10_+_71389983 | 1.21 |
ENST00000373279.4 |
C10orf35 |
chromosome 10 open reading frame 35 |
| chr8_+_124194875 | 1.20 |
ENST00000522648.1 ENST00000276699.6 |
FAM83A |
family with sequence similarity 83, member A |
| chr11_-_119991589 | 1.17 |
ENST00000526881.1 |
TRIM29 |
tripartite motif containing 29 |
| chr19_+_45312347 | 1.16 |
ENST00000270233.6 ENST00000591520.1 |
BCAM |
basal cell adhesion molecule (Lutheran blood group) |
| chr5_+_68788594 | 1.16 |
ENST00000396442.2 ENST00000380766.2 |
OCLN |
occludin |
| chr14_+_61788429 | 1.14 |
ENST00000332981.5 |
PRKCH |
protein kinase C, eta |
| chr1_+_26869597 | 1.14 |
ENST00000530003.1 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr4_-_90758227 | 1.14 |
ENST00000506691.1 ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr14_-_94856987 | 1.13 |
ENST00000449399.3 ENST00000404814.4 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr1_+_233765353 | 1.12 |
ENST00000366620.1 |
KCNK1 |
potassium channel, subfamily K, member 1 |
| chr6_-_32784687 | 1.11 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr10_-_116164239 | 1.10 |
ENST00000419268.1 ENST00000304129.4 ENST00000545353.1 |
AFAP1L2 |
actin filament associated protein 1-like 2 |
| chr16_+_57653854 | 1.10 |
ENST00000568908.1 ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr7_+_69064300 | 1.10 |
ENST00000342771.4 |
AUTS2 |
autism susceptibility candidate 2 |
| chr16_+_57653989 | 1.09 |
ENST00000567835.1 ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr1_-_201368707 | 1.09 |
ENST00000391967.2 |
LAD1 |
ladinin 1 |
| chr8_+_124194752 | 1.08 |
ENST00000318462.6 |
FAM83A |
family with sequence similarity 83, member A |
| chr13_-_20767037 | 1.06 |
ENST00000382848.4 |
GJB2 |
gap junction protein, beta 2, 26kDa |
| chr9_-_27529726 | 1.05 |
ENST00000262244.5 |
MOB3B |
MOB kinase activator 3B |
| chr6_-_46703069 | 1.02 |
ENST00000538237.1 ENST00000274793.7 |
PLA2G7 |
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr17_-_39928106 | 1.01 |
ENST00000540235.1 |
JUP |
junction plakoglobin |
| chr7_-_994302 | 1.01 |
ENST00000265846.5 |
ADAP1 |
ArfGAP with dual PH domains 1 |
| chr20_-_6104191 | 1.00 |
ENST00000217289.4 |
FERMT1 |
fermitin family member 1 |
| chr17_-_34122596 | 1.00 |
ENST00000250144.8 |
MMP28 |
matrix metallopeptidase 28 |
| chr15_-_72490114 | 0.96 |
ENST00000309731.7 |
GRAMD2 |
GRAM domain containing 2 |
| chrX_+_135618258 | 0.94 |
ENST00000440515.1 ENST00000456412.1 |
VGLL1 |
vestigial like 1 (Drosophila) |
| chr1_+_156123359 | 0.92 |
ENST00000368284.1 ENST00000368286.2 ENST00000438830.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr15_+_40532058 | 0.91 |
ENST00000260404.4 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr1_-_85358850 | 0.91 |
ENST00000370611.3 |
LPAR3 |
lysophosphatidic acid receptor 3 |
| chr10_-_116164450 | 0.90 |
ENST00000369271.3 |
AFAP1L2 |
actin filament associated protein 1-like 2 |
| chr8_+_21916710 | 0.90 |
ENST00000523266.1 ENST00000519907.1 |
DMTN |
dematin actin binding protein |
| chr17_+_73750699 | 0.89 |
ENST00000584939.1 |
ITGB4 |
integrin, beta 4 |
| chr8_+_98900132 | 0.89 |
ENST00000520016.1 |
MATN2 |
matrilin 2 |
| chr9_+_35673853 | 0.89 |
ENST00000378357.4 |
CA9 |
carbonic anhydrase IX |
| chr2_+_241625749 | 0.87 |
ENST00000407635.2 |
AC011298.2 |
AC011298.2 |
| chr11_-_88070920 | 0.86 |
ENST00000524463.1 ENST00000227266.5 |
CTSC |
cathepsin C |
| chr18_+_59992514 | 0.86 |
ENST00000269485.7 |
TNFRSF11A |
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
| chrX_+_37545012 | 0.86 |
ENST00000378616.3 |
XK |
X-linked Kx blood group (McLeod syndrome) |
| chr16_+_3115611 | 0.86 |
ENST00000530890.1 ENST00000444393.3 ENST00000533097.2 ENST00000008180.9 ENST00000396890.2 ENST00000525228.1 ENST00000548652.1 ENST00000525377.2 ENST00000530538.2 ENST00000549213.1 ENST00000552936.1 ENST00000548476.1 ENST00000552664.1 ENST00000552356.1 ENST00000551513.1 ENST00000382213.3 ENST00000548246.1 |
IL32 |
interleukin 32 |
| chr8_+_21916680 | 0.85 |
ENST00000358242.3 ENST00000415253.1 |
DMTN |
dematin actin binding protein |
| chr22_-_20255212 | 0.85 |
ENST00000416372.1 |
RTN4R |
reticulon 4 receptor |
| chr19_+_18284477 | 0.84 |
ENST00000407280.3 |
IFI30 |
interferon, gamma-inducible protein 30 |
| chr1_+_156123318 | 0.84 |
ENST00000368285.3 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr6_-_46703430 | 0.83 |
ENST00000537365.1 |
PLA2G7 |
phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) |
| chr15_+_45422178 | 0.82 |
ENST00000389037.3 ENST00000558322.1 |
DUOX1 |
dual oxidase 1 |
| chr8_-_17555164 | 0.82 |
ENST00000297488.6 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr3_-_128840604 | 0.82 |
ENST00000476465.1 ENST00000315150.5 ENST00000393304.1 ENST00000393308.1 ENST00000393307.1 ENST00000393305.1 |
RAB43 |
RAB43, member RAS oncogene family |
| chr18_+_59992527 | 0.81 |
ENST00000586569.1 |
TNFRSF11A |
tumor necrosis factor receptor superfamily, member 11a, NFKB activator |
| chr20_-_50808236 | 0.81 |
ENST00000361387.2 |
ZFP64 |
ZFP64 zinc finger protein |
| chr8_-_144660771 | 0.81 |
ENST00000449291.2 |
NAPRT1 |
nicotinate phosphoribosyltransferase domain containing 1 |
| chr1_+_152881014 | 0.79 |
ENST00000368764.3 ENST00000392667.2 |
IVL |
involucrin |
| chr1_+_15479054 | 0.79 |
ENST00000376014.3 ENST00000451326.2 |
TMEM51 |
transmembrane protein 51 |
| chr15_+_45422131 | 0.79 |
ENST00000321429.4 |
DUOX1 |
dual oxidase 1 |
| chr14_-_94857004 | 0.79 |
ENST00000557492.1 ENST00000448921.1 ENST00000437397.1 ENST00000355814.4 ENST00000393088.4 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr3_-_196756646 | 0.77 |
ENST00000439320.1 ENST00000296351.4 ENST00000296350.5 |
MFI2 |
antigen p97 (melanoma associated) identified by monoclonal antibodies 133.2 and 96.5 |
| chr20_+_49348081 | 0.77 |
ENST00000371610.2 |
PARD6B |
par-6 family cell polarity regulator beta |
| chr9_-_5304432 | 0.75 |
ENST00000416837.1 ENST00000308420.3 |
RLN2 |
relaxin 2 |
| chr1_+_43735646 | 0.74 |
ENST00000439858.1 |
TMEM125 |
transmembrane protein 125 |
| chr1_+_43735678 | 0.73 |
ENST00000432792.2 |
TMEM125 |
transmembrane protein 125 |
| chr14_-_94856951 | 0.73 |
ENST00000553327.1 ENST00000556955.1 ENST00000557118.1 ENST00000440909.1 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr1_+_11724167 | 0.72 |
ENST00000376753.4 |
FBXO6 |
F-box protein 6 |
| chr3_+_136537911 | 0.71 |
ENST00000393079.3 |
SLC35G2 |
solute carrier family 35, member G2 |
| chrX_-_78622805 | 0.71 |
ENST00000373298.2 |
ITM2A |
integral membrane protein 2A |
| chr1_+_15480197 | 0.71 |
ENST00000400796.3 ENST00000434578.2 ENST00000376008.2 |
TMEM51 |
transmembrane protein 51 |
| chr20_+_62185491 | 0.70 |
ENST00000370097.1 |
C20orf195 |
chromosome 20 open reading frame 195 |
| chr3_+_186648307 | 0.70 |
ENST00000457772.2 ENST00000455441.1 ENST00000427315.1 |
ST6GAL1 |
ST6 beta-galactosamide alpha-2,6-sialyltranferase 1 |
| chr1_-_153433120 | 0.69 |
ENST00000368723.3 |
S100A7 |
S100 calcium binding protein A7 |
| chr17_+_15848231 | 0.69 |
ENST00000304222.2 |
ADORA2B |
adenosine A2b receptor |
| chr19_+_41281416 | 0.68 |
ENST00000597140.1 |
MIA |
melanoma inhibitory activity |
| chr19_-_2041159 | 0.68 |
ENST00000589441.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chr16_+_30194916 | 0.68 |
ENST00000570045.1 ENST00000565497.1 ENST00000570244.1 |
CORO1A |
coronin, actin binding protein, 1A |
| chr1_+_15479021 | 0.68 |
ENST00000428417.1 |
TMEM51 |
transmembrane protein 51 |
| chr6_+_43739697 | 0.68 |
ENST00000230480.6 |
VEGFA |
vascular endothelial growth factor A |
| chr7_-_108096822 | 0.67 |
ENST00000379028.3 ENST00000413765.2 ENST00000379022.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr4_-_48908737 | 0.67 |
ENST00000381464.2 |
OCIAD2 |
OCIA domain containing 2 |
| chr15_+_81591757 | 0.66 |
ENST00000558332.1 |
IL16 |
interleukin 16 |
| chr6_-_134373732 | 0.65 |
ENST00000275230.5 |
SLC2A12 |
solute carrier family 2 (facilitated glucose transporter), member 12 |
| chr20_-_6103666 | 0.65 |
ENST00000536936.1 |
FERMT1 |
fermitin family member 1 |
| chr7_-_24797546 | 0.63 |
ENST00000414428.1 ENST00000419307.1 ENST00000342947.3 |
DFNA5 |
deafness, autosomal dominant 5 |
| chr10_-_105615164 | 0.63 |
ENST00000355946.2 ENST00000369774.4 |
SH3PXD2A |
SH3 and PX domains 2A |
| chr19_+_41281282 | 0.62 |
ENST00000263369.3 |
MIA |
melanoma inhibitory activity |
| chr11_-_88070896 | 0.62 |
ENST00000529974.1 ENST00000527018.1 |
CTSC |
cathepsin C |
| chr20_-_1306351 | 0.61 |
ENST00000381812.1 |
SDCBP2 |
syndecan binding protein (syntenin) 2 |
| chr19_+_41281060 | 0.61 |
ENST00000594436.1 ENST00000597784.1 |
MIA |
melanoma inhibitory activity |
| chr4_-_89080003 | 0.61 |
ENST00000237612.3 |
ABCG2 |
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chrX_+_51149767 | 0.60 |
ENST00000342995.2 |
CXorf67 |
chromosome X open reading frame 67 |
| chr6_-_33756867 | 0.59 |
ENST00000293760.5 |
LEMD2 |
LEM domain containing 2 |
| chr5_+_65892174 | 0.58 |
ENST00000404260.3 ENST00000403625.2 ENST00000406374.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr1_+_40420802 | 0.58 |
ENST00000372811.5 ENST00000420632.2 ENST00000434861.1 ENST00000372809.5 |
MFSD2A |
major facilitator superfamily domain containing 2A |
| chr14_-_92302784 | 0.58 |
ENST00000340892.5 ENST00000360594.5 |
TC2N |
tandem C2 domains, nuclear |
| chr1_-_62785054 | 0.58 |
ENST00000371153.4 |
KANK4 |
KN motif and ankyrin repeat domains 4 |
| chr19_-_35981358 | 0.58 |
ENST00000484218.2 ENST00000338897.3 |
KRTDAP |
keratinocyte differentiation-associated protein |
| chr8_+_110374683 | 0.57 |
ENST00000378402.5 |
PKHD1L1 |
polycystic kidney and hepatic disease 1 (autosomal recessive)-like 1 |
| chr2_-_241080069 | 0.56 |
ENST00000319460.1 |
OTOS |
otospiralin |
| chr20_-_1306391 | 0.56 |
ENST00000339987.3 |
SDCBP2 |
syndecan binding protein (syntenin) 2 |
| chr11_+_121461097 | 0.56 |
ENST00000527934.1 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr12_-_54778471 | 0.55 |
ENST00000550120.1 ENST00000394313.2 ENST00000547210.1 |
ZNF385A |
zinc finger protein 385A |
| chr12_+_7055767 | 0.55 |
ENST00000447931.2 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr2_+_131769256 | 0.55 |
ENST00000355771.3 |
ARHGEF4 |
Rho guanine nucleotide exchange factor (GEF) 4 |
| chr7_-_108096765 | 0.53 |
ENST00000379024.4 ENST00000351718.4 |
NRCAM |
neuronal cell adhesion molecule |
| chr19_+_45417812 | 0.53 |
ENST00000592535.1 |
APOC1 |
apolipoprotein C-I |
| chr1_+_13910194 | 0.53 |
ENST00000376057.4 ENST00000510906.1 |
PDPN |
podoplanin |
| chr4_-_48908822 | 0.52 |
ENST00000508632.1 |
OCIAD2 |
OCIA domain containing 2 |
| chr18_+_11752040 | 0.52 |
ENST00000423027.3 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr3_-_48470838 | 0.52 |
ENST00000358459.4 ENST00000358536.4 |
PLXNB1 |
plexin B1 |
| chr17_-_39274606 | 0.52 |
ENST00000391413.2 |
KRTAP4-11 |
keratin associated protein 4-11 |
| chr12_+_57984965 | 0.52 |
ENST00000540759.2 ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C |
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr4_-_48908805 | 0.50 |
ENST00000273860.4 |
OCIAD2 |
OCIA domain containing 2 |
| chr1_-_1356628 | 0.49 |
ENST00000442470.1 ENST00000537107.1 |
ANKRD65 |
ankyrin repeat domain 65 |
| chr12_+_113354341 | 0.48 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_-_38273840 | 0.48 |
ENST00000373044.2 |
YRDC |
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr8_-_17533838 | 0.47 |
ENST00000400046.1 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr19_-_16653226 | 0.47 |
ENST00000198939.6 |
CHERP |
calcium homeostasis endoplasmic reticulum protein |
| chr21_-_27945562 | 0.47 |
ENST00000299340.4 ENST00000435845.2 |
CYYR1 |
cysteine/tyrosine-rich 1 |
| chr17_-_4463856 | 0.46 |
ENST00000574584.1 ENST00000381550.3 ENST00000301395.3 |
GGT6 |
gamma-glutamyltransferase 6 |
| chr16_+_2521500 | 0.46 |
ENST00000293973.1 |
NTN3 |
netrin 3 |
| chr16_+_27325202 | 0.46 |
ENST00000395762.2 ENST00000562142.1 ENST00000561742.1 ENST00000543915.2 ENST00000449195.1 ENST00000380922.3 ENST00000563002.1 |
IL4R |
interleukin 4 receptor |
| chrX_+_106449862 | 0.46 |
ENST00000372453.3 ENST00000535523.1 |
PIH1D3 |
PIH1 domain containing 3 |
| chr6_-_105627735 | 0.45 |
ENST00000254765.3 |
POPDC3 |
popeye domain containing 3 |
| chr4_-_38666430 | 0.44 |
ENST00000436901.1 |
AC021860.1 |
Uncharacterized protein |
| chr19_+_45417921 | 0.44 |
ENST00000252491.4 ENST00000592885.1 ENST00000589781.1 |
APOC1 |
apolipoprotein C-I |
| chr19_-_51472031 | 0.44 |
ENST00000391808.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chrX_-_100914781 | 0.44 |
ENST00000431597.1 ENST00000458024.1 ENST00000413506.1 ENST00000440675.1 ENST00000328766.5 ENST00000356824.4 |
ARMCX2 |
armadillo repeat containing, X-linked 2 |
| chr12_-_125348329 | 0.44 |
ENST00000546215.1 ENST00000415380.2 ENST00000261693.6 ENST00000376788.1 ENST00000545493.1 |
SCARB1 |
scavenger receptor class B, member 1 |
| chr2_+_192109911 | 0.44 |
ENST00000418908.1 ENST00000339514.4 ENST00000392318.3 |
MYO1B |
myosin IB |
| chr12_-_125348448 | 0.44 |
ENST00000339570.5 |
SCARB1 |
scavenger receptor class B, member 1 |
| chr19_-_16653325 | 0.44 |
ENST00000546361.2 |
CHERP |
calcium homeostasis endoplasmic reticulum protein |
| chr19_+_45417504 | 0.44 |
ENST00000588750.1 ENST00000588802.1 |
APOC1 |
apolipoprotein C-I |
| chr20_-_22566089 | 0.43 |
ENST00000377115.4 |
FOXA2 |
forkhead box A2 |
| chr2_+_238768187 | 0.43 |
ENST00000254661.4 ENST00000409726.1 |
RAMP1 |
receptor (G protein-coupled) activity modifying protein 1 |
| chr20_+_55204351 | 0.43 |
ENST00000201031.2 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr9_+_124413873 | 0.43 |
ENST00000408936.3 |
DAB2IP |
DAB2 interacting protein |
| chr17_+_38333263 | 0.42 |
ENST00000456989.2 ENST00000543876.1 ENST00000544503.1 ENST00000264644.6 ENST00000538884.1 |
RAPGEFL1 |
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr7_-_24797032 | 0.42 |
ENST00000409970.1 ENST00000409775.3 |
DFNA5 |
deafness, autosomal dominant 5 |
| chr19_-_11689752 | 0.42 |
ENST00000592659.1 ENST00000592828.1 ENST00000218758.5 ENST00000412435.2 |
ACP5 |
acid phosphatase 5, tartrate resistant |
| chr17_+_7531281 | 0.42 |
ENST00000575729.1 ENST00000340624.5 |
SHBG |
sex hormone-binding globulin |
| chr8_-_145331153 | 0.42 |
ENST00000377412.4 |
KM-PA-2 |
KM-PA-2 protein; Uncharacterized protein |
| chr14_+_105147464 | 0.42 |
ENST00000540171.2 |
RP11-982M15.6 |
RP11-982M15.6 |
| chr12_-_323689 | 0.41 |
ENST00000428720.1 |
SLC6A12 |
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr19_+_38810447 | 0.41 |
ENST00000263372.3 |
KCNK6 |
potassium channel, subfamily K, member 6 |
| chr19_-_2050852 | 0.40 |
ENST00000541165.1 ENST00000591601.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chr6_-_17987694 | 0.40 |
ENST00000378814.5 ENST00000378843.2 ENST00000378826.2 ENST00000378816.5 ENST00000259711.6 ENST00000502704.1 |
KIF13A |
kinesin family member 13A |
| chr7_-_112430427 | 0.40 |
ENST00000449743.1 ENST00000441474.1 ENST00000454074.1 ENST00000447395.1 |
TMEM168 |
transmembrane protein 168 |
| chr17_-_79620721 | 0.39 |
ENST00000571004.1 |
PDE6G |
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr1_-_205782304 | 0.39 |
ENST00000367137.3 |
SLC41A1 |
solute carrier family 41 (magnesium transporter), member 1 |
| chr12_+_7055631 | 0.39 |
ENST00000543115.1 ENST00000399448.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr9_-_5339873 | 0.39 |
ENST00000223862.1 ENST00000223858.4 |
RLN1 |
relaxin 1 |
| chr14_-_21566731 | 0.38 |
ENST00000360947.3 |
ZNF219 |
zinc finger protein 219 |
| chr19_+_45418067 | 0.38 |
ENST00000589078.1 ENST00000586638.1 |
APOC1 |
apolipoprotein C-I |
| chr22_+_31644388 | 0.38 |
ENST00000333611.4 ENST00000340552.4 |
LIMK2 |
LIM domain kinase 2 |
| chr2_-_96781984 | 0.38 |
ENST00000409345.3 |
ADRA2B |
adrenoceptor alpha 2B |
| chr19_-_20844343 | 0.38 |
ENST00000595405.1 |
ZNF626 |
zinc finger protein 626 |
| chr2_+_238767517 | 0.38 |
ENST00000404910.2 |
RAMP1 |
receptor (G protein-coupled) activity modifying protein 1 |
| chr19_-_38746979 | 0.37 |
ENST00000591291.1 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr8_+_103563792 | 0.37 |
ENST00000285402.3 |
ODF1 |
outer dense fiber of sperm tails 1 |
| chr20_-_50722183 | 0.37 |
ENST00000371523.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr21_-_38445470 | 0.37 |
ENST00000399098.1 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr20_-_22565101 | 0.37 |
ENST00000419308.2 |
FOXA2 |
forkhead box A2 |
| chr5_+_6583380 | 0.37 |
ENST00000507582.1 |
LINC01018 |
long intergenic non-protein coding RNA 1018 |
| chr3_-_59035673 | 0.37 |
ENST00000491845.1 ENST00000472469.1 ENST00000471288.1 ENST00000295966.7 |
C3orf67 |
chromosome 3 open reading frame 67 |
| chr1_-_161014731 | 0.36 |
ENST00000368020.1 |
USF1 |
upstream transcription factor 1 |
| chr4_-_7069760 | 0.36 |
ENST00000264954.4 |
GRPEL1 |
GrpE-like 1, mitochondrial (E. coli) |
| chr15_-_76005170 | 0.36 |
ENST00000308508.5 |
CSPG4 |
chondroitin sulfate proteoglycan 4 |
| chr8_-_142011036 | 0.36 |
ENST00000520892.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr1_-_20834586 | 0.36 |
ENST00000264198.3 |
MUL1 |
mitochondrial E3 ubiquitin protein ligase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 8.5 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 1.3 | 3.8 | GO:0060901 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.8 | 1.7 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.8 | 3.1 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.6 | 1.7 | GO:0100009 | circadian temperature homeostasis(GO:0060086) regulation of fever generation by regulation of prostaglandin secretion(GO:0071810) positive regulation of fever generation by positive regulation of prostaglandin secretion(GO:0071812) positive regulation of ERK1 and ERK2 cascade via TNFSF11-mediated signaling(GO:0071848) regulation of fever generation by prostaglandin secretion(GO:0100009) |
| 0.4 | 1.8 | GO:0010900 | negative regulation of phosphatidylcholine catabolic process(GO:0010900) |
| 0.4 | 1.3 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.4 | 1.9 | GO:0097021 | lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.4 | 1.1 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.4 | 2.2 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.4 | 1.1 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.4 | 1.1 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.4 | 1.8 | GO:1902162 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.3 | 2.9 | GO:0071680 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.3 | 1.3 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.3 | 9.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.3 | 1.2 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.3 | 1.8 | GO:0034441 | plasma lipoprotein particle oxidation(GO:0034441) |
| 0.3 | 0.9 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.3 | 1.8 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.3 | 0.9 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.3 | 0.8 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.3 | 1.6 | GO:0042335 | cuticle development(GO:0042335) |
| 0.3 | 0.8 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.3 | 1.1 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.2 | 0.9 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.2 | 1.1 | GO:0051621 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 0.7 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.2 | 1.2 | GO:0061502 | early endosome to recycling endosome transport(GO:0061502) |
| 0.2 | 1.4 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.2 | 1.0 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 | 1.7 | GO:2001268 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:2001268) |
| 0.2 | 0.5 | GO:1904328 | positive regulation of platelet aggregation(GO:1901731) regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.2 | 0.5 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.2 | 1.6 | GO:0015705 | iodide transport(GO:0015705) |
| 0.2 | 0.7 | GO:0002545 | chronic inflammatory response to non-antigenic stimulus(GO:0002545) regulation of chronic inflammatory response to non-antigenic stimulus(GO:0002880) |
| 0.2 | 1.1 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.2 | 0.5 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.2 | 0.3 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.1 | 0.7 | GO:0002317 | plasma cell differentiation(GO:0002317) |
| 0.1 | 2.6 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.1 | 1.5 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.5 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.3 | GO:1903970 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) |
| 0.1 | 0.6 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.1 | 0.3 | GO:0048597 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.1 | 1.0 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.1 | 0.3 | GO:0030264 | nuclear fragmentation involved in apoptotic nuclear change(GO:0030264) |
| 0.1 | 0.4 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.1 | 0.6 | GO:0072675 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 0.3 | GO:0051795 | positive regulation of catagen(GO:0051795) |
| 0.1 | 1.4 | GO:0070863 | positive regulation of protein exit from endoplasmic reticulum(GO:0070863) |
| 0.1 | 0.8 | GO:0050689 | negative regulation of defense response to virus by host(GO:0050689) |
| 0.1 | 0.3 | GO:1904580 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.1 | 1.4 | GO:0045162 | clustering of voltage-gated sodium channels(GO:0045162) |
| 0.1 | 0.8 | GO:0035771 | interleukin-4-mediated signaling pathway(GO:0035771) |
| 0.1 | 0.1 | GO:0014839 | myoblast migration involved in skeletal muscle regeneration(GO:0014839) |
| 0.1 | 0.4 | GO:0035625 | negative regulation of epinephrine secretion(GO:0032811) epidermal growth factor-activated receptor transactivation by G-protein coupled receptor signaling pathway(GO:0035625) |
| 0.1 | 0.3 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.9 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.1 | 0.8 | GO:0097646 | calcitonin family receptor signaling pathway(GO:0097646) amylin receptor signaling pathway(GO:0097647) |
| 0.1 | 2.9 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.1 | 0.4 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.8 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 0.4 | GO:0010961 | cellular magnesium ion homeostasis(GO:0010961) |
| 0.1 | 0.5 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.1 | 0.2 | GO:0006788 | heme oxidation(GO:0006788) |
| 0.1 | 0.6 | GO:0044821 | meiotic telomere tethering at nuclear periphery(GO:0044821) meiotic attachment of telomere to nuclear envelope(GO:0070197) chromosome attachment to the nuclear envelope(GO:0097240) |
| 0.1 | 0.4 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.1 | 0.2 | GO:2000979 | hepatocyte cell migration(GO:0002194) otic placode formation(GO:0043049) branching involved in pancreas morphogenesis(GO:0061114) acinar cell differentiation(GO:0090425) positive regulation of forebrain neuron differentiation(GO:2000979) |
| 0.1 | 0.2 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.1 | 0.2 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.1 | 0.3 | GO:0000432 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.1 | 0.7 | GO:0010820 | positive regulation of T cell chemotaxis(GO:0010820) |
| 0.1 | 1.2 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.1 | 0.4 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.5 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.1 | 1.6 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.1 | 0.6 | GO:0043542 | endothelial cell migration(GO:0043542) |
| 0.1 | 0.2 | GO:1904199 | positive regulation of regulation of vascular smooth muscle cell membrane depolarization(GO:1904199) regulation of vascular smooth muscle cell membrane depolarization(GO:1990736) |
| 0.1 | 0.2 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.2 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.1 | 0.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 0.4 | GO:0070649 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.1 | 0.2 | GO:0051039 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 0.2 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.1 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.1 | 0.1 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 1.5 | GO:0045063 | T-helper 1 cell differentiation(GO:0045063) |
| 0.1 | 0.5 | GO:0019344 | cysteine biosynthetic process(GO:0019344) |
| 0.1 | 0.2 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.1 | 0.2 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.1 | 0.3 | GO:0033133 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.3 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.7 | GO:0006054 | N-acetylneuraminate metabolic process(GO:0006054) |
| 0.0 | 0.1 | GO:2001301 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.1 | GO:1901301 | regulation of cargo loading into COPII-coated vesicle(GO:1901301) |
| 0.0 | 0.2 | GO:1903412 | response to bile acid(GO:1903412) |
| 0.0 | 0.1 | GO:1901856 | negative regulation of cellular respiration(GO:1901856) |
| 0.0 | 0.2 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.0 | 0.3 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.0 | 2.5 | GO:0045742 | positive regulation of epidermal growth factor receptor signaling pathway(GO:0045742) |
| 0.0 | 0.2 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.5 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0006423 | cysteinyl-tRNA aminoacylation(GO:0006423) |
| 0.0 | 0.2 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.5 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.2 | GO:0071051 | polyadenylation-dependent snoRNA 3'-end processing(GO:0071051) |
| 0.0 | 0.2 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 1.5 | GO:0060445 | branching involved in salivary gland morphogenesis(GO:0060445) |
| 0.0 | 0.3 | GO:0061042 | vascular wound healing(GO:0061042) |
| 0.0 | 0.2 | GO:2000051 | Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.2 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.1 | GO:0014016 | neuroblast differentiation(GO:0014016) |
| 0.0 | 0.1 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 0.1 | GO:0002625 | regulation of T cell antigen processing and presentation(GO:0002625) |
| 0.0 | 0.2 | GO:0051562 | negative regulation of mitochondrial calcium ion concentration(GO:0051562) |
| 0.0 | 0.2 | GO:0021769 | orbitofrontal cortex development(GO:0021769) glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.0 | 0.6 | GO:0060856 | establishment of blood-brain barrier(GO:0060856) |
| 0.0 | 0.2 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.4 | GO:0034625 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0086048 | membrane depolarization during bundle of His cell action potential(GO:0086048) |
| 0.0 | 0.2 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.2 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.0 | 1.9 | GO:0001913 | T cell mediated cytotoxicity(GO:0001913) |
| 0.0 | 0.2 | GO:1901535 | regulation of DNA demethylation(GO:1901535) |
| 0.0 | 0.1 | GO:2000282 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) regulation of cellular amino acid biosynthetic process(GO:2000282) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.0 | 0.2 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.0 | 0.5 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.2 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 1.9 | GO:0045104 | intermediate filament cytoskeleton organization(GO:0045104) |
| 0.0 | 0.1 | GO:0032079 | positive regulation of endodeoxyribonuclease activity(GO:0032079) |
| 0.0 | 0.1 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.2 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.4 | GO:0038007 | netrin-activated signaling pathway(GO:0038007) |
| 0.0 | 0.1 | GO:0090267 | positive regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090267) |
| 0.0 | 0.1 | GO:0030187 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 0.1 | GO:0050882 | voluntary musculoskeletal movement(GO:0050882) |
| 0.0 | 0.3 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.7 | GO:0006516 | glycoprotein catabolic process(GO:0006516) |
| 0.0 | 0.2 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.1 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.3 | GO:0051151 | negative regulation of smooth muscle cell differentiation(GO:0051151) |
| 0.0 | 0.1 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 1.4 | GO:0090004 | positive regulation of establishment of protein localization to plasma membrane(GO:0090004) |
| 0.0 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) positive regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000969) |
| 0.0 | 0.2 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.0 | GO:0031338 | regulation of vesicle fusion(GO:0031338) |
| 0.0 | 0.1 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.1 | GO:0032661 | regulation of interleukin-18 production(GO:0032661) |
| 0.0 | 0.5 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.3 | GO:0032926 | negative regulation of activin receptor signaling pathway(GO:0032926) |
| 0.0 | 0.6 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.2 | GO:0070315 | G1 to G0 transition involved in cell differentiation(GO:0070315) |
| 0.0 | 0.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 1.6 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.2 | GO:0032310 | prostaglandin secretion(GO:0032310) |
| 0.0 | 0.5 | GO:0048681 | negative regulation of axon regeneration(GO:0048681) |
| 0.0 | 0.1 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.3 | GO:0097012 | cellular response to granulocyte macrophage colony-stimulating factor stimulus(GO:0097011) response to granulocyte macrophage colony-stimulating factor(GO:0097012) |
| 0.0 | 1.8 | GO:0006953 | acute-phase response(GO:0006953) |
| 0.0 | 0.6 | GO:0002523 | leukocyte migration involved in inflammatory response(GO:0002523) |
| 0.0 | 0.1 | GO:0019747 | regulation of isoprenoid metabolic process(GO:0019747) |
| 0.0 | 0.5 | GO:0060294 | cilium movement involved in cell motility(GO:0060294) |
| 0.0 | 0.1 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 1.7 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.3 | GO:0097500 | receptor localization to nonmotile primary cilium(GO:0097500) protein localization to photoreceptor outer segment(GO:1903546) |
| 0.0 | 0.1 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.2 | GO:0010310 | regulation of hydrogen peroxide metabolic process(GO:0010310) |
| 0.0 | 0.2 | GO:0051131 | chaperone-mediated protein complex assembly(GO:0051131) |
| 0.0 | 0.1 | GO:0071442 | positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.4 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0007256 | activation of JNKK activity(GO:0007256) |
| 0.0 | 0.1 | GO:0034653 | diterpenoid catabolic process(GO:0016103) retinoic acid catabolic process(GO:0034653) |
| 0.0 | 0.0 | GO:0046855 | phosphorylated carbohydrate dephosphorylation(GO:0046838) inositol phosphate dephosphorylation(GO:0046855) |
| 0.0 | 0.3 | GO:1990403 | embryonic brain development(GO:1990403) |
| 0.0 | 0.3 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.2 | GO:0070236 | regulation of activation-induced cell death of T cells(GO:0070235) negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.6 | GO:0090023 | positive regulation of neutrophil chemotaxis(GO:0090023) |
| 0.0 | 0.1 | GO:0016199 | axon midline choice point recognition(GO:0016199) |
| 0.0 | 0.2 | GO:1903898 | negative regulation of PERK-mediated unfolded protein response(GO:1903898) |
| 0.0 | 0.1 | GO:0006121 | mitochondrial electron transport, succinate to ubiquinone(GO:0006121) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0098914 | membrane repolarization during atrial cardiac muscle cell action potential(GO:0098914) |
| 0.0 | 0.2 | GO:0060836 | lymphatic endothelial cell differentiation(GO:0060836) |
| 0.0 | 0.5 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.3 | GO:0002862 | negative regulation of inflammatory response to antigenic stimulus(GO:0002862) |
| 0.0 | 0.2 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0006864 | pyrimidine nucleotide transport(GO:0006864) mitochondrial pyrimidine nucleotide import(GO:1990519) |
| 0.0 | 0.3 | GO:1900864 | mitochondrial tRNA modification(GO:0070900) mitochondrial RNA modification(GO:1900864) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.2 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.0 | 0.2 | GO:0007341 | penetration of zona pellucida(GO:0007341) |
| 0.0 | 0.4 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.2 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.2 | GO:0042476 | odontogenesis(GO:0042476) |
| 0.0 | 0.2 | GO:0072540 | T-helper 17 cell lineage commitment(GO:0072540) |
| 0.0 | 0.1 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:0019896 | axonal transport of mitochondrion(GO:0019896) |
| 0.0 | 0.1 | GO:0033629 | negative regulation of cell adhesion mediated by integrin(GO:0033629) |
| 0.0 | 0.1 | GO:0045799 | positive regulation of chromatin assembly or disassembly(GO:0045799) |
| 0.0 | 0.1 | GO:0097501 | detoxification of inorganic compound(GO:0061687) stress response to metal ion(GO:0097501) |
| 0.0 | 1.1 | GO:0006730 | one-carbon metabolic process(GO:0006730) |
| 0.0 | 0.4 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:0098989 | NMDA selective glutamate receptor signaling pathway(GO:0098989) |
| 0.0 | 0.2 | GO:0016127 | cholesterol catabolic process(GO:0006707) sterol catabolic process(GO:0016127) |
| 0.0 | 0.1 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.0 | 0.2 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.9 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.0 | 0.2 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.0 | 1.1 | GO:0008347 | glial cell migration(GO:0008347) |
| 0.0 | 1.0 | GO:0006370 | 7-methylguanosine mRNA capping(GO:0006370) |
| 0.0 | 0.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.3 | GO:0048096 | chromatin-mediated maintenance of transcription(GO:0048096) histone H3-K4 trimethylation(GO:0080182) |
| 0.0 | 0.1 | GO:0000422 | mitophagy(GO:0000422) mitochondrion disassembly(GO:0061726) |
| 0.0 | 0.1 | GO:0009452 | 7-methylguanosine RNA capping(GO:0009452) RNA capping(GO:0036260) |
| 0.0 | 0.2 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.2 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0046707 | IDP metabolic process(GO:0046707) IDP catabolic process(GO:0046709) |
| 0.0 | 0.1 | GO:0018199 | peptidyl-glutamine modification(GO:0018199) |
| 0.0 | 0.4 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.0 | GO:0015701 | bicarbonate transport(GO:0015701) |
| 0.0 | 0.2 | GO:0006703 | estrogen biosynthetic process(GO:0006703) |
| 0.0 | 0.1 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.0 | 0.1 | GO:0000727 | double-strand break repair via break-induced replication(GO:0000727) |
| 0.0 | 0.4 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:1902570 | protein localization to nucleolus(GO:1902570) |
| 0.0 | 0.0 | GO:1903897 | regulation of PERK-mediated unfolded protein response(GO:1903897) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0010748 | regulation of plasma membrane long-chain fatty acid transport(GO:0010746) negative regulation of plasma membrane long-chain fatty acid transport(GO:0010748) |
| 0.0 | 0.1 | GO:0042350 | GDP-L-fucose biosynthetic process(GO:0042350) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0060482 | lobar bronchus epithelium development(GO:0060481) lobar bronchus development(GO:0060482) |
| 0.0 | 0.1 | GO:0072642 | type I interferon secretion(GO:0072641) interferon-alpha secretion(GO:0072642) regulation of interferon-alpha secretion(GO:1902739) positive regulation of interferon-alpha secretion(GO:1902741) |
| 0.0 | 2.3 | GO:0046330 | positive regulation of JNK cascade(GO:0046330) |
| 0.0 | 0.1 | GO:0016584 | nucleosome positioning(GO:0016584) |
| 0.0 | 0.3 | GO:0000183 | chromatin silencing at rDNA(GO:0000183) |
| 0.0 | 0.0 | GO:0050822 | peptide stabilization(GO:0050822) peptide antigen stabilization(GO:0050823) |
| 0.0 | 0.2 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.6 | GO:0006506 | GPI anchor biosynthetic process(GO:0006506) |
| 0.0 | 0.1 | GO:0021683 | cerebellar granular layer morphogenesis(GO:0021683) |
| 0.0 | 0.5 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.2 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.2 | GO:0000466 | maturation of 5.8S rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000466) |
| 0.0 | 0.4 | GO:0007214 | gamma-aminobutyric acid signaling pathway(GO:0007214) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.5 | GO:0007411 | axon guidance(GO:0007411) |
| 0.0 | 0.0 | GO:0071140 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.1 | GO:0072383 | plus-end-directed vesicle transport along microtubule(GO:0072383) plus-end-directed organelle transport along microtubule(GO:0072386) |
| 0.0 | 0.0 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.2 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.2 | GO:1903204 | negative regulation of oxidative stress-induced neuron death(GO:1903204) |
| 0.0 | 0.1 | GO:0015014 | heparan sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0015014) |
| 0.0 | 0.1 | GO:0044342 | type B pancreatic cell proliferation(GO:0044342) |
| 0.0 | 0.1 | GO:0016553 | base conversion or substitution editing(GO:0016553) |
| 0.0 | 0.3 | GO:0050812 | regulation of acyl-CoA biosynthetic process(GO:0050812) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.0 | GO:2000118 | regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.0 | 0.3 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.2 | GO:0032098 | regulation of appetite(GO:0032098) |
| 0.0 | 0.1 | GO:0006465 | signal peptide processing(GO:0006465) |
| 0.0 | 0.1 | GO:0048548 | regulation of pinocytosis(GO:0048548) positive regulation of pinocytosis(GO:0048549) |
| 0.0 | 0.2 | GO:0006265 | DNA topological change(GO:0006265) |
| 0.0 | 0.3 | GO:0045948 | positive regulation of translational initiation(GO:0045948) |
| 0.0 | 0.2 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 1.6 | GO:0060333 | interferon-gamma-mediated signaling pathway(GO:0060333) |
| 0.0 | 0.0 | GO:0033967 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.9 | GO:0060113 | inner ear receptor cell differentiation(GO:0060113) |
| 0.0 | 0.1 | GO:0060285 | cilium-dependent cell motility(GO:0060285) |
| 0.0 | 0.0 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.1 | GO:0007352 | zygotic specification of dorsal/ventral axis(GO:0007352) |
| 0.0 | 0.2 | GO:0006346 | methylation-dependent chromatin silencing(GO:0006346) |
| 0.0 | 0.1 | GO:0043045 | DNA methylation involved in embryo development(GO:0043045) changes to DNA methylation involved in embryo development(GO:1901538) |
| 0.0 | 0.3 | GO:0070286 | axonemal dynein complex assembly(GO:0070286) |
| 0.0 | 0.1 | GO:0071285 | cellular response to lithium ion(GO:0071285) |
| 0.0 | 1.3 | GO:1903779 | regulation of cardiac conduction(GO:1903779) |
| 0.0 | 0.0 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.2 | GO:0001522 | pseudouridine synthesis(GO:0001522) |
| 0.0 | 0.3 | GO:0036150 | phosphatidylserine acyl-chain remodeling(GO:0036150) |
| 0.0 | 0.1 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 1.2 | GO:0007173 | epidermal growth factor receptor signaling pathway(GO:0007173) |
| 0.0 | 0.1 | GO:0034199 | activation of protein kinase A activity(GO:0034199) negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.2 | GO:0097034 | mitochondrial respiratory chain complex IV assembly(GO:0033617) mitochondrial respiratory chain complex IV biogenesis(GO:0097034) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 8.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.9 | 2.8 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.7 | 5.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 1.3 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 2.9 | GO:0016342 | catenin complex(GO:0016342) |
| 0.2 | 2.0 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.2 | 1.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.2 | 4.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 1.8 | GO:0042627 | chylomicron(GO:0042627) |
| 0.1 | 0.4 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.1 | 0.8 | GO:1903440 | calcitonin family receptor complex(GO:1903439) amylin receptor complex(GO:1903440) |
| 0.1 | 9.5 | GO:0045095 | keratin filament(GO:0045095) |
| 0.1 | 2.1 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.6 | GO:0034688 | integrin alphaM-beta2 complex(GO:0034688) |
| 0.1 | 0.9 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.1 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.1 | 0.6 | GO:0098560 | cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.1 | 2.2 | GO:0034362 | low-density lipoprotein particle(GO:0034362) |
| 0.1 | 0.4 | GO:0001520 | outer dense fiber(GO:0001520) |
| 0.1 | 0.4 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 2.0 | GO:0031528 | microvillus membrane(GO:0031528) |
| 0.1 | 0.2 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.1 | 0.2 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.1 | 1.2 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.1 | 0.2 | GO:0016938 | kinesin I complex(GO:0016938) |
| 0.1 | 0.3 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.1 | 1.3 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.0 | 0.1 | GO:0042022 | interleukin-12 receptor complex(GO:0042022) |
| 0.0 | 1.0 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.1 | GO:0030895 | apolipoprotein B mRNA editing enzyme complex(GO:0030895) |
| 0.0 | 0.9 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.9 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.3 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 2.5 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 1.1 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 1.0 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 3.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 4.8 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.8 | GO:0031362 | anchored component of external side of plasma membrane(GO:0031362) |
| 0.0 | 0.3 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 1.6 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.5 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.4 | GO:0016589 | NURF complex(GO:0016589) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 1.3 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.4 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0045273 | mitochondrial respiratory chain complex II, succinate dehydrogenase complex (ubiquinone)(GO:0005749) succinate dehydrogenase complex (ubiquinone)(GO:0045257) respiratory chain complex II(GO:0045273) succinate dehydrogenase complex(GO:0045281) fumarate reductase complex(GO:0045283) |
| 0.0 | 0.5 | GO:0042599 | lamellar body(GO:0042599) |
| 0.0 | 0.8 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 0.1 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.6 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.2 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:0072379 | BAT3 complex(GO:0071818) ER membrane insertion complex(GO:0072379) |
| 0.0 | 0.2 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
| 0.0 | 0.1 | GO:0090571 | RNA polymerase II transcription repressor complex(GO:0090571) |
| 0.0 | 0.2 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 2.2 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 0.3 | GO:0097381 | photoreceptor disc membrane(GO:0097381) |
| 0.0 | 1.1 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.7 | GO:0000178 | exosome (RNase complex)(GO:0000178) |
| 0.0 | 0.1 | GO:0060053 | neurofilament cytoskeleton(GO:0060053) |
| 0.0 | 0.1 | GO:0033655 | host cell cytoplasm(GO:0030430) host cell cytoplasm part(GO:0033655) |
| 0.0 | 0.1 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0031229 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) nuclear membrane part(GO:0044453) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 0.2 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.2 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0036038 | MKS complex(GO:0036038) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 0.9 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 0.1 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.8 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 1.0 | GO:0019005 | SCF ubiquitin ligase complex(GO:0019005) |
| 0.0 | 0.3 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.2 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 2.2 | GO:0001650 | fibrillar center(GO:0001650) |
| 0.0 | 0.8 | GO:0030670 | phagocytic vesicle membrane(GO:0030670) |
| 0.0 | 0.2 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.0 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.4 | GO:0030140 | trans-Golgi network transport vesicle(GO:0030140) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.6 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0097255 | R2TP complex(GO:0097255) |
| 0.0 | 0.1 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.3 | 0.9 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.3 | 0.8 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.2 | 1.2 | GO:0015111 | iodide transmembrane transporter activity(GO:0015111) |
| 0.2 | 1.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.2 | 1.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 1.8 | GO:0003847 | 1-alkyl-2-acetylglycerophosphocholine esterase activity(GO:0003847) |
| 0.2 | 0.8 | GO:0031716 | calcitonin receptor activity(GO:0004948) calcitonin receptor binding(GO:0031716) |
| 0.2 | 0.7 | GO:0003835 | beta-galactoside alpha-2,6-sialyltransferase activity(GO:0003835) |
| 0.2 | 0.5 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.2 | 2.7 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.2 | 1.6 | GO:0060228 | phosphatidylcholine-sterol O-acyltransferase activator activity(GO:0060228) |
| 0.1 | 2.6 | GO:0086080 | protein binding involved in heterotypic cell-cell adhesion(GO:0086080) |
| 0.1 | 0.4 | GO:0004938 | alpha2-adrenergic receptor activity(GO:0004938) |
| 0.1 | 1.4 | GO:0043125 | ErbB-3 class receptor binding(GO:0043125) |
| 0.1 | 1.7 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.1 | 1.0 | GO:0043533 | inositol 1,3,4,5 tetrakisphosphate binding(GO:0043533) |
| 0.1 | 1.6 | GO:0016174 | NAD(P)H oxidase activity(GO:0016174) |
| 0.1 | 0.3 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.1 | 0.3 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.1 | 0.3 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.1 | 4.0 | GO:0005031 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 0.6 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 1.9 | GO:0045295 | gamma-catenin binding(GO:0045295) |
| 0.1 | 0.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.3 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.1 | 1.2 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.1 | 1.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.1 | 0.2 | GO:0047025 | 3-oxoacyl-[acyl-carrier-protein] reductase (NADH) activity(GO:0047025) |
| 0.1 | 1.7 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.3 | GO:0004966 | galanin receptor activity(GO:0004966) |
| 0.1 | 0.3 | GO:0030899 | calcium-dependent ATPase activity(GO:0030899) |
| 0.1 | 0.2 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.5 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.1 | 1.4 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.1 | 1.7 | GO:0030296 | protein tyrosine kinase activator activity(GO:0030296) |
| 0.1 | 0.4 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.1 | 0.9 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.6 | GO:0030369 | ICAM-3 receptor activity(GO:0030369) |
| 0.1 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.6 | GO:0008503 | benzodiazepine receptor activity(GO:0008503) |
| 0.1 | 0.3 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.4 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.1 | 0.3 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.2 | GO:0008097 | 5S rRNA binding(GO:0008097) |
| 0.1 | 1.1 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.1 | 0.7 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.1 | 0.9 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.1 | 0.2 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.1 | 0.5 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.3 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.1 | 0.3 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.1 | 0.2 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.1 | 0.6 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.1 | 0.2 | GO:0019862 | IgA binding(GO:0019862) |
| 0.1 | 0.2 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.3 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 1.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.1 | 0.2 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.6 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.1 | 0.2 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.1 | 0.4 | GO:0005497 | androgen binding(GO:0005497) |
| 0.1 | 0.2 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.1 | 0.2 | GO:0052871 | tocopherol omega-hydroxylase activity(GO:0052870) alpha-tocopherol omega-hydroxylase activity(GO:0052871) 20-hydroxy-leukotriene B4 omega oxidase activity(GO:0097258) 20-aldehyde-leukotriene B4 20-monooxygenase activity(GO:0097259) |
| 0.1 | 0.2 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.3 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.0 | 0.7 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.6 | GO:0046703 | natural killer cell lectin-like receptor binding(GO:0046703) |
| 0.0 | 0.1 | GO:0016517 | interleukin-12 receptor activity(GO:0016517) |
| 0.0 | 0.1 | GO:0047977 | linoleate 13S-lipoxygenase activity(GO:0016165) hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 1.0 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.9 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.1 | GO:0004817 | cysteine-tRNA ligase activity(GO:0004817) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.4 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.0 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.5 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.8 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.0 | 0.1 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.2 | GO:0045174 | oxidoreductase activity, acting on a sulfur group of donors, quinone or similar compound as acceptor(GO:0016672) glutathione dehydrogenase (ascorbate) activity(GO:0045174) methylarsonate reductase activity(GO:0050610) |
| 0.0 | 0.4 | GO:0102336 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 1.2 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 2.2 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.2 | GO:0052834 | inositol monophosphate 1-phosphatase activity(GO:0008934) inositol monophosphate 3-phosphatase activity(GO:0052832) inositol monophosphate 4-phosphatase activity(GO:0052833) inositol monophosphate phosphatase activity(GO:0052834) |
| 0.0 | 0.2 | GO:0051916 | granulocyte colony-stimulating factor binding(GO:0051916) |
| 0.0 | 0.1 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.0 | 0.5 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0019828 | aspartic-type endopeptidase inhibitor activity(GO:0019828) |
| 0.0 | 0.2 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.2 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 1.3 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.6 | GO:0032036 | myosin heavy chain binding(GO:0032036) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0042019 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 1.3 | GO:0043027 | cysteine-type endopeptidase inhibitor activity involved in apoptotic process(GO:0043027) |
| 0.0 | 0.1 | GO:0004119 | cGMP-inhibited cyclic-nucleotide phosphodiesterase activity(GO:0004119) |
| 0.0 | 0.9 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.4 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 2.6 | GO:0048365 | Rac GTPase binding(GO:0048365) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.1 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.7 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0030107 | HLA-A specific inhibitory MHC class I receptor activity(GO:0030107) |
| 0.0 | 0.5 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 0.2 | GO:0047493 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 10.2 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.3 | GO:0003810 | protein-glutamine gamma-glutamyltransferase activity(GO:0003810) |
| 0.0 | 0.3 | GO:0003917 | DNA topoisomerase type I activity(GO:0003917) |
| 0.0 | 0.2 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.2 | GO:0004950 | G-protein coupled chemoattractant receptor activity(GO:0001637) chemokine receptor activity(GO:0004950) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.2 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.7 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.2 | GO:0005113 | patched binding(GO:0005113) |
| 0.0 | 0.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 1.2 | GO:0043236 | laminin binding(GO:0043236) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.2 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0050253 | retinyl-palmitate esterase activity(GO:0050253) |
| 0.0 | 0.3 | GO:0031624 | ubiquitin conjugating enzyme binding(GO:0031624) |
| 0.0 | 0.1 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.2 | GO:0004875 | complement receptor activity(GO:0004875) |
| 0.0 | 0.2 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.0 | 0.0 | GO:0004439 | phosphatidylinositol-4,5-bisphosphate 5-phosphatase activity(GO:0004439) inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol phosphate 5-phosphatase activity(GO:0034595) inositol trisphosphate phosphatase activity(GO:0046030) |
| 0.0 | 0.1 | GO:0047685 | amine sulfotransferase activity(GO:0047685) |
| 0.0 | 0.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.2 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.4 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0015218 | pyrimidine nucleotide transmembrane transporter activity(GO:0015218) |
| 0.0 | 0.2 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.2 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.3 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 0.2 | GO:0035500 | MH2 domain binding(GO:0035500) |
| 0.0 | 0.6 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.3 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.2 | GO:0015269 | calcium-activated potassium channel activity(GO:0015269) |
| 0.0 | 0.2 | GO:0032453 | histone demethylase activity (H3-K4 specific)(GO:0032453) |
| 0.0 | 3.3 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 2.4 | GO:0003730 | mRNA 3'-UTR binding(GO:0003730) |
| 0.0 | 1.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 2.6 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.4 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.2 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0016603 | glutaminyl-peptide cyclotransferase activity(GO:0016603) |
| 0.0 | 0.1 | GO:0005244 | voltage-gated ion channel activity(GO:0005244) voltage-gated channel activity(GO:0022832) |
| 0.0 | 0.3 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.0 | 0.2 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) |
| 0.0 | 0.1 | GO:0003746 | translation elongation factor activity(GO:0003746) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.3 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
| 0.0 | 0.1 | GO:0004351 | glutamate decarboxylase activity(GO:0004351) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.2 | GO:0009982 | pseudouridine synthase activity(GO:0009982) |
| 0.0 | 0.8 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.0 | 0.0 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.7 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.2 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.0 | 0.2 | GO:0008199 | ferric iron binding(GO:0008199) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.0 | GO:0036009 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.3 | GO:0042800 | histone methyltransferase activity (H3-K4 specific)(GO:0042800) |
| 0.0 | 0.1 | GO:0004301 | epoxide hydrolase activity(GO:0004301) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0030267 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.4 | GO:0070577 | lysine-acetylated histone binding(GO:0070577) |
| 0.0 | 0.7 | GO:0005548 | phospholipid transporter activity(GO:0005548) |
| 0.0 | 0.1 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.3 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0004859 | phospholipase inhibitor activity(GO:0004859) |
| 0.0 | 0.2 | GO:0003841 | 1-acylglycerol-3-phosphate O-acyltransferase activity(GO:0003841) |
| 0.0 | 0.6 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.1 | GO:0042923 | neuropeptide binding(GO:0042923) |
| 0.0 | 0.0 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 9.1 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 5.3 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.2 | SIG PIP3 SIGNALING IN CARDIAC MYOCTES | Genes related to PIP3 signaling in cardiac myocytes |
| 0.0 | 1.9 | PID LIS1 PATHWAY | Lissencephaly gene (LIS1) in neuronal migration and development |
| 0.0 | 1.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.9 | PID SYNDECAN 4 PATHWAY | Syndecan-4-mediated signaling events |
| 0.0 | 0.8 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 1.2 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 3.1 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 1.3 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | PID S1P S1P1 PATHWAY | S1P1 pathway |
| 0.0 | 1.9 | SIG REGULATION OF THE ACTIN CYTOSKELETON BY RHO GTPASES | Genes related to regulation of the actin cytoskeleton |
| 0.0 | 1.3 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.5 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 2.1 | ST FAS SIGNALING PATHWAY | Fas Signaling Pathway |
| 0.0 | 0.3 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 2.1 | PID HIF1 TFPATHWAY | HIF-1-alpha transcription factor network |
| 0.0 | 0.7 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.1 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 0.7 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.3 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.4 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.7 | PID ARF6 PATHWAY | Arf6 signaling events |
| 0.0 | 0.7 | PID ENDOTHELIN PATHWAY | Endothelins |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID IL8 CXCR1 PATHWAY | IL8- and CXCR1-mediated signaling events |
| 0.0 | 0.5 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.6 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.7 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.0 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.2 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.6 | PID RAC1 PATHWAY | RAC1 signaling pathway |
| 0.0 | 0.2 | PID A6B1 A6B4 INTEGRIN PATHWAY | a6b1 and a6b4 Integrin signaling |
| 0.0 | 0.1 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.1 | PID AVB3 OPN PATHWAY | Osteopontin-mediated events |
| 0.0 | 0.3 | PID ERBB1 DOWNSTREAM PATHWAY | ErbB1 downstream signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 7.1 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.2 | 4.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 12.4 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.8 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.1 | 1.3 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 2.3 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.1 | 1.7 | REACTOME SYNTHESIS SECRETION AND DEACYLATION OF GHRELIN | Genes involved in Synthesis, Secretion, and Deacylation of Ghrelin |
| 0.1 | 1.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 1.1 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.1 | 1.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.1 | 0.9 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.1 | 1.3 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.2 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.0 | 0.6 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 1.0 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 1.0 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 1.0 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 1.1 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.4 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.5 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PI | Genes involved in Acyl chain remodelling of PI |
| 0.0 | 0.7 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME FGFR4 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR4 ligand binding and activation |
| 0.0 | 0.6 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 1.1 | REACTOME ASSOCIATION OF TRIC CCT WITH TARGET PROTEINS DURING BIOSYNTHESIS | Genes involved in Association of TriC/CCT with target proteins during biosynthesis |
| 0.0 | 2.6 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.4 | REACTOME LATENT INFECTION OF HOMO SAPIENS WITH MYCOBACTERIUM TUBERCULOSIS | Genes involved in Latent infection of Homo sapiens with Mycobacterium tuberculosis |
| 0.0 | 0.9 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.6 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 1.5 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.2 | REACTOME ENDOGENOUS STEROLS | Genes involved in Endogenous sterols |
| 0.0 | 0.7 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.7 | REACTOME IRON UPTAKE AND TRANSPORT | Genes involved in Iron uptake and transport |
| 0.0 | 0.2 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.0 | 0.4 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.5 | REACTOME GRB2 SOS PROVIDES LINKAGE TO MAPK SIGNALING FOR INTERGRINS | Genes involved in GRB2:SOS provides linkage to MAPK signaling for Intergrins |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.7 | REACTOME SEMA4D INDUCED CELL MIGRATION AND GROWTH CONE COLLAPSE | Genes involved in Sema4D induced cell migration and growth-cone collapse |
| 0.0 | 0.2 | REACTOME MRNA DECAY BY 3 TO 5 EXORIBONUCLEASE | Genes involved in mRNA Decay by 3' to 5' Exoribonuclease |
| 0.0 | 0.8 | REACTOME CGMP EFFECTS | Genes involved in cGMP effects |
| 0.0 | 0.2 | REACTOME VIF MEDIATED DEGRADATION OF APOBEC3G | Genes involved in Vif-mediated degradation of APOBEC3G |
| 0.0 | 0.4 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.3 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.0 | 0.1 | REACTOME FGFR1 LIGAND BINDING AND ACTIVATION | Genes involved in FGFR1 ligand binding and activation |
| 0.0 | 1.0 | REACTOME NRAGE SIGNALS DEATH THROUGH JNK | Genes involved in NRAGE signals death through JNK |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.3 | REACTOME RECYCLING PATHWAY OF L1 | Genes involved in Recycling pathway of L1 |
| 0.0 | 1.1 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.7 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.0 | REACTOME AMYLOIDS | Genes involved in Amyloids |
| 0.0 | 1.3 | REACTOME MHC CLASS II ANTIGEN PRESENTATION | Genes involved in MHC class II antigen presentation |