Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for HBP1
Z-value: 0.72
Transcription factors associated with HBP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HBP1
|
ENSG00000105856.9 | HBP1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HBP1 | hg19_v2_chr7_+_106809406_106809460 | -0.49 | 2.2e-01 | Click! |
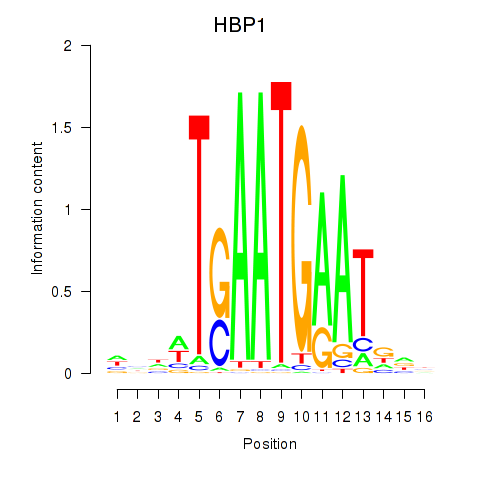
Activity profile of HBP1 motif
Sorted Z-values of HBP1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HBP1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_10697895 | 1.09 |
ENST00000591240.1 ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr2_-_161056762 | 0.67 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr2_-_161056802 | 0.60 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr12_-_53320245 | 0.58 |
ENST00000552150.1 |
KRT8 |
keratin 8 |
| chr19_-_52034971 | 0.53 |
ENST00000346477.3 |
SIGLEC6 |
sialic acid binding Ig-like lectin 6 |
| chr19_-_6690723 | 0.49 |
ENST00000601008.1 |
C3 |
complement component 3 |
| chr2_-_113594279 | 0.38 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr15_+_57884117 | 0.36 |
ENST00000267853.5 |
MYZAP |
myocardial zonula adherens protein |
| chr2_+_173292280 | 0.36 |
ENST00000264107.7 |
ITGA6 |
integrin, alpha 6 |
| chr2_+_173292390 | 0.36 |
ENST00000442250.1 ENST00000458358.1 ENST00000409080.1 |
ITGA6 |
integrin, alpha 6 |
| chr19_-_38747172 | 0.33 |
ENST00000347262.4 ENST00000591585.1 ENST00000301242.4 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr19_-_52035044 | 0.32 |
ENST00000359982.4 ENST00000436458.1 ENST00000425629.3 ENST00000391797.3 ENST00000343300.4 |
SIGLEC6 |
sialic acid binding Ig-like lectin 6 |
| chr2_+_173292301 | 0.31 |
ENST00000264106.6 ENST00000375221.2 ENST00000343713.4 |
ITGA6 |
integrin, alpha 6 |
| chr7_-_100808843 | 0.31 |
ENST00000249330.2 |
VGF |
VGF nerve growth factor inducible |
| chr15_-_72490114 | 0.29 |
ENST00000309731.7 |
GRAMD2 |
GRAM domain containing 2 |
| chr1_+_65613340 | 0.28 |
ENST00000546702.1 |
AK4 |
adenylate kinase 4 |
| chr15_+_57884086 | 0.28 |
ENST00000380569.2 ENST00000380561.2 ENST00000574161.1 ENST00000572390.1 ENST00000396180.1 ENST00000380560.2 |
GCOM1 |
GRINL1A complex locus 1 |
| chr11_+_66824276 | 0.27 |
ENST00000308831.2 |
RHOD |
ras homolog family member D |
| chr3_-_52090461 | 0.27 |
ENST00000296483.6 ENST00000495880.1 |
DUSP7 |
dual specificity phosphatase 7 |
| chr4_+_86525299 | 0.26 |
ENST00000512201.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr19_-_45657028 | 0.24 |
ENST00000429338.1 ENST00000589776.1 |
NKPD1 |
NTPase, KAP family P-loop domain containing 1 |
| chr13_-_20077417 | 0.23 |
ENST00000382978.1 ENST00000400230.2 ENST00000255310.6 |
TPTE2 |
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr11_+_66824346 | 0.23 |
ENST00000532559.1 |
RHOD |
ras homolog family member D |
| chr7_+_29874341 | 0.22 |
ENST00000409290.1 ENST00000242140.5 |
WIPF3 |
WAS/WASL interacting protein family, member 3 |
| chr6_+_30614779 | 0.21 |
ENST00000293604.6 ENST00000376473.5 |
C6orf136 |
chromosome 6 open reading frame 136 |
| chr6_+_30614886 | 0.21 |
ENST00000376471.4 |
C6orf136 |
chromosome 6 open reading frame 136 |
| chr12_-_89746173 | 0.21 |
ENST00000308385.6 |
DUSP6 |
dual specificity phosphatase 6 |
| chr3_+_111805182 | 0.21 |
ENST00000430855.1 ENST00000431717.2 ENST00000264848.5 |
C3orf52 |
chromosome 3 open reading frame 52 |
| chr15_+_75074385 | 0.20 |
ENST00000220003.9 |
CSK |
c-src tyrosine kinase |
| chr1_+_65613217 | 0.20 |
ENST00000545314.1 |
AK4 |
adenylate kinase 4 |
| chr7_+_18535786 | 0.19 |
ENST00000406072.1 |
HDAC9 |
histone deacetylase 9 |
| chr4_+_159236462 | 0.19 |
ENST00000460056.2 |
RXFP1 |
relaxin/insulin-like family peptide receptor 1 |
| chr15_+_57884199 | 0.19 |
ENST00000587652.1 ENST00000380568.3 ENST00000380565.4 ENST00000380563.2 |
GCOM1 MYZAP POLR2M |
GRINL1A complex locus 1 myocardial zonula adherens protein polymerase (RNA) II (DNA directed) polypeptide M |
| chr1_-_25291475 | 0.18 |
ENST00000338888.3 ENST00000399916.1 |
RUNX3 |
runt-related transcription factor 3 |
| chr19_-_38746979 | 0.18 |
ENST00000591291.1 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr19_+_49055332 | 0.18 |
ENST00000201586.2 |
SULT2B1 |
sulfotransferase family, cytosolic, 2B, member 1 |
| chr1_-_24126892 | 0.17 |
ENST00000374497.3 ENST00000425913.1 |
GALE |
UDP-galactose-4-epimerase |
| chr1_+_13516066 | 0.17 |
ENST00000332192.6 |
PRAMEF21 |
PRAME family member 21 |
| chr20_+_45523227 | 0.17 |
ENST00000327619.5 ENST00000357410.3 |
EYA2 |
eyes absent homolog 2 (Drosophila) |
| chr5_+_149737202 | 0.16 |
ENST00000451292.1 ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1 |
Treacher Collins-Franceschetti syndrome 1 |
| chr1_+_13736907 | 0.16 |
ENST00000316412.5 |
PRAMEF20 |
PRAME family member 20 |
| chr9_+_78505554 | 0.16 |
ENST00000545128.1 |
PCSK5 |
proprotein convertase subtilisin/kexin type 5 |
| chr3_+_44626446 | 0.16 |
ENST00000441021.1 ENST00000322734.2 |
ZNF660 |
zinc finger protein 660 |
| chr5_-_78809950 | 0.16 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr1_-_24127256 | 0.15 |
ENST00000418277.1 |
GALE |
UDP-galactose-4-epimerase |
| chr1_+_158149737 | 0.15 |
ENST00000368171.3 |
CD1D |
CD1d molecule |
| chr1_+_92632542 | 0.15 |
ENST00000409154.4 ENST00000370378.4 |
KIAA1107 |
KIAA1107 |
| chr12_-_11139511 | 0.15 |
ENST00000506868.1 |
TAS2R50 |
taste receptor, type 2, member 50 |
| chr11_-_47574664 | 0.15 |
ENST00000310513.5 ENST00000531165.1 |
CELF1 |
CUGBP, Elav-like family member 1 |
| chrX_+_150151752 | 0.15 |
ENST00000325307.7 |
HMGB3 |
high mobility group box 3 |
| chr1_-_31769595 | 0.15 |
ENST00000263694.4 |
SNRNP40 |
small nuclear ribonucleoprotein 40kDa (U5) |
| chr1_+_110577229 | 0.14 |
ENST00000369795.3 ENST00000369794.2 |
STRIP1 |
striatin interacting protein 1 |
| chr10_+_13203543 | 0.14 |
ENST00000378714.3 ENST00000479669.1 ENST00000484800.2 |
MCM10 |
minichromosome maintenance complex component 10 |
| chr17_-_16256718 | 0.14 |
ENST00000476243.1 ENST00000299736.4 |
CENPV |
centromere protein V |
| chr3_+_184279566 | 0.14 |
ENST00000330394.2 |
EPHB3 |
EPH receptor B3 |
| chr1_+_202172848 | 0.14 |
ENST00000255432.7 |
LGR6 |
leucine-rich repeat containing G protein-coupled receptor 6 |
| chr13_-_44735393 | 0.13 |
ENST00000400419.1 |
SMIM2 |
small integral membrane protein 2 |
| chrX_+_150151824 | 0.13 |
ENST00000455596.1 ENST00000448905.2 |
HMGB3 |
high mobility group box 3 |
| chr11_-_65667884 | 0.12 |
ENST00000448083.2 ENST00000531493.1 ENST00000532401.1 |
FOSL1 |
FOS-like antigen 1 |
| chr2_-_40739501 | 0.12 |
ENST00000403092.1 ENST00000402441.1 ENST00000448531.1 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr19_+_39390587 | 0.12 |
ENST00000572515.1 ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chrX_+_100075368 | 0.12 |
ENST00000415585.2 ENST00000372972.2 ENST00000413437.1 |
CSTF2 |
cleavage stimulation factor, 3' pre-RNA, subunit 2, 64kDa |
| chr20_-_48330377 | 0.12 |
ENST00000371711.4 |
B4GALT5 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 5 |
| chr14_+_78266408 | 0.12 |
ENST00000238561.5 |
ADCK1 |
aarF domain containing kinase 1 |
| chr12_-_110271178 | 0.12 |
ENST00000261740.2 ENST00000392719.2 ENST00000346520.2 |
TRPV4 |
transient receptor potential cation channel, subfamily V, member 4 |
| chr11_-_89653576 | 0.12 |
ENST00000420869.1 |
TRIM49D1 |
tripartite motif containing 49D1 |
| chr15_+_75074410 | 0.11 |
ENST00000439220.2 |
CSK |
c-src tyrosine kinase |
| chr14_+_78266436 | 0.11 |
ENST00000557501.1 ENST00000341211.5 |
ADCK1 |
aarF domain containing kinase 1 |
| chr10_+_82300575 | 0.11 |
ENST00000313455.4 |
SH2D4B |
SH2 domain containing 4B |
| chr2_-_220264703 | 0.11 |
ENST00000519905.1 ENST00000523282.1 ENST00000434339.1 ENST00000457935.1 |
DNPEP |
aspartyl aminopeptidase |
| chr2_-_73053126 | 0.11 |
ENST00000272427.6 ENST00000410104.1 |
EXOC6B |
exocyst complex component 6B |
| chr6_+_30297306 | 0.11 |
ENST00000420746.1 ENST00000513556.1 |
TRIM39 TRIM39-RPP21 |
tripartite motif containing 39 TRIM39-RPP21 readthrough |
| chr7_-_105319536 | 0.11 |
ENST00000477775.1 |
ATXN7L1 |
ataxin 7-like 1 |
| chr19_-_3500635 | 0.11 |
ENST00000250937.3 |
DOHH |
deoxyhypusine hydroxylase/monooxygenase |
| chr3_+_184032419 | 0.11 |
ENST00000352767.3 ENST00000427141.2 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_+_23345943 | 0.11 |
ENST00000400181.4 ENST00000542151.1 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr11_-_65667997 | 0.11 |
ENST00000312562.2 ENST00000534222.1 |
FOSL1 |
FOS-like antigen 1 |
| chr8_-_130951940 | 0.10 |
ENST00000522250.1 ENST00000522941.1 ENST00000522746.1 ENST00000520204.1 ENST00000519070.1 ENST00000520254.1 ENST00000519824.2 ENST00000519540.1 |
FAM49B |
family with sequence similarity 49, member B |
| chr1_+_23345930 | 0.10 |
ENST00000356634.3 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr11_+_73087309 | 0.10 |
ENST00000064780.2 ENST00000545687.1 |
RELT |
RELT tumor necrosis factor receptor |
| chr6_+_149539767 | 0.10 |
ENST00000606202.1 ENST00000536230.1 ENST00000445901.1 |
TAB2 RP1-111D6.3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 2 RP1-111D6.3 |
| chr3_-_197026152 | 0.10 |
ENST00000419354.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr3_+_184032283 | 0.10 |
ENST00000346169.2 ENST00000414031.1 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr1_+_44435646 | 0.10 |
ENST00000255108.3 ENST00000412950.2 ENST00000396758.2 |
DPH2 |
DPH2 homolog (S. cerevisiae) |
| chr19_+_41698927 | 0.10 |
ENST00000310054.4 |
CYP2S1 |
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr11_-_65363259 | 0.10 |
ENST00000342202.4 |
KCNK7 |
potassium channel, subfamily K, member 7 |
| chr2_+_65663812 | 0.09 |
ENST00000606978.1 ENST00000377977.3 ENST00000536804.1 |
AC074391.1 |
AC074391.1 |
| chr8_-_8751068 | 0.09 |
ENST00000276282.6 |
MFHAS1 |
malignant fibrous histiocytoma amplified sequence 1 |
| chr19_+_39390320 | 0.09 |
ENST00000576510.1 |
NFKBIB |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr3_+_184032313 | 0.09 |
ENST00000392537.2 ENST00000444134.1 ENST00000450424.1 ENST00000421110.1 ENST00000382330.3 ENST00000426123.1 ENST00000350481.5 ENST00000455679.1 ENST00000440448.1 |
EIF4G1 |
eukaryotic translation initiation factor 4 gamma, 1 |
| chr7_+_39125365 | 0.09 |
ENST00000559001.1 ENST00000464276.2 |
POU6F2 |
POU class 6 homeobox 2 |
| chr5_+_72469014 | 0.09 |
ENST00000296776.5 |
TMEM174 |
transmembrane protein 174 |
| chr2_-_69180083 | 0.09 |
ENST00000328895.4 |
GKN2 |
gastrokine 2 |
| chr3_-_42452050 | 0.09 |
ENST00000441172.1 ENST00000287748.3 |
LYZL4 |
lysozyme-like 4 |
| chr6_-_44233361 | 0.09 |
ENST00000275015.5 |
NFKBIE |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, epsilon |
| chr19_+_19144384 | 0.09 |
ENST00000392335.2 ENST00000535612.1 ENST00000537263.1 ENST00000540707.1 ENST00000541725.1 ENST00000269932.6 ENST00000546344.1 ENST00000540792.1 ENST00000536098.1 ENST00000541898.1 ENST00000543877.1 |
ARMC6 |
armadillo repeat containing 6 |
| chr17_+_36584662 | 0.09 |
ENST00000431231.2 ENST00000437668.3 |
ARHGAP23 |
Rho GTPase activating protein 23 |
| chr16_+_57769635 | 0.08 |
ENST00000379661.3 ENST00000562592.1 ENST00000566726.1 |
KATNB1 |
katanin p80 (WD repeat containing) subunit B 1 |
| chrX_-_131262048 | 0.08 |
ENST00000298542.4 |
FRMD7 |
FERM domain containing 7 |
| chr12_+_41221794 | 0.08 |
ENST00000547849.1 |
CNTN1 |
contactin 1 |
| chrX_+_100663243 | 0.08 |
ENST00000316594.5 |
HNRNPH2 |
heterogeneous nuclear ribonucleoprotein H2 (H') |
| chr19_+_840963 | 0.08 |
ENST00000234347.5 |
PRTN3 |
proteinase 3 |
| chr7_-_135612198 | 0.07 |
ENST00000589735.1 |
LUZP6 |
leucine zipper protein 6 |
| chr20_+_37554955 | 0.07 |
ENST00000217429.4 |
FAM83D |
family with sequence similarity 83, member D |
| chr19_+_17830051 | 0.07 |
ENST00000594625.1 ENST00000324096.4 ENST00000600186.1 ENST00000597735.1 |
MAP1S |
microtubule-associated protein 1S |
| chr6_-_30640811 | 0.07 |
ENST00000376442.3 |
DHX16 |
DEAH (Asp-Glu-Ala-His) box polypeptide 16 |
| chr1_-_26394114 | 0.07 |
ENST00000374272.3 |
TRIM63 |
tripartite motif containing 63, E3 ubiquitin protein ligase |
| chr12_-_99548270 | 0.07 |
ENST00000546568.1 ENST00000332712.7 ENST00000546960.1 |
ANKS1B |
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr13_-_46626847 | 0.06 |
ENST00000242848.4 ENST00000282007.3 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
| chr16_-_3661578 | 0.06 |
ENST00000294008.3 |
SLX4 |
SLX4 structure-specific endonuclease subunit |
| chr22_+_22988816 | 0.06 |
ENST00000480559.1 ENST00000448514.1 |
GGTLC2 |
gamma-glutamyltransferase light chain 2 |
| chr7_-_56101826 | 0.06 |
ENST00000421626.1 |
PSPH |
phosphoserine phosphatase |
| chr18_+_18943554 | 0.06 |
ENST00000580732.2 |
GREB1L |
growth regulation by estrogen in breast cancer-like |
| chr11_-_46867780 | 0.06 |
ENST00000529230.1 ENST00000415402.1 ENST00000312055.5 |
CKAP5 |
cytoskeleton associated protein 5 |
| chr6_-_32339671 | 0.06 |
ENST00000442822.2 ENST00000375015.4 ENST00000533191.1 |
C6orf10 |
chromosome 6 open reading frame 10 |
| chr9_+_127624387 | 0.06 |
ENST00000353214.2 |
ARPC5L |
actin related protein 2/3 complex, subunit 5-like |
| chr9_-_128246769 | 0.06 |
ENST00000444226.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr9_-_25678856 | 0.06 |
ENST00000358022.3 |
TUSC1 |
tumor suppressor candidate 1 |
| chr2_+_207630081 | 0.06 |
ENST00000236980.6 ENST00000418289.1 ENST00000402774.3 ENST00000403094.3 |
FASTKD2 |
FAST kinase domains 2 |
| chr10_-_120101752 | 0.06 |
ENST00000369170.4 |
FAM204A |
family with sequence similarity 204, member A |
| chr4_+_186347388 | 0.06 |
ENST00000511138.1 ENST00000511581.1 |
C4orf47 |
chromosome 4 open reading frame 47 |
| chr12_+_110562135 | 0.06 |
ENST00000361948.4 ENST00000552912.1 ENST00000242591.5 ENST00000546374.1 |
IFT81 |
intraflagellar transport 81 homolog (Chlamydomonas) |
| chr2_-_239197201 | 0.05 |
ENST00000254658.3 |
PER2 |
period circadian clock 2 |
| chr14_-_94759361 | 0.05 |
ENST00000393096.1 |
SERPINA10 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr6_-_32339649 | 0.05 |
ENST00000527965.1 ENST00000532023.1 ENST00000447241.2 ENST00000375007.4 ENST00000534588.1 |
C6orf10 |
chromosome 6 open reading frame 10 |
| chr1_+_153175900 | 0.05 |
ENST00000368747.1 |
LELP1 |
late cornified envelope-like proline-rich 1 |
| chr11_+_43702322 | 0.05 |
ENST00000395700.4 |
HSD17B12 |
hydroxysteroid (17-beta) dehydrogenase 12 |
| chr4_-_165305086 | 0.05 |
ENST00000507270.1 ENST00000514618.1 ENST00000503008.1 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr2_-_74375081 | 0.05 |
ENST00000327428.5 |
BOLA3 |
bolA family member 3 |
| chr22_+_25615489 | 0.05 |
ENST00000398215.2 |
CRYBB2 |
crystallin, beta B2 |
| chr3_+_51575596 | 0.05 |
ENST00000409535.2 |
RAD54L2 |
RAD54-like 2 (S. cerevisiae) |
| chrX_-_23926004 | 0.05 |
ENST00000379226.4 ENST00000379220.3 |
APOO |
apolipoprotein O |
| chr15_-_86338134 | 0.05 |
ENST00000337975.5 |
KLHL25 |
kelch-like family member 25 |
| chr12_+_124118366 | 0.05 |
ENST00000539994.1 ENST00000538845.1 ENST00000228955.7 ENST00000543341.2 ENST00000536375.1 |
GTF2H3 |
general transcription factor IIH, polypeptide 3, 34kDa |
| chr3_-_197025447 | 0.05 |
ENST00000346964.2 ENST00000357674.4 ENST00000314062.3 ENST00000448528.2 ENST00000419553.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr14_-_94759408 | 0.05 |
ENST00000554723.1 |
SERPINA10 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 10 |
| chr3_+_126423045 | 0.05 |
ENST00000290913.3 ENST00000508789.1 |
CHCHD6 |
coiled-coil-helix-coiled-coil-helix domain containing 6 |
| chr16_-_28223166 | 0.05 |
ENST00000304658.5 |
XPO6 |
exportin 6 |
| chr4_-_47465666 | 0.05 |
ENST00000381571.4 |
COMMD8 |
COMM domain containing 8 |
| chrX_+_23925918 | 0.05 |
ENST00000379211.3 |
CXorf58 |
chromosome X open reading frame 58 |
| chr6_-_9933500 | 0.05 |
ENST00000492169.1 |
OFCC1 |
orofacial cleft 1 candidate 1 |
| chr10_+_104263743 | 0.05 |
ENST00000369902.3 ENST00000369899.2 ENST00000423559.2 |
SUFU |
suppressor of fused homolog (Drosophila) |
| chr16_+_31366455 | 0.05 |
ENST00000268296.4 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr20_-_1373682 | 0.04 |
ENST00000381724.3 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chr17_-_41856305 | 0.04 |
ENST00000397937.2 ENST00000226004.3 |
DUSP3 |
dual specificity phosphatase 3 |
| chr8_-_86290333 | 0.04 |
ENST00000521846.1 ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1 |
carbonic anhydrase I |
| chr22_-_20367797 | 0.04 |
ENST00000424787.2 |
GGTLC3 |
gamma-glutamyltransferase light chain 3 |
| chr17_+_33895090 | 0.04 |
ENST00000592381.1 |
RP11-1094M14.11 |
RP11-1094M14.11 |
| chr10_+_24528108 | 0.04 |
ENST00000438429.1 |
KIAA1217 |
KIAA1217 |
| chr2_+_74425689 | 0.04 |
ENST00000394053.2 ENST00000409804.1 ENST00000264090.4 ENST00000394050.3 ENST00000409601.1 |
MTHFD2 |
methylenetetrahydrofolate dehydrogenase (NADP+ dependent) 2, methenyltetrahydrofolate cyclohydrolase |
| chr7_+_95115210 | 0.04 |
ENST00000428113.1 ENST00000325885.5 |
ASB4 |
ankyrin repeat and SOCS box containing 4 |
| chr11_-_62457371 | 0.04 |
ENST00000317449.4 |
LRRN4CL |
LRRN4 C-terminal like |
| chr22_-_20368028 | 0.04 |
ENST00000404912.1 |
GGTLC3 |
gamma-glutamyltransferase light chain 3 |
| chr10_-_120101804 | 0.04 |
ENST00000369183.4 ENST00000369172.4 |
FAM204A |
family with sequence similarity 204, member A |
| chr9_+_131217459 | 0.04 |
ENST00000497812.2 ENST00000393533.2 |
ODF2 |
outer dense fiber of sperm tails 2 |
| chr12_+_57984965 | 0.04 |
ENST00000540759.2 ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C |
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chr10_+_6130966 | 0.04 |
ENST00000437845.2 ENST00000432931.1 |
RBM17 |
RNA binding motif protein 17 |
| chrX_-_54824673 | 0.04 |
ENST00000218436.6 |
ITIH6 |
inter-alpha-trypsin inhibitor heavy chain family, member 6 |
| chr2_+_87808725 | 0.04 |
ENST00000413202.1 |
LINC00152 |
long intergenic non-protein coding RNA 152 |
| chr17_-_1588101 | 0.04 |
ENST00000577001.1 ENST00000572621.1 ENST00000304992.6 |
PRPF8 |
pre-mRNA processing factor 8 |
| chr8_+_63161491 | 0.04 |
ENST00000523211.1 ENST00000524201.1 |
NKAIN3 |
Na+/K+ transporting ATPase interacting 3 |
| chr4_-_47983519 | 0.04 |
ENST00000358519.4 ENST00000544810.1 ENST00000402813.3 |
CNGA1 |
cyclic nucleotide gated channel alpha 1 |
| chr19_+_10713112 | 0.04 |
ENST00000590382.1 ENST00000407327.4 |
SLC44A2 |
solute carrier family 44 (choline transporter), member 2 |
| chr13_+_34392200 | 0.04 |
ENST00000434425.1 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
| chr6_+_140175987 | 0.04 |
ENST00000414038.1 ENST00000431609.1 |
RP5-899B16.1 |
RP5-899B16.1 |
| chr18_-_19283649 | 0.04 |
ENST00000584464.1 ENST00000578270.1 |
ABHD3 |
abhydrolase domain containing 3 |
| chr2_-_74374995 | 0.04 |
ENST00000295326.4 |
BOLA3 |
bolA family member 3 |
| chr2_+_12246664 | 0.04 |
ENST00000449986.1 |
AC096559.1 |
AC096559.1 |
| chr16_+_22517166 | 0.03 |
ENST00000356156.3 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr17_+_75369167 | 0.03 |
ENST00000423034.2 |
SEPT9 |
septin 9 |
| chr12_-_89919765 | 0.03 |
ENST00000541909.1 ENST00000313546.3 |
POC1B |
POC1 centriolar protein B |
| chr6_-_8102279 | 0.03 |
ENST00000488226.2 |
EEF1E1 |
eukaryotic translation elongation factor 1 epsilon 1 |
| chr1_-_25747283 | 0.03 |
ENST00000346452.4 ENST00000340849.4 ENST00000349438.4 ENST00000294413.7 ENST00000413854.1 ENST00000455194.1 ENST00000243186.6 ENST00000425135.1 |
RHCE |
Rh blood group, CcEe antigens |
| chr10_+_6130946 | 0.03 |
ENST00000379888.4 |
RBM17 |
RNA binding motif protein 17 |
| chr2_+_219433588 | 0.03 |
ENST00000295701.5 |
RQCD1 |
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr6_-_133035185 | 0.03 |
ENST00000367928.4 |
VNN1 |
vanin 1 |
| chr16_+_31366536 | 0.03 |
ENST00000562522.1 |
ITGAX |
integrin, alpha X (complement component 3 receptor 4 subunit) |
| chr10_-_60027642 | 0.03 |
ENST00000373935.3 |
IPMK |
inositol polyphosphate multikinase |
| chr15_-_56535464 | 0.03 |
ENST00000559447.2 ENST00000422057.1 ENST00000317318.6 ENST00000423270.1 |
RFX7 |
regulatory factor X, 7 |
| chr2_+_27886330 | 0.03 |
ENST00000326019.6 |
SLC4A1AP |
solute carrier family 4 (anion exchanger), member 1, adaptor protein |
| chr12_+_123868320 | 0.03 |
ENST00000402868.3 ENST00000330479.4 |
SETD8 |
SET domain containing (lysine methyltransferase) 8 |
| chr2_+_242498135 | 0.03 |
ENST00000318407.3 |
BOK |
BCL2-related ovarian killer |
| chr15_+_38226827 | 0.03 |
ENST00000559502.1 ENST00000558148.1 ENST00000558158.1 |
TMCO5A |
transmembrane and coiled-coil domains 5A |
| chr11_+_57435219 | 0.03 |
ENST00000527985.1 ENST00000287169.3 |
ZDHHC5 |
zinc finger, DHHC-type containing 5 |
| chr6_+_42531798 | 0.03 |
ENST00000372903.2 ENST00000372899.1 ENST00000372901.1 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr11_+_69061594 | 0.03 |
ENST00000441339.2 ENST00000308946.3 ENST00000535407.1 |
MYEOV |
myeloma overexpressed |
| chr1_+_182584314 | 0.03 |
ENST00000566297.1 |
RP11-317P15.4 |
RP11-317P15.4 |
| chr2_-_239197238 | 0.03 |
ENST00000254657.3 |
PER2 |
period circadian clock 2 |
| chr1_-_13117736 | 0.03 |
ENST00000376192.5 ENST00000376182.1 |
PRAMEF6 |
PRAME family member 6 |
| chr1_+_25598872 | 0.03 |
ENST00000328664.4 |
RHD |
Rh blood group, D antigen |
| chr1_+_240255166 | 0.03 |
ENST00000319653.9 |
FMN2 |
formin 2 |
| chr10_-_73976884 | 0.03 |
ENST00000317126.4 ENST00000545550.1 |
ASCC1 |
activating signal cointegrator 1 complex subunit 1 |
| chr9_-_39288092 | 0.03 |
ENST00000323947.7 ENST00000297668.6 ENST00000377656.2 ENST00000377659.1 |
CNTNAP3 |
contactin associated protein-like 3 |
| chr1_-_13115578 | 0.03 |
ENST00000414205.2 |
PRAMEF6 |
PRAME family member 6 |
| chr8_+_23104130 | 0.03 |
ENST00000313219.7 ENST00000519984.1 |
CHMP7 |
charged multivesicular body protein 7 |
| chr17_-_39165366 | 0.03 |
ENST00000391588.1 |
KRTAP3-1 |
keratin associated protein 3-1 |
| chr3_+_186288454 | 0.03 |
ENST00000265028.3 |
DNAJB11 |
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr17_+_8152590 | 0.03 |
ENST00000584044.1 ENST00000314666.6 ENST00000545834.1 ENST00000581242.1 |
PFAS |
phosphoribosylformylglycinamidine synthase |
| chr19_-_18508396 | 0.03 |
ENST00000595840.1 ENST00000339007.3 |
LRRC25 |
leucine rich repeat containing 25 |
| chr7_-_83278322 | 0.03 |
ENST00000307792.3 |
SEMA3E |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3E |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.4 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 1.3 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 1.0 | GO:0010668 | ectodermal cell differentiation(GO:0010668) |
| 0.1 | 0.2 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.1 | 0.2 | GO:0051685 | maintenance of ER location(GO:0051685) |
| 0.1 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 0.2 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.5 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.6 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.3 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.2 | GO:0007296 | vitellogenesis(GO:0007296) |
| 0.0 | 0.2 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.2 | GO:0032455 | nerve growth factor processing(GO:0032455) |
| 0.0 | 0.1 | GO:1990523 | bone regeneration(GO:1990523) |
| 0.0 | 0.3 | GO:0002191 | cap-dependent translational initiation(GO:0002191) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.3 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0000706 | meiotic DNA double-strand break processing(GO:0000706) |
| 0.0 | 0.2 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 1.0 | GO:0050690 | regulation of defense response to virus by virus(GO:0050690) |
| 0.0 | 0.1 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:0051941 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.0 | 0.1 | GO:0014878 | response to electrical stimulus involved in regulation of muscle adaptation(GO:0014878) |
| 0.0 | 0.0 | GO:1902995 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.0 | 0.2 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0097029 | mature conventional dendritic cell differentiation(GO:0097029) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.1 | GO:1902775 | mitochondrial large ribosomal subunit assembly(GO:1902775) |
| 0.0 | 0.2 | GO:0016576 | histone dephosphorylation(GO:0016576) |
| 0.0 | 0.1 | GO:1900383 | regulation of synaptic plasticity by receptor localization to synapse(GO:1900383) |
| 0.0 | 0.3 | GO:0002021 | response to dietary excess(GO:0002021) |
| 0.0 | 0.0 | GO:0097360 | chorionic trophoblast cell proliferation(GO:0097360) regulation of chorionic trophoblast cell proliferation(GO:1901382) |
| 0.0 | 0.1 | GO:0098789 | pre-mRNA cleavage required for polyadenylation(GO:0098789) |
| 0.0 | 0.0 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.3 | 1.0 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.0 | 1.1 | GO:0030131 | clathrin adaptor complex(GO:0030131) |
| 0.0 | 0.6 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.0 | GO:0000438 | core TFIIH complex portion of holo TFIIH complex(GO:0000438) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.0 | GO:0031372 | UBC13-MMS2 complex(GO:0031372) |
| 0.0 | 0.3 | GO:0016281 | eukaryotic translation initiation factor 4F complex(GO:0016281) |
| 0.0 | 0.2 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0031298 | replication fork protection complex(GO:0031298) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.1 | 0.3 | GO:0003978 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 1.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.2 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 0.2 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0015275 | stretch-activated, cation-selective, calcium channel activity(GO:0015275) |
| 0.0 | 0.2 | GO:0050294 | steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.5 | GO:0033549 | MAP kinase phosphatase activity(GO:0033549) |
| 0.0 | 0.1 | GO:0042835 | BRE binding(GO:0042835) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0023025 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 0.4 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.3 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.2 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.0 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.2 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.0 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.0 | 0.1 | GO:0017108 | 5'-flap endonuclease activity(GO:0017108) |
| 0.0 | 0.3 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 1.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0003831 | beta-N-acetylglucosaminylglycopeptide beta-1,4-galactosyltransferase activity(GO:0003831) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.2 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.5 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.4 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.5 | REACTOME ERKS ARE INACTIVATED | Genes involved in ERKs are inactivated |
| 0.0 | 0.5 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.3 | REACTOME MTORC1 MEDIATED SIGNALLING | Genes involved in mTORC1-mediated signalling |
| 0.0 | 0.3 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |