Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for HES1
Z-value: 0.87
Transcription factors associated with HES1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HES1
|
ENSG00000114315.3 | HES1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HES1 | hg19_v2_chr3_+_193853927_193853944 | 0.58 | 1.4e-01 | Click! |
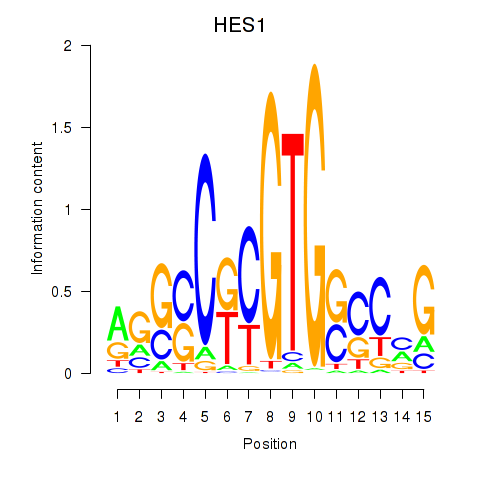
Activity profile of HES1 motif
Sorted Z-values of HES1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HES1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_-_68269971 | 1.47 |
ENST00000565858.1 |
ESRP2 |
epithelial splicing regulatory protein 2 |
| chr18_+_33877654 | 1.39 |
ENST00000257209.4 ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr10_-_123357598 | 1.24 |
ENST00000358487.5 ENST00000369058.3 ENST00000369060.4 ENST00000359354.2 |
FGFR2 |
fibroblast growth factor receptor 2 |
| chr8_+_95653373 | 1.15 |
ENST00000358397.5 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr8_+_95653427 | 1.14 |
ENST00000454170.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr8_+_95653302 | 1.14 |
ENST00000423620.2 ENST00000433389.2 |
ESRP1 |
epithelial splicing regulatory protein 1 |
| chr1_-_9189229 | 1.10 |
ENST00000377411.4 |
GPR157 |
G protein-coupled receptor 157 |
| chr10_-_123357910 | 0.95 |
ENST00000336553.6 ENST00000457416.2 ENST00000360144.3 ENST00000369059.1 ENST00000356226.4 ENST00000351936.6 |
FGFR2 |
fibroblast growth factor receptor 2 |
| chr14_-_105635090 | 0.88 |
ENST00000331782.3 ENST00000347004.2 |
JAG2 |
jagged 2 |
| chr20_+_58152524 | 0.87 |
ENST00000359926.3 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr17_+_78075361 | 0.76 |
ENST00000577106.1 ENST00000390015.3 |
GAA |
glucosidase, alpha; acid |
| chr7_-_98030360 | 0.70 |
ENST00000005260.8 |
BAIAP2L1 |
BAI1-associated protein 2-like 1 |
| chr17_-_3599696 | 0.69 |
ENST00000225328.5 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr19_+_38755203 | 0.69 |
ENST00000587090.1 ENST00000454580.3 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
| chr19_+_38755042 | 0.68 |
ENST00000301244.7 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
| chr8_+_124194752 | 0.67 |
ENST00000318462.6 |
FAM83A |
family with sequence similarity 83, member A |
| chr15_+_41136216 | 0.67 |
ENST00000562057.1 ENST00000344051.4 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr1_-_205290865 | 0.66 |
ENST00000367157.3 |
NUAK2 |
NUAK family, SNF1-like kinase, 2 |
| chr7_-_44365020 | 0.63 |
ENST00000395747.2 ENST00000347193.4 ENST00000346990.4 ENST00000258682.6 ENST00000353625.4 ENST00000421607.1 ENST00000424197.1 ENST00000502837.2 ENST00000350811.3 ENST00000395749.2 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
| chr17_-_42402138 | 0.62 |
ENST00000592857.1 ENST00000586016.1 ENST00000590194.1 ENST00000377095.5 ENST00000588049.1 ENST00000586633.1 ENST00000537904.2 ENST00000585636.1 ENST00000585523.1 ENST00000225308.8 |
SLC25A39 |
solute carrier family 25, member 39 |
| chr17_-_3599327 | 0.62 |
ENST00000551178.1 ENST00000552276.1 ENST00000547178.1 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr1_+_44401479 | 0.61 |
ENST00000438616.3 |
ARTN |
artemin |
| chr17_+_78075498 | 0.61 |
ENST00000302262.3 |
GAA |
glucosidase, alpha; acid |
| chr17_-_3599492 | 0.61 |
ENST00000435558.1 ENST00000345901.3 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr12_-_53614155 | 0.60 |
ENST00000543726.1 |
RARG |
retinoic acid receptor, gamma |
| chr7_-_44365216 | 0.60 |
ENST00000358707.3 ENST00000457475.1 ENST00000440254.2 |
CAMK2B |
calcium/calmodulin-dependent protein kinase II beta |
| chr8_+_28351707 | 0.60 |
ENST00000537916.1 ENST00000523546.1 ENST00000240093.3 |
FZD3 |
frizzled family receptor 3 |
| chr12_-_53614043 | 0.60 |
ENST00000338561.5 |
RARG |
retinoic acid receptor, gamma |
| chr20_+_6748311 | 0.58 |
ENST00000378827.4 |
BMP2 |
bone morphogenetic protein 2 |
| chr19_+_34287174 | 0.56 |
ENST00000587559.1 ENST00000588637.1 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
| chr9_+_133971863 | 0.54 |
ENST00000372309.3 |
AIF1L |
allograft inflammatory factor 1-like |
| chr2_-_73511559 | 0.53 |
ENST00000521871.1 |
FBXO41 |
F-box protein 41 |
| chr9_+_133971909 | 0.52 |
ENST00000247291.3 ENST00000372302.1 ENST00000372300.1 ENST00000372298.1 |
AIF1L |
allograft inflammatory factor 1-like |
| chr11_+_124932955 | 0.50 |
ENST00000403796.2 |
SLC37A2 |
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr9_-_139948487 | 0.49 |
ENST00000355097.2 |
ENTPD2 |
ectonucleoside triphosphate diphosphohydrolase 2 |
| chr1_-_231175964 | 0.49 |
ENST00000366654.4 |
FAM89A |
family with sequence similarity 89, member A |
| chr15_+_89182178 | 0.49 |
ENST00000559876.1 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr2_+_18059906 | 0.49 |
ENST00000304101.4 |
KCNS3 |
potassium voltage-gated channel, delayed-rectifier, subfamily S, member 3 |
| chr1_-_19283163 | 0.48 |
ENST00000455833.2 |
IFFO2 |
intermediate filament family orphan 2 |
| chr11_+_124932986 | 0.47 |
ENST00000407458.1 ENST00000298280.5 |
SLC37A2 |
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr8_-_142011036 | 0.46 |
ENST00000520892.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr5_+_140220769 | 0.46 |
ENST00000531613.1 ENST00000378123.3 |
PCDHA8 |
protocadherin alpha 8 |
| chr11_+_124933191 | 0.46 |
ENST00000532000.1 ENST00000308074.4 |
SLC37A2 |
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr1_-_175162048 | 0.45 |
ENST00000444639.1 |
KIAA0040 |
KIAA0040 |
| chr2_+_106361333 | 0.44 |
ENST00000233154.4 ENST00000451463.2 |
NCK2 |
NCK adaptor protein 2 |
| chrX_+_105969893 | 0.44 |
ENST00000255499.2 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr17_+_54671047 | 0.43 |
ENST00000332822.4 |
NOG |
noggin |
| chr14_-_100070363 | 0.43 |
ENST00000380243.4 |
CCDC85C |
coiled-coil domain containing 85C |
| chr15_+_89181974 | 0.43 |
ENST00000306072.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr9_-_126030817 | 0.43 |
ENST00000348403.5 ENST00000447404.2 ENST00000360998.3 |
STRBP |
spermatid perinuclear RNA binding protein |
| chr20_+_35201857 | 0.42 |
ENST00000373874.2 |
TGIF2 |
TGFB-induced factor homeobox 2 |
| chr18_+_11752040 | 0.42 |
ENST00000423027.3 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr8_-_66754172 | 0.41 |
ENST00000401827.3 |
PDE7A |
phosphodiesterase 7A |
| chr15_+_89182156 | 0.41 |
ENST00000379224.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr16_-_4401258 | 0.40 |
ENST00000577031.1 |
PAM16 |
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr16_-_402639 | 0.39 |
ENST00000262320.3 |
AXIN1 |
axin 1 |
| chr5_+_149737202 | 0.39 |
ENST00000451292.1 ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1 |
Treacher Collins-Franceschetti syndrome 1 |
| chr1_-_25291475 | 0.39 |
ENST00000338888.3 ENST00000399916.1 |
RUNX3 |
runt-related transcription factor 3 |
| chr2_-_96811170 | 0.38 |
ENST00000288943.4 |
DUSP2 |
dual specificity phosphatase 2 |
| chr1_-_167522982 | 0.38 |
ENST00000370509.4 |
CREG1 |
cellular repressor of E1A-stimulated genes 1 |
| chr12_+_122064673 | 0.38 |
ENST00000537188.1 |
ORAI1 |
ORAI calcium release-activated calcium modulator 1 |
| chr8_+_32406179 | 0.38 |
ENST00000405005.3 |
NRG1 |
neuregulin 1 |
| chr16_-_4401284 | 0.37 |
ENST00000318059.3 |
PAM16 |
presequence translocase-associated motor 16 homolog (S. cerevisiae) |
| chr14_+_67999999 | 0.36 |
ENST00000329153.5 |
PLEKHH1 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr20_+_61273797 | 0.36 |
ENST00000217159.1 |
SLCO4A1 |
solute carrier organic anion transporter family, member 4A1 |
| chr20_+_35201993 | 0.36 |
ENST00000373872.4 |
TGIF2 |
TGFB-induced factor homeobox 2 |
| chr1_+_152881014 | 0.36 |
ENST00000368764.3 ENST00000392667.2 |
IVL |
involucrin |
| chr8_+_104513086 | 0.36 |
ENST00000406091.3 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr15_-_63674034 | 0.36 |
ENST00000344366.3 ENST00000422263.2 |
CA12 |
carbonic anhydrase XII |
| chr16_-_4466565 | 0.35 |
ENST00000572467.1 ENST00000423908.2 ENST00000572044.1 ENST00000571052.1 |
CORO7-PAM16 CORO7 |
CORO7-PAM16 readthrough coronin 7 |
| chr19_-_46272106 | 0.35 |
ENST00000560168.1 |
SIX5 |
SIX homeobox 5 |
| chr16_+_577697 | 0.35 |
ENST00000562370.1 ENST00000568988.1 ENST00000219611.2 |
CAPN15 |
calpain 15 |
| chr14_-_91526462 | 0.35 |
ENST00000536315.2 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr2_+_64681641 | 0.34 |
ENST00000409537.2 |
LGALSL |
lectin, galactoside-binding-like |
| chr8_+_145734433 | 0.34 |
ENST00000301327.4 |
MFSD3 |
major facilitator superfamily domain containing 3 |
| chr21_-_42219065 | 0.34 |
ENST00000400454.1 |
DSCAM |
Down syndrome cell adhesion molecule |
| chrX_+_152907913 | 0.34 |
ENST00000370167.4 |
DUSP9 |
dual specificity phosphatase 9 |
| chr2_+_97426631 | 0.33 |
ENST00000377075.2 |
CNNM4 |
cyclin M4 |
| chr6_+_43738444 | 0.33 |
ENST00000324450.6 ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA |
vascular endothelial growth factor A |
| chr1_-_117210290 | 0.32 |
ENST00000369483.1 ENST00000369486.3 |
IGSF3 |
immunoglobulin superfamily, member 3 |
| chr8_-_28243590 | 0.32 |
ENST00000523095.1 ENST00000522795.1 |
ZNF395 |
zinc finger protein 395 |
| chr8_-_145515055 | 0.32 |
ENST00000526552.1 ENST00000529231.1 ENST00000307404.5 |
BOP1 |
block of proliferation 1 |
| chr20_-_50384864 | 0.31 |
ENST00000311637.5 ENST00000402822.1 |
ATP9A |
ATPase, class II, type 9A |
| chr11_+_68080077 | 0.31 |
ENST00000294304.7 |
LRP5 |
low density lipoprotein receptor-related protein 5 |
| chr1_-_32801825 | 0.31 |
ENST00000329421.7 |
MARCKSL1 |
MARCKS-like 1 |
| chr16_+_2205755 | 0.31 |
ENST00000326181.6 |
TRAF7 |
TNF receptor-associated factor 7, E3 ubiquitin protein ligase |
| chr15_-_63674218 | 0.30 |
ENST00000178638.3 |
CA12 |
carbonic anhydrase XII |
| chr17_+_29718642 | 0.30 |
ENST00000325874.8 |
RAB11FIP4 |
RAB11 family interacting protein 4 (class II) |
| chr2_+_203499901 | 0.30 |
ENST00000303116.6 ENST00000392238.2 |
FAM117B |
family with sequence similarity 117, member B |
| chr10_-_105615164 | 0.30 |
ENST00000355946.2 ENST00000369774.4 |
SH3PXD2A |
SH3 and PX domains 2A |
| chr8_+_31496809 | 0.30 |
ENST00000518104.1 ENST00000519301.1 |
NRG1 |
neuregulin 1 |
| chr10_+_71561630 | 0.29 |
ENST00000398974.3 ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr9_-_124976185 | 0.29 |
ENST00000464484.2 |
LHX6 |
LIM homeobox 6 |
| chr14_-_91526922 | 0.29 |
ENST00000418736.2 ENST00000261991.3 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr10_+_71561649 | 0.29 |
ENST00000398978.3 ENST00000354547.3 ENST00000357811.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr7_-_140340576 | 0.29 |
ENST00000275884.6 ENST00000475837.1 |
DENND2A |
DENN/MADD domain containing 2A |
| chr18_-_74207146 | 0.29 |
ENST00000443185.2 |
ZNF516 |
zinc finger protein 516 |
| chr15_+_74908147 | 0.28 |
ENST00000568139.1 ENST00000563297.1 ENST00000568488.1 ENST00000352989.5 ENST00000348245.3 |
CLK3 |
CDC-like kinase 3 |
| chr7_-_139876812 | 0.28 |
ENST00000397560.2 |
JHDM1D |
lysine (K)-specific demethylase 7A |
| chr6_-_19804973 | 0.28 |
ENST00000457670.1 ENST00000607810.1 ENST00000606628.1 |
RP4-625H18.2 |
RP4-625H18.2 |
| chr1_+_227127981 | 0.28 |
ENST00000366778.1 ENST00000366777.3 ENST00000458507.2 |
ADCK3 |
aarF domain containing kinase 3 |
| chr15_-_67813924 | 0.28 |
ENST00000559298.1 |
IQCH-AS1 |
IQCH antisense RNA 1 |
| chrX_-_7066159 | 0.27 |
ENST00000486446.2 ENST00000412827.2 ENST00000424830.2 ENST00000381077.5 ENST00000540122.1 |
HDHD1 |
haloacid dehalogenase-like hydrolase domain containing 1 |
| chr8_-_28243934 | 0.27 |
ENST00000521185.1 ENST00000520290.1 ENST00000344423.5 |
ZNF395 |
zinc finger protein 395 |
| chr11_+_64058820 | 0.26 |
ENST00000422670.2 |
KCNK4 |
potassium channel, subfamily K, member 4 |
| chr6_-_143771799 | 0.26 |
ENST00000237283.8 |
ADAT2 |
adenosine deaminase, tRNA-specific 2 |
| chr7_-_994302 | 0.26 |
ENST00000265846.5 |
ADAP1 |
ArfGAP with dual PH domains 1 |
| chr14_-_81687575 | 0.26 |
ENST00000434192.2 |
GTF2A1 |
general transcription factor IIA, 1, 19/37kDa |
| chr1_-_217262969 | 0.25 |
ENST00000361525.3 |
ESRRG |
estrogen-related receptor gamma |
| chr11_+_64058758 | 0.25 |
ENST00000538767.1 |
KCNK4 |
potassium channel, subfamily K, member 4 |
| chr2_-_101925055 | 0.25 |
ENST00000295317.3 |
RNF149 |
ring finger protein 149 |
| chr17_+_30813576 | 0.25 |
ENST00000313401.3 |
CDK5R1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr21_+_45527171 | 0.25 |
ENST00000291576.7 ENST00000456705.1 |
PWP2 |
PWP2 periodic tryptophan protein homolog (yeast) |
| chr13_-_44735393 | 0.25 |
ENST00000400419.1 |
SMIM2 |
small integral membrane protein 2 |
| chr15_+_42120283 | 0.25 |
ENST00000542534.2 ENST00000397299.4 ENST00000408047.1 ENST00000431823.1 ENST00000382448.4 ENST00000342159.4 |
PLA2G4B JMJD7 JMJD7-PLA2G4B |
phospholipase A2, group IVB (cytosolic) jumonji domain containing 7 JMJD7-PLA2G4B readthrough |
| chr21_+_46359907 | 0.24 |
ENST00000291634.6 ENST00000397826.3 ENST00000458015.1 |
FAM207A |
family with sequence similarity 207, member A |
| chr1_+_44445549 | 0.24 |
ENST00000356836.6 |
B4GALT2 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr10_-_135379132 | 0.24 |
ENST00000343131.5 |
SYCE1 |
synaptonemal complex central element protein 1 |
| chr10_+_134210672 | 0.24 |
ENST00000305233.5 ENST00000368609.4 |
PWWP2B |
PWWP domain containing 2B |
| chr3_-_185542817 | 0.24 |
ENST00000382199.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr1_-_85514120 | 0.24 |
ENST00000370589.2 ENST00000341115.4 ENST00000370587.1 |
MCOLN3 |
mucolipin 3 |
| chr3_+_184081175 | 0.24 |
ENST00000452961.1 ENST00000296223.3 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr1_-_38273840 | 0.24 |
ENST00000373044.2 |
YRDC |
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr19_+_797443 | 0.24 |
ENST00000394601.4 ENST00000589575.1 |
PTBP1 |
polypyrimidine tract binding protein 1 |
| chr19_+_8274204 | 0.23 |
ENST00000561053.1 ENST00000251363.5 ENST00000559450.1 ENST00000559336.1 |
CERS4 |
ceramide synthase 4 |
| chr8_-_102218292 | 0.23 |
ENST00000518336.1 ENST00000520454.1 |
ZNF706 |
zinc finger protein 706 |
| chr9_-_124976154 | 0.23 |
ENST00000482062.1 |
LHX6 |
LIM homeobox 6 |
| chr3_+_184081213 | 0.23 |
ENST00000429568.1 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr3_-_138763734 | 0.23 |
ENST00000413199.1 ENST00000502927.2 |
PRR23C |
proline rich 23C |
| chr1_-_217262933 | 0.23 |
ENST00000359162.2 |
ESRRG |
estrogen-related receptor gamma |
| chr11_+_46354455 | 0.23 |
ENST00000343674.6 |
DGKZ |
diacylglycerol kinase, zeta |
| chr8_+_145515263 | 0.23 |
ENST00000528838.1 |
HSF1 |
heat shock transcription factor 1 |
| chr10_-_99094458 | 0.23 |
ENST00000371019.2 |
FRAT2 |
frequently rearranged in advanced T-cell lymphomas 2 |
| chr19_+_8274185 | 0.23 |
ENST00000558268.1 ENST00000558331.1 |
CERS4 |
ceramide synthase 4 |
| chr14_-_93582214 | 0.22 |
ENST00000556603.2 ENST00000354313.3 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
| chr7_+_24613034 | 0.22 |
ENST00000409761.1 ENST00000396475.2 |
MPP6 |
membrane protein, palmitoylated 6 (MAGUK p55 subfamily member 6) |
| chr6_+_43739697 | 0.22 |
ENST00000230480.6 |
VEGFA |
vascular endothelial growth factor A |
| chr3_-_185542761 | 0.22 |
ENST00000457616.2 ENST00000346192.3 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr3_+_121554046 | 0.22 |
ENST00000273668.2 ENST00000451944.2 |
EAF2 |
ELL associated factor 2 |
| chr10_+_74033672 | 0.22 |
ENST00000307365.3 |
DDIT4 |
DNA-damage-inducible transcript 4 |
| chr19_-_3478477 | 0.22 |
ENST00000591708.1 |
C19orf77 |
chromosome 19 open reading frame 77 |
| chr20_-_50385138 | 0.22 |
ENST00000338821.5 |
ATP9A |
ATPase, class II, type 9A |
| chr3_+_51428704 | 0.22 |
ENST00000323686.4 |
RBM15B |
RNA binding motif protein 15B |
| chr12_+_122064398 | 0.22 |
ENST00000330079.7 |
ORAI1 |
ORAI calcium release-activated calcium modulator 1 |
| chr22_+_38302285 | 0.22 |
ENST00000215957.6 |
MICALL1 |
MICAL-like 1 |
| chr19_-_42724261 | 0.22 |
ENST00000595337.1 |
DEDD2 |
death effector domain containing 2 |
| chr6_-_105850937 | 0.21 |
ENST00000369110.3 |
PREP |
prolyl endopeptidase |
| chr19_-_14629224 | 0.21 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr7_-_558876 | 0.21 |
ENST00000354513.5 ENST00000402802.3 |
PDGFA |
platelet-derived growth factor alpha polypeptide |
| chr9_-_139137648 | 0.21 |
ENST00000358701.5 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
| chr22_+_29279552 | 0.21 |
ENST00000544604.2 |
ZNRF3 |
zinc and ring finger 3 |
| chr5_-_78809950 | 0.21 |
ENST00000334082.6 |
HOMER1 |
homer homolog 1 (Drosophila) |
| chr14_-_77279153 | 0.21 |
ENST00000251089.2 |
ANGEL1 |
angel homolog 1 (Drosophila) |
| chr19_+_5823813 | 0.21 |
ENST00000303212.2 |
NRTN |
neurturin |
| chr2_-_46769694 | 0.21 |
ENST00000522587.1 |
ATP6V1E2 |
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr2_-_73053126 | 0.20 |
ENST00000272427.6 ENST00000410104.1 |
EXOC6B |
exocyst complex component 6B |
| chr17_+_7348658 | 0.20 |
ENST00000570557.1 ENST00000536404.2 ENST00000576360.1 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr18_-_77793891 | 0.20 |
ENST00000592957.1 ENST00000585474.1 |
TXNL4A |
thioredoxin-like 4A |
| chr1_+_44444865 | 0.20 |
ENST00000372324.1 |
B4GALT2 |
UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 2 |
| chr9_+_101867359 | 0.20 |
ENST00000374994.4 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
| chr12_+_53574464 | 0.20 |
ENST00000416904.3 |
ZNF740 |
zinc finger protein 740 |
| chr22_+_46731596 | 0.20 |
ENST00000381019.3 |
TRMU |
tRNA 5-methylaminomethyl-2-thiouridylate methyltransferase |
| chr1_-_46152174 | 0.20 |
ENST00000290795.3 ENST00000355105.3 |
GPBP1L1 |
GC-rich promoter binding protein 1-like 1 |
| chr1_-_200379129 | 0.20 |
ENST00000367353.1 |
ZNF281 |
zinc finger protein 281 |
| chr18_-_33077556 | 0.20 |
ENST00000589273.1 ENST00000586489.1 |
INO80C |
INO80 complex subunit C |
| chr17_+_73257945 | 0.19 |
ENST00000579002.1 |
MRPS7 |
mitochondrial ribosomal protein S7 |
| chr14_-_93581615 | 0.19 |
ENST00000555553.1 ENST00000555495.1 ENST00000554999.1 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
| chr16_-_2059797 | 0.19 |
ENST00000563630.1 |
ZNF598 |
zinc finger protein 598 |
| chr20_-_43280325 | 0.19 |
ENST00000537820.1 |
ADA |
adenosine deaminase |
| chr11_-_113746212 | 0.19 |
ENST00000537642.1 ENST00000537706.1 ENST00000544750.1 ENST00000260188.5 ENST00000540925.1 |
USP28 |
ubiquitin specific peptidase 28 |
| chr2_+_26915584 | 0.19 |
ENST00000302909.3 |
KCNK3 |
potassium channel, subfamily K, member 3 |
| chr19_+_38397839 | 0.19 |
ENST00000222345.6 |
SIPA1L3 |
signal-induced proliferation-associated 1 like 3 |
| chr3_+_184081137 | 0.19 |
ENST00000443489.1 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr16_+_66914264 | 0.19 |
ENST00000311765.2 ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr16_-_4466622 | 0.19 |
ENST00000570645.1 ENST00000574025.1 ENST00000572898.1 ENST00000537233.2 ENST00000571059.1 ENST00000251166.4 |
CORO7 |
coronin 7 |
| chr9_-_140484917 | 0.19 |
ENST00000298585.2 |
ZMYND19 |
zinc finger, MYND-type containing 19 |
| chr2_+_27237615 | 0.19 |
ENST00000458529.1 ENST00000402218.1 |
MAPRE3 |
microtubule-associated protein, RP/EB family, member 3 |
| chr1_+_41707996 | 0.19 |
ENST00000425554.1 |
RP11-399E6.1 |
RP11-399E6.1 |
| chr12_-_124018252 | 0.19 |
ENST00000376874.4 |
RILPL1 |
Rab interacting lysosomal protein-like 1 |
| chr6_+_89855765 | 0.19 |
ENST00000275072.4 |
PM20D2 |
peptidase M20 domain containing 2 |
| chr2_-_25142708 | 0.18 |
ENST00000260600.5 ENST00000435135.1 |
ADCY3 |
adenylate cyclase 3 |
| chr1_-_200379180 | 0.18 |
ENST00000294740.3 |
ZNF281 |
zinc finger protein 281 |
| chr17_-_2614927 | 0.18 |
ENST00000435359.1 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
| chr9_+_101867387 | 0.18 |
ENST00000374990.2 ENST00000552516.1 |
TGFBR1 |
transforming growth factor, beta receptor 1 |
| chrX_+_21392529 | 0.18 |
ENST00000425654.2 ENST00000543067.1 |
CNKSR2 |
connector enhancer of kinase suppressor of Ras 2 |
| chr17_+_7348374 | 0.18 |
ENST00000306071.2 ENST00000572857.1 |
CHRNB1 |
cholinergic receptor, nicotinic, beta 1 (muscle) |
| chr5_+_612387 | 0.18 |
ENST00000264935.5 ENST00000444221.1 |
CEP72 |
centrosomal protein 72kDa |
| chr19_-_19051927 | 0.18 |
ENST00000600077.1 |
HOMER3 |
homer homolog 3 (Drosophila) |
| chr6_-_13711773 | 0.18 |
ENST00000011619.3 |
RANBP9 |
RAN binding protein 9 |
| chr14_-_51562745 | 0.17 |
ENST00000298355.3 |
TRIM9 |
tripartite motif containing 9 |
| chr11_-_113746277 | 0.17 |
ENST00000003302.4 ENST00000545540.1 |
USP28 |
ubiquitin specific peptidase 28 |
| chr3_+_10857885 | 0.17 |
ENST00000254488.2 ENST00000454147.1 |
SLC6A11 |
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr4_+_144257915 | 0.17 |
ENST00000262995.4 |
GAB1 |
GRB2-associated binding protein 1 |
| chr8_+_25042192 | 0.17 |
ENST00000410074.1 |
DOCK5 |
dedicator of cytokinesis 5 |
| chr14_-_93582148 | 0.17 |
ENST00000267615.6 ENST00000553452.1 |
ITPK1 |
inositol-tetrakisphosphate 1-kinase |
| chr5_+_44809027 | 0.17 |
ENST00000507110.1 |
MRPS30 |
mitochondrial ribosomal protein S30 |
| chr19_+_984313 | 0.17 |
ENST00000251289.5 ENST00000587001.2 ENST00000607440.1 |
WDR18 |
WD repeat domain 18 |
| chr14_+_104604916 | 0.17 |
ENST00000423312.2 |
KIF26A |
kinesin family member 26A |
| chr14_-_24780573 | 0.17 |
ENST00000336557.5 ENST00000258807.5 |
CIDEB |
cell death-inducing DFFA-like effector b |
| chr3_-_179169181 | 0.17 |
ENST00000497513.1 |
GNB4 |
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.2 | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.3 | 1.4 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.3 | 1.4 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.3 | 1.3 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 1.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) |
| 0.2 | 0.6 | GO:0045976 | negative regulation of mitotic cell cycle, embryonic(GO:0045976) |
| 0.2 | 0.6 | GO:0060129 | thyroid-stimulating hormone-secreting cell differentiation(GO:0060129) |
| 0.2 | 0.6 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.7 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.1 | 0.4 | GO:0071789 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 0.4 | GO:0048320 | axial mesoderm formation(GO:0048320) canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.1 | 1.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.5 | GO:0071469 | detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.6 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.4 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.1 | 0.7 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.3 | GO:0060086 | circadian temperature homeostasis(GO:0060086) |
| 0.1 | 0.3 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.1 | 0.7 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.3 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 1.4 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.4 | GO:2000313 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.3 | GO:0015920 | lipopolysaccharide transport(GO:0015920) |
| 0.1 | 0.3 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.1 | 0.2 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.5 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.1 | 0.9 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 0.2 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.1 | 0.3 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.1 | 0.4 | GO:1905073 | occluding junction disassembly(GO:1905071) regulation of occluding junction disassembly(GO:1905073) positive regulation of occluding junction disassembly(GO:1905075) |
| 0.1 | 0.2 | GO:1900365 | positive regulation of mRNA polyadenylation(GO:1900365) positive regulation of apoptotic DNA fragmentation(GO:1902512) |
| 0.1 | 0.2 | GO:1904886 | beta-catenin destruction complex disassembly(GO:1904886) |
| 0.1 | 0.2 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.1 | 0.4 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.1 | 1.2 | GO:0051823 | regulation of synapse structural plasticity(GO:0051823) |
| 0.1 | 0.3 | GO:0046102 | inosine metabolic process(GO:0046102) |
| 0.1 | 0.2 | GO:0061198 | fungiform papilla formation(GO:0061198) |
| 0.1 | 0.2 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.1 | 1.1 | GO:0060019 | radial glial cell differentiation(GO:0060019) |
| 0.1 | 0.2 | GO:0051780 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.1 | 0.2 | GO:0002143 | tRNA wobble position uridine thiolation(GO:0002143) |
| 0.1 | 0.2 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.1 | 0.3 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.1 | 0.2 | GO:0008355 | olfactory learning(GO:0008355) |
| 0.1 | 0.4 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.2 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.1 | 0.2 | GO:0061188 | negative regulation of chromatin silencing at rDNA(GO:0061188) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.7 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.1 | 0.2 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.1 | 0.5 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.3 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.0 | 0.2 | GO:1902498 | regulation of protein autoubiquitination(GO:1902498) |
| 0.0 | 1.9 | GO:0035590 | purinergic nucleotide receptor signaling pathway(GO:0035590) |
| 0.0 | 0.2 | GO:1903445 | intermembrane transport(GO:0046909) protein transport from ciliary membrane to plasma membrane(GO:1903445) |
| 0.0 | 0.2 | GO:1904751 | positive regulation of protein localization to nucleolus(GO:1904751) |
| 0.0 | 0.1 | GO:1903526 | negative regulation of membrane tubulation(GO:1903526) |
| 0.0 | 0.4 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.2 | GO:0010512 | negative regulation of phosphatidylinositol biosynthetic process(GO:0010512) |
| 0.0 | 0.2 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.0 | 0.7 | GO:0032957 | inositol trisphosphate metabolic process(GO:0032957) |
| 0.0 | 0.4 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:2001226 | negative regulation of chloride transport(GO:2001226) |
| 0.0 | 0.1 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.2 | GO:1901097 | negative regulation of autophagosome maturation(GO:1901097) |
| 0.0 | 0.1 | GO:0097069 | cellular response to thyroxine stimulus(GO:0097069) cellular response to L-phenylalanine derivative(GO:1904387) |
| 0.0 | 0.8 | GO:0038092 | nodal signaling pathway(GO:0038092) |
| 0.0 | 0.1 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.0 | 0.5 | GO:0014029 | neural crest formation(GO:0014029) |
| 0.0 | 0.2 | GO:0032526 | response to retinoic acid(GO:0032526) |
| 0.0 | 0.2 | GO:0045213 | neurotransmitter receptor metabolic process(GO:0045213) |
| 0.0 | 1.5 | GO:0060441 | epithelial tube branching involved in lung morphogenesis(GO:0060441) |
| 0.0 | 0.8 | GO:0032780 | protein import into mitochondrial matrix(GO:0030150) negative regulation of ATPase activity(GO:0032780) |
| 0.0 | 0.3 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.4 | GO:0035095 | behavioral response to nicotine(GO:0035095) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) canonical Wnt signaling pathway involved in midbrain dopaminergic neuron differentiation(GO:1904954) |
| 0.0 | 0.3 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.5 | GO:0007172 | signal complex assembly(GO:0007172) |
| 0.0 | 0.7 | GO:2000251 | positive regulation of actin cytoskeleton reorganization(GO:2000251) |
| 0.0 | 0.2 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.2 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.3 | GO:0070166 | enamel mineralization(GO:0070166) |
| 0.0 | 0.1 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.1 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) regulation of melanosome transport(GO:1902908) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.4 | GO:0002115 | store-operated calcium entry(GO:0002115) |
| 0.0 | 0.1 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.0 | 0.1 | GO:0005988 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.1 | GO:0072364 | regulation of cellular ketone metabolic process by regulation of transcription from RNA polymerase II promoter(GO:0072364) |
| 0.0 | 0.1 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.3 | GO:0033313 | meiotic cell cycle checkpoint(GO:0033313) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0018076 | N-terminal peptidyl-lysine acetylation(GO:0018076) |
| 0.0 | 0.1 | GO:0006238 | CMP salvage(GO:0006238) CMP biosynthetic process(GO:0009224) CMP metabolic process(GO:0046035) |
| 0.0 | 0.1 | GO:0019242 | methylglyoxal biosynthetic process(GO:0019242) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.2 | GO:0060770 | negative regulation of epithelial cell proliferation involved in prostate gland development(GO:0060770) |
| 0.0 | 0.3 | GO:0000463 | maturation of LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000463) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.6 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.6 | GO:0006895 | Golgi to endosome transport(GO:0006895) |
| 0.0 | 0.1 | GO:0036518 | chemorepulsion of dopaminergic neuron axon(GO:0036518) |
| 0.0 | 0.1 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.0 | 0.8 | GO:0021680 | cerebellar Purkinje cell layer development(GO:0021680) |
| 0.0 | 0.2 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.5 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.1 | GO:0021555 | midbrain-hindbrain boundary morphogenesis(GO:0021555) |
| 0.0 | 0.2 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:2000182 | regulation of progesterone biosynthetic process(GO:2000182) |
| 0.0 | 3.2 | GO:0008543 | fibroblast growth factor receptor signaling pathway(GO:0008543) |
| 0.0 | 0.0 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.1 | GO:0042214 | terpene metabolic process(GO:0042214) phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.1 | GO:0010641 | positive regulation of platelet-derived growth factor receptor signaling pathway(GO:0010641) |
| 0.0 | 0.8 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.2 | GO:0035093 | spermatogenesis, exchange of chromosomal proteins(GO:0035093) |
| 0.0 | 0.1 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.0 | 0.1 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.0 | GO:0048541 | mucosal-associated lymphoid tissue development(GO:0048537) Peyer's patch development(GO:0048541) |
| 0.0 | 0.5 | GO:0045332 | phospholipid translocation(GO:0045332) |
| 0.0 | 0.1 | GO:1901906 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.2 | GO:0045820 | negative regulation of glycolytic process(GO:0045820) negative regulation of cofactor metabolic process(GO:0051195) negative regulation of coenzyme metabolic process(GO:0051198) |
| 0.0 | 0.1 | GO:0019605 | butyrate metabolic process(GO:0019605) |
| 0.0 | 0.1 | GO:0000187 | activation of MAPK activity(GO:0000187) |
| 0.0 | 0.1 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.0 | 0.1 | GO:1902595 | regulation of DNA replication origin binding(GO:1902595) |
| 0.0 | 0.2 | GO:1904779 | regulation of protein localization to centrosome(GO:1904779) |
| 0.0 | 0.1 | GO:2000234 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.2 | GO:0019375 | galactosylceramide biosynthetic process(GO:0006682) galactolipid biosynthetic process(GO:0019375) |
| 0.0 | 0.6 | GO:0001958 | endochondral ossification(GO:0001958) replacement ossification(GO:0036075) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.1 | GO:2001295 | malonyl-CoA biosynthetic process(GO:2001295) |
| 0.0 | 0.4 | GO:0007638 | mechanosensory behavior(GO:0007638) |
| 0.0 | 0.0 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.4 | GO:0007216 | G-protein coupled glutamate receptor signaling pathway(GO:0007216) |
| 0.0 | 0.4 | GO:0007191 | adenylate cyclase-activating dopamine receptor signaling pathway(GO:0007191) |
| 0.0 | 0.1 | GO:0043696 | dedifferentiation(GO:0043696) cell dedifferentiation(GO:0043697) |
| 0.0 | 0.5 | GO:0006783 | heme biosynthetic process(GO:0006783) |
| 0.0 | 0.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.2 | GO:0006686 | sphingomyelin biosynthetic process(GO:0006686) |
| 0.0 | 0.0 | GO:0006384 | transcription initiation from RNA polymerase III promoter(GO:0006384) |
| 0.0 | 0.3 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.3 | GO:0035563 | positive regulation of chromatin binding(GO:0035563) |
| 0.0 | 0.1 | GO:0016557 | peroxisome membrane biogenesis(GO:0016557) |
| 0.0 | 0.1 | GO:0032511 | late endosome to vacuole transport via multivesicular body sorting pathway(GO:0032511) |
| 0.0 | 0.1 | GO:0042699 | follicle-stimulating hormone signaling pathway(GO:0042699) |
| 0.0 | 0.4 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.0 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.1 | GO:0033152 | immunoglobulin V(D)J recombination(GO:0033152) |
| 0.0 | 0.2 | GO:0002097 | tRNA wobble base modification(GO:0002097) |
| 0.0 | 0.1 | GO:0060052 | neurofilament cytoskeleton organization(GO:0060052) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.3 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 0.2 | GO:0034134 | toll-like receptor 2 signaling pathway(GO:0034134) |
| 0.0 | 0.2 | GO:0051481 | negative regulation of cytosolic calcium ion concentration(GO:0051481) |
| 0.0 | 0.1 | GO:0007351 | tripartite regional subdivision(GO:0007351) anterior/posterior axis specification, embryo(GO:0008595) |
| 0.0 | 0.1 | GO:2000049 | positive regulation of cell-cell adhesion mediated by cadherin(GO:2000049) |
| 0.0 | 0.0 | GO:0098905 | regulation of bundle of His cell action potential(GO:0098905) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.0 | GO:1903243 | negative regulation of cardiac muscle adaptation(GO:0010616) lung vasculature development(GO:0060426) negative regulation of cardiac muscle hypertrophy in response to stress(GO:1903243) |
| 0.0 | 0.1 | GO:1905146 | lysosomal protein catabolic process(GO:1905146) |
| 0.0 | 0.2 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.0 | 0.0 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.0 | GO:0006597 | spermine biosynthetic process(GO:0006597) |
| 0.0 | 0.2 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.7 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.1 | GO:0007406 | negative regulation of neuroblast proliferation(GO:0007406) |
| 0.0 | 0.1 | GO:1904321 | response to forskolin(GO:1904321) cellular response to forskolin(GO:1904322) |
| 0.0 | 0.4 | GO:0050873 | brown fat cell differentiation(GO:0050873) |
| 0.0 | 0.0 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.2 | GO:0016075 | rRNA catabolic process(GO:0016075) |
| 0.0 | 0.3 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.1 | 0.6 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.4 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 0.3 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.1 | 2.0 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.1 | 0.2 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.2 | GO:0005953 | CAAX-protein geranylgeranyltransferase complex(GO:0005953) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.4 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.4 | GO:0070022 | transforming growth factor beta receptor homodimeric complex(GO:0070022) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.3 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.8 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.1 | GO:1990742 | microvesicle(GO:1990742) |
| 0.0 | 0.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.6 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.1 | GO:0000938 | GARP complex(GO:0000938) |
| 0.0 | 1.4 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.4 | GO:0030877 | beta-catenin destruction complex(GO:0030877) |
| 0.0 | 0.2 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.0 | 1.3 | GO:0033017 | sarcoplasmic reticulum membrane(GO:0033017) |
| 0.0 | 0.2 | GO:0097470 | ribbon synapse(GO:0097470) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0000799 | nuclear condensin complex(GO:0000799) |
| 0.0 | 0.3 | GO:0033391 | chromatoid body(GO:0033391) |
| 0.0 | 0.3 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
| 0.0 | 0.1 | GO:0071001 | U4/U6 snRNP(GO:0071001) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.4 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.1 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.3 | GO:0017101 | aminoacyl-tRNA synthetase multienzyme complex(GO:0017101) |
| 0.0 | 0.1 | GO:0038038 | G-protein coupled receptor homodimeric complex(GO:0038038) |
| 0.0 | 0.4 | GO:0033202 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.0 | GO:0035102 | PRC1 complex(GO:0035102) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 1.3 | GO:0101003 | ficolin-1-rich granule membrane(GO:0101003) |
| 0.0 | 0.5 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.1 | GO:0042272 | nuclear RNA export factor complex(GO:0042272) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.4 | 1.3 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.2 | 1.4 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 0.6 | GO:0052831 | inositol tetrakisphosphate 1-kinase activity(GO:0047325) inositol-1,3,4-trisphosphate 6-kinase activity(GO:0052725) inositol-1,3,4-trisphosphate 5-kinase activity(GO:0052726) inositol-1,3,4,5,6-pentakisphosphate 1-phosphatase activity(GO:0052825) inositol-1,3,4,6-tetrakisphosphate 6-phosphatase activity(GO:0052830) inositol-1,3,4,6-tetrakisphosphate 1-phosphatase activity(GO:0052831) inositol-3,4,6-trisphosphate 1-kinase activity(GO:0052835) |
| 0.2 | 1.9 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.2 | 0.5 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.1 | 0.4 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.1 | 0.4 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.1 | 1.1 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 1.7 | GO:0003708 | retinoic acid receptor activity(GO:0003708) |
| 0.1 | 0.3 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.1 | 0.2 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.5 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.1 | 0.2 | GO:0070025 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.1 | 0.3 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.1 | 0.2 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.1 | 0.7 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 0.2 | GO:0004662 | CAAX-protein geranylgeranyltransferase activity(GO:0004662) |
| 0.1 | 0.3 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.2 | GO:0035650 | AP-1 adaptor complex binding(GO:0035650) |
| 0.1 | 0.2 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.1 | 0.3 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.1 | 0.2 | GO:0003945 | N-acetyllactosamine synthase activity(GO:0003945) |
| 0.1 | 0.6 | GO:0019211 | phosphatase activator activity(GO:0019211) |
| 0.1 | 0.8 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.0 | 0.1 | GO:0098808 | mRNA cap binding(GO:0098808) |
| 0.0 | 0.5 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.4 | GO:0071936 | coreceptor activity involved in Wnt signaling pathway(GO:0071936) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.1 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.8 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.5 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 1.0 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.8 | GO:0070411 | I-SMAD binding(GO:0070411) |
| 0.0 | 0.1 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.0 | 0.7 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0017082 | mineralocorticoid receptor activity(GO:0017082) |
| 0.0 | 0.2 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.3 | GO:0005087 | Ran guanyl-nucleotide exchange factor activity(GO:0005087) |
| 0.0 | 0.5 | GO:0035256 | G-protein coupled glutamate receptor binding(GO:0035256) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.1 | GO:0004731 | purine-nucleoside phosphorylase activity(GO:0004731) |
| 0.0 | 0.2 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.3 | GO:0000978 | RNA polymerase II core promoter proximal region sequence-specific DNA binding(GO:0000978) |
| 0.0 | 0.9 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.9 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.5 | GO:0017110 | nucleoside-diphosphatase activity(GO:0017110) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.5 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.1 | GO:0004347 | glucose-6-phosphate isomerase activity(GO:0004347) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
| 0.0 | 0.4 | GO:0015279 | store-operated calcium channel activity(GO:0015279) |
| 0.0 | 0.2 | GO:0016618 | hydroxypyruvate reductase activity(GO:0016618) glyoxylate reductase (NADP) activity(GO:0030267) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.2 | GO:0004704 | NF-kappaB-inducing kinase activity(GO:0004704) |
| 0.0 | 0.2 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.3 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 1.2 | GO:0004683 | calmodulin-dependent protein kinase activity(GO:0004683) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0022897 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.5 | GO:0004012 | phospholipid-translocating ATPase activity(GO:0004012) |
| 0.0 | 0.1 | GO:0017050 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.1 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.0 | 0.1 | GO:0034432 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0035033 | histone deacetylase regulator activity(GO:0035033) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.3 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.7 | GO:0030159 | receptor signaling complex scaffold activity(GO:0030159) |
| 0.0 | 0.1 | GO:0030151 | molybdenum ion binding(GO:0030151) |
| 0.0 | 0.1 | GO:0003989 | acetyl-CoA carboxylase activity(GO:0003989) |
| 0.0 | 0.4 | GO:0022848 | acetylcholine-gated cation channel activity(GO:0022848) |
| 0.0 | 0.1 | GO:0098518 | polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 5.2 | GO:0044822 | mRNA binding(GO:0003729) poly(A) RNA binding(GO:0044822) |
| 0.0 | 0.4 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0016316 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.0 | 2.0 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.2 | GO:0035240 | dopamine binding(GO:0035240) |
| 0.0 | 0.2 | GO:0043021 | ribonucleoprotein complex binding(GO:0043021) |
| 0.0 | 0.5 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.2 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.5 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 0.0 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0004652 | polynucleotide adenylyltransferase activity(GO:0004652) |
| 0.0 | 0.5 | GO:0017025 | TBP-class protein binding(GO:0017025) |
| 0.0 | 0.1 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.1 | GO:0022851 | benzodiazepine receptor activity(GO:0008503) GABA-gated chloride ion channel activity(GO:0022851) |
| 0.0 | 0.3 | GO:0030515 | snoRNA binding(GO:0030515) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0097157 | pre-mRNA intronic binding(GO:0097157) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0019826 | oxygen sensor activity(GO:0019826) |
| 0.0 | 0.2 | GO:0016783 | sulfurtransferase activity(GO:0016783) |
| 0.0 | 0.2 | GO:0031078 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.1 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0035242 | protein-arginine omega-N asymmetric methyltransferase activity(GO:0035242) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.0 | GO:0032408 | MutLbeta complex binding(GO:0032406) MutSbeta complex binding(GO:0032408) |
| 0.0 | 0.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.1 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 0.1 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 1.5 | PID RETINOIC ACID PATHWAY | Retinoic acid receptors-mediated signaling |
| 0.0 | 1.1 | ST WNT CA2 CYCLIC GMP PATHWAY | Wnt/Ca2+/cyclic GMP signaling. |
| 0.0 | 0.5 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.9 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.3 | PID BMP PATHWAY | BMP receptor signaling |
| 0.0 | 0.5 | PID EPHA2 FWD PATHWAY | EPHA2 forward signaling |
| 0.0 | 1.4 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.6 | ST ERK1 ERK2 MAPK PATHWAY | ERK1/ERK2 MAPK Pathway |
| 0.0 | 0.9 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.1 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | PID NOTCH PATHWAY | Notch signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 0.9 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.6 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.0 | 1.2 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.6 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.7 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.7 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.8 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.9 | REACTOME DOWNREGULATION OF SMAD2 3 SMAD4 TRANSCRIPTIONAL ACTIVITY | Genes involved in Downregulation of SMAD2/3:SMAD4 transcriptional activity |
| 0.0 | 0.6 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.7 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 1.1 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.7 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.6 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.4 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.8 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |
| 0.0 | 0.5 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.5 | REACTOME DCC MEDIATED ATTRACTIVE SIGNALING | Genes involved in DCC mediated attractive signaling |
| 0.0 | 0.4 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 1.5 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.6 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.8 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.7 | REACTOME ION TRANSPORT BY P TYPE ATPASES | Genes involved in Ion transport by P-type ATPases |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |
| 0.0 | 0.7 | REACTOME MITOCHONDRIAL PROTEIN IMPORT | Genes involved in Mitochondrial Protein Import |