Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for HIVEP1
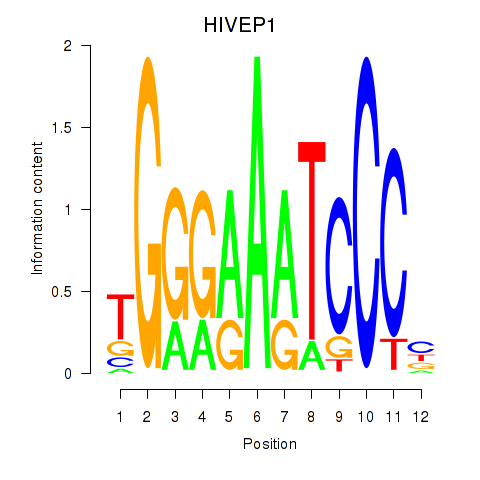
Z-value: 1.31
Transcription factors associated with HIVEP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HIVEP1
|
ENSG00000095951.12 | HIVEP1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HIVEP1 | hg19_v2_chr6_+_12012536_12012571 | 0.87 | 5.3e-03 | Click! |
Activity profile of HIVEP1 motif
Sorted Z-values of HIVEP1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HIVEP1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_-_209824643 | 3.23 |
ENST00000391911.1 ENST00000415782.1 |
LAMB3 |
laminin, beta 3 |
| chr6_+_30850697 | 2.58 |
ENST00000509639.1 ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr19_-_51522955 | 2.44 |
ENST00000358789.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr1_+_209602771 | 2.03 |
ENST00000440276.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr1_-_242612779 | 1.96 |
ENST00000427495.1 |
PLD5 |
phospholipase D family, member 5 |
| chr7_+_69064300 | 1.96 |
ENST00000342771.4 |
AUTS2 |
autism susceptibility candidate 2 |
| chr1_+_46640750 | 1.79 |
ENST00000372003.1 |
TSPAN1 |
tetraspanin 1 |
| chr1_-_209979375 | 1.76 |
ENST00000367021.3 |
IRF6 |
interferon regulatory factor 6 |
| chr16_+_22825475 | 1.70 |
ENST00000261374.3 |
HS3ST2 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 2 |
| chr7_-_22396533 | 1.35 |
ENST00000344041.6 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr15_-_72490114 | 1.32 |
ENST00000309731.7 |
GRAMD2 |
GRAM domain containing 2 |
| chr19_-_51523412 | 1.31 |
ENST00000391805.1 ENST00000599077.1 |
KLK10 |
kallikrein-related peptidase 10 |
| chr11_+_1860682 | 1.26 |
ENST00000381906.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr19_-_51523275 | 1.23 |
ENST00000309958.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr11_+_18287721 | 1.22 |
ENST00000356524.4 |
SAA1 |
serum amyloid A1 |
| chr11_+_18287801 | 1.22 |
ENST00000532858.1 ENST00000405158.2 |
SAA1 |
serum amyloid A1 |
| chr11_-_72492903 | 1.19 |
ENST00000537947.1 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr17_+_48610074 | 1.13 |
ENST00000503690.1 ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3 |
epsin 3 |
| chr12_+_53342625 | 1.12 |
ENST00000388837.2 ENST00000550600.1 ENST00000388835.3 |
KRT18 |
keratin 18 |
| chr19_+_35739897 | 1.10 |
ENST00000605618.1 ENST00000427250.1 ENST00000601623.1 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr3_-_69435428 | 1.10 |
ENST00000542259.1 |
FRMD4B |
FERM domain containing 4B |
| chr3_-_69435224 | 1.06 |
ENST00000398540.3 |
FRMD4B |
FERM domain containing 4B |
| chr2_+_163175394 | 1.04 |
ENST00000446271.1 ENST00000429691.2 |
GCA |
grancalcin, EF-hand calcium binding protein |
| chr11_-_119999539 | 0.99 |
ENST00000541857.1 |
TRIM29 |
tripartite motif containing 29 |
| chr12_-_53320245 | 0.99 |
ENST00000552150.1 |
KRT8 |
keratin 8 |
| chr11_+_1860200 | 0.95 |
ENST00000381911.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr11_-_119999611 | 0.93 |
ENST00000529044.1 |
TRIM29 |
tripartite motif containing 29 |
| chr11_-_72492878 | 0.86 |
ENST00000535054.1 ENST00000545082.1 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr4_+_155665123 | 0.83 |
ENST00000336356.3 |
LRAT |
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase) |
| chr3_-_124839648 | 0.82 |
ENST00000430155.2 |
SLC12A8 |
solute carrier family 12, member 8 |
| chr8_-_15095832 | 0.81 |
ENST00000382080.1 |
SGCZ |
sarcoglycan, zeta |
| chrX_-_117107680 | 0.79 |
ENST00000447671.2 ENST00000262820.3 |
KLHL13 |
kelch-like family member 13 |
| chr15_+_90728145 | 0.75 |
ENST00000561085.1 ENST00000379122.3 ENST00000332496.6 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr1_+_156119466 | 0.71 |
ENST00000414683.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr1_+_24645865 | 0.71 |
ENST00000342072.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr2_-_235405679 | 0.71 |
ENST00000390645.2 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr11_+_121447469 | 0.63 |
ENST00000532694.1 ENST00000534286.1 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chrX_-_117107542 | 0.62 |
ENST00000371878.1 |
KLHL13 |
kelch-like family member 13 |
| chr6_+_116782527 | 0.60 |
ENST00000368606.3 ENST00000368605.1 |
FAM26F |
family with sequence similarity 26, member F |
| chr2_+_64681641 | 0.59 |
ENST00000409537.2 |
LGALSL |
lectin, galactoside-binding-like |
| chr1_-_153066998 | 0.57 |
ENST00000368750.3 |
SPRR2E |
small proline-rich protein 2E |
| chr19_+_45504688 | 0.57 |
ENST00000221452.8 ENST00000540120.1 ENST00000505236.1 |
RELB |
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr9_+_72002837 | 0.56 |
ENST00000377216.3 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr8_-_17533838 | 0.56 |
ENST00000400046.1 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr2_-_237412331 | 0.56 |
ENST00000418802.1 |
IQCA1 |
IQ motif containing with AAA domain 1 |
| chr12_+_7055631 | 0.56 |
ENST00000543115.1 ENST00000399448.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr11_+_34642656 | 0.55 |
ENST00000257831.3 ENST00000450654.2 |
EHF |
ets homologous factor |
| chr2_+_61108771 | 0.55 |
ENST00000394479.3 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr3_+_98482175 | 0.55 |
ENST00000485391.1 ENST00000492254.1 |
ST3GAL6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr2_+_26568965 | 0.55 |
ENST00000260585.7 ENST00000447170.1 |
EPT1 |
ethanolaminephosphotransferase 1 (CDP-ethanolamine-specific) |
| chr4_-_56502451 | 0.54 |
ENST00000511469.1 ENST00000264218.3 |
NMU |
neuromedin U |
| chr1_+_24645807 | 0.54 |
ENST00000361548.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr19_+_35485682 | 0.54 |
ENST00000599564.1 |
GRAMD1A |
GRAM domain containing 1A |
| chr1_+_156123359 | 0.53 |
ENST00000368284.1 ENST00000368286.2 ENST00000438830.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr1_+_156123318 | 0.52 |
ENST00000368285.3 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr1_+_212738676 | 0.52 |
ENST00000366981.4 ENST00000366987.2 |
ATF3 |
activating transcription factor 3 |
| chr2_-_163175133 | 0.51 |
ENST00000421365.2 ENST00000263642.2 |
IFIH1 |
interferon induced with helicase C domain 1 |
| chr6_+_138188551 | 0.51 |
ENST00000237289.4 ENST00000433680.1 |
TNFAIP3 |
tumor necrosis factor, alpha-induced protein 3 |
| chr15_-_75017711 | 0.51 |
ENST00000567032.1 ENST00000564596.1 ENST00000566503.1 ENST00000395049.4 ENST00000395048.2 ENST00000379727.3 |
CYP1A1 |
cytochrome P450, family 1, subfamily A, polypeptide 1 |
| chr19_-_51466681 | 0.50 |
ENST00000456750.2 |
KLK6 |
kallikrein-related peptidase 6 |
| chr2_+_106468204 | 0.50 |
ENST00000425756.1 ENST00000393349.2 |
NCK2 |
NCK adaptor protein 2 |
| chr17_-_18908040 | 0.49 |
ENST00000388995.6 |
FAM83G |
family with sequence similarity 83, member G |
| chr4_+_155484155 | 0.48 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr2_+_61108650 | 0.48 |
ENST00000295025.8 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr12_-_48298785 | 0.48 |
ENST00000550325.1 ENST00000546653.1 ENST00000549336.1 ENST00000535672.1 ENST00000229022.3 ENST00000548664.1 |
VDR |
vitamin D (1,25- dihydroxyvitamin D3) receptor |
| chr4_-_76944621 | 0.48 |
ENST00000306602.1 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
| chr3_-_111852128 | 0.46 |
ENST00000308910.4 |
GCSAM |
germinal center-associated, signaling and motility |
| chr4_-_80994210 | 0.46 |
ENST00000403729.2 |
ANTXR2 |
anthrax toxin receptor 2 |
| chr1_+_155051305 | 0.46 |
ENST00000368408.3 |
EFNA3 |
ephrin-A3 |
| chr10_+_129705299 | 0.45 |
ENST00000254667.3 |
PTPRE |
protein tyrosine phosphatase, receptor type, E |
| chr6_+_160327974 | 0.45 |
ENST00000252660.4 |
MAS1 |
MAS1 oncogene |
| chr2_-_74781061 | 0.45 |
ENST00000264094.3 ENST00000393937.2 ENST00000409986.1 |
LOXL3 |
lysyl oxidase-like 3 |
| chr5_+_150591678 | 0.44 |
ENST00000523466.1 |
GM2A |
GM2 ganglioside activator |
| chr14_+_77648167 | 0.43 |
ENST00000554346.1 ENST00000298351.4 |
TMEM63C |
transmembrane protein 63C |
| chr1_+_156117149 | 0.43 |
ENST00000435124.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr1_-_116383322 | 0.43 |
ENST00000429731.1 |
NHLH2 |
nescient helix loop helix 2 |
| chr21_+_44589118 | 0.42 |
ENST00000291554.2 |
CRYAA |
crystallin, alpha A |
| chr14_-_35873856 | 0.42 |
ENST00000553342.1 ENST00000216797.5 ENST00000557140.1 |
NFKBIA |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, alpha |
| chr1_+_24117627 | 0.41 |
ENST00000400061.1 |
LYPLA2 |
lysophospholipase II |
| chr5_-_35230434 | 0.39 |
ENST00000504500.1 |
PRLR |
prolactin receptor |
| chr1_+_37940153 | 0.39 |
ENST00000373087.6 |
ZC3H12A |
zinc finger CCCH-type containing 12A |
| chr6_-_116381918 | 0.38 |
ENST00000606080.1 |
FRK |
fyn-related kinase |
| chr22_+_20748405 | 0.38 |
ENST00000400451.2 ENST00000403682.3 ENST00000357502.5 |
ZNF74 |
zinc finger protein 74 |
| chr1_-_200379129 | 0.36 |
ENST00000367353.1 |
ZNF281 |
zinc finger protein 281 |
| chr6_+_29691198 | 0.36 |
ENST00000440587.2 ENST00000434407.2 |
HLA-F |
major histocompatibility complex, class I, F |
| chr2_-_73511559 | 0.36 |
ENST00000521871.1 |
FBXO41 |
F-box protein 41 |
| chr8_+_86121448 | 0.36 |
ENST00000520225.1 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr15_+_91411810 | 0.35 |
ENST00000268171.3 |
FURIN |
furin (paired basic amino acid cleaving enzyme) |
| chr6_-_4079334 | 0.35 |
ENST00000492651.1 ENST00000498677.1 ENST00000274673.3 |
FAM217A |
family with sequence similarity 217, member A |
| chr15_-_90222642 | 0.35 |
ENST00000430628.2 |
PLIN1 |
perilipin 1 |
| chr19_-_33555780 | 0.35 |
ENST00000254260.3 ENST00000400226.4 |
RHPN2 |
rhophilin, Rho GTPase binding protein 2 |
| chr15_+_73976715 | 0.34 |
ENST00000558689.1 ENST00000560786.2 ENST00000561213.1 ENST00000563584.1 ENST00000561416.1 |
CD276 |
CD276 molecule |
| chr15_-_90222610 | 0.34 |
ENST00000300055.5 |
PLIN1 |
perilipin 1 |
| chr16_+_50730910 | 0.34 |
ENST00000300589.2 |
NOD2 |
nucleotide-binding oligomerization domain containing 2 |
| chr15_+_74908147 | 0.34 |
ENST00000568139.1 ENST00000563297.1 ENST00000568488.1 ENST00000352989.5 ENST00000348245.3 |
CLK3 |
CDC-like kinase 3 |
| chr6_-_10694766 | 0.34 |
ENST00000460742.2 ENST00000259983.3 ENST00000379586.1 |
C6orf52 |
chromosome 6 open reading frame 52 |
| chr5_+_140579162 | 0.34 |
ENST00000536699.1 ENST00000354757.3 |
PCDHB11 |
protocadherin beta 11 |
| chr13_-_46756351 | 0.33 |
ENST00000323076.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr1_-_155959853 | 0.33 |
ENST00000462460.2 ENST00000368316.1 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr1_+_15272271 | 0.33 |
ENST00000400797.3 |
KAZN |
kazrin, periplakin interacting protein |
| chr3_-_119396193 | 0.32 |
ENST00000484810.1 ENST00000497116.1 ENST00000261070.2 |
COX17 |
COX17 cytochrome c oxidase copper chaperone |
| chr12_+_72056773 | 0.32 |
ENST00000308086.2 |
THAP2 |
THAP domain containing, apoptosis associated protein 2 |
| chr22_-_38380543 | 0.32 |
ENST00000396884.2 |
SOX10 |
SRY (sex determining region Y)-box 10 |
| chr1_-_94079648 | 0.32 |
ENST00000370247.3 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr3_+_97868170 | 0.32 |
ENST00000437310.1 |
OR5H14 |
olfactory receptor, family 5, subfamily H, member 14 |
| chr4_-_80993854 | 0.32 |
ENST00000346652.6 |
ANTXR2 |
anthrax toxin receptor 2 |
| chr1_-_200379180 | 0.32 |
ENST00000294740.3 |
ZNF281 |
zinc finger protein 281 |
| chr6_-_31324943 | 0.31 |
ENST00000412585.2 ENST00000434333.1 |
HLA-B |
major histocompatibility complex, class I, B |
| chr19_-_42894420 | 0.31 |
ENST00000597255.1 ENST00000222032.5 |
CNFN |
cornifelin |
| chr9_+_131644388 | 0.31 |
ENST00000372600.4 |
LRRC8A |
leucine rich repeat containing 8 family, member A |
| chr10_-_120925054 | 0.31 |
ENST00000419372.1 ENST00000369131.4 ENST00000330036.6 ENST00000355697.2 |
SFXN4 |
sideroflexin 4 |
| chr1_-_47069886 | 0.31 |
ENST00000371946.4 ENST00000371945.4 ENST00000428112.2 ENST00000529170.1 |
MKNK1 |
MAP kinase interacting serine/threonine kinase 1 |
| chr17_+_38024417 | 0.30 |
ENST00000348931.4 ENST00000583811.1 ENST00000584588.1 ENST00000377940.3 |
ZPBP2 |
zona pellucida binding protein 2 |
| chr17_-_39093672 | 0.30 |
ENST00000209718.3 ENST00000436344.3 ENST00000485751.1 |
KRT23 |
keratin 23 (histone deacetylase inducible) |
| chr11_-_75062829 | 0.30 |
ENST00000393505.4 |
ARRB1 |
arrestin, beta 1 |
| chr11_+_71846764 | 0.30 |
ENST00000456237.1 ENST00000442948.2 ENST00000546166.1 |
FOLR3 |
folate receptor 3 (gamma) |
| chr2_+_11752379 | 0.30 |
ENST00000396123.1 |
GREB1 |
growth regulation by estrogen in breast cancer 1 |
| chr7_-_98741714 | 0.30 |
ENST00000361125.1 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
| chr1_+_22333943 | 0.29 |
ENST00000400271.2 |
CELA3A |
chymotrypsin-like elastase family, member 3A |
| chr6_+_29691056 | 0.29 |
ENST00000414333.1 ENST00000334668.4 ENST00000259951.7 |
HLA-F |
major histocompatibility complex, class I, F |
| chr9_-_114090713 | 0.29 |
ENST00000302681.1 ENST00000374428.1 |
OR2K2 |
olfactory receptor, family 2, subfamily K, member 2 |
| chr6_+_129204337 | 0.29 |
ENST00000421865.2 |
LAMA2 |
laminin, alpha 2 |
| chr7_-_98741642 | 0.29 |
ENST00000361368.2 |
SMURF1 |
SMAD specific E3 ubiquitin protein ligase 1 |
| chr6_+_43484760 | 0.29 |
ENST00000372389.3 ENST00000372344.2 ENST00000304004.3 ENST00000423780.1 |
POLR1C |
polymerase (RNA) I polypeptide C, 30kDa |
| chr1_-_47069955 | 0.29 |
ENST00000341183.5 ENST00000496619.1 |
MKNK1 |
MAP kinase interacting serine/threonine kinase 1 |
| chr1_+_111770232 | 0.29 |
ENST00000369744.2 |
CHI3L2 |
chitinase 3-like 2 |
| chr1_+_111770278 | 0.29 |
ENST00000369748.4 |
CHI3L2 |
chitinase 3-like 2 |
| chr14_-_22005062 | 0.29 |
ENST00000317492.5 |
SALL2 |
spalt-like transcription factor 2 |
| chr20_+_3024266 | 0.29 |
ENST00000245983.2 ENST00000359100.2 ENST00000359987.1 |
GNRH2 |
gonadotropin-releasing hormone 2 |
| chr9_+_21409146 | 0.28 |
ENST00000380205.1 |
IFNA8 |
interferon, alpha 8 |
| chr3_+_127634069 | 0.28 |
ENST00000405109.1 |
KBTBD12 |
kelch repeat and BTB (POZ) domain containing 12 |
| chr21_-_43373999 | 0.28 |
ENST00000380486.3 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
| chr11_+_71846748 | 0.28 |
ENST00000445078.2 |
FOLR3 |
folate receptor 3 (gamma) |
| chr3_+_118905564 | 0.27 |
ENST00000460625.1 |
UPK1B |
uroplakin 1B |
| chr10_-_100027943 | 0.27 |
ENST00000260702.3 |
LOXL4 |
lysyl oxidase-like 4 |
| chr12_+_53662110 | 0.27 |
ENST00000552462.1 |
ESPL1 |
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr16_-_11681316 | 0.27 |
ENST00000571688.1 |
LITAF |
lipopolysaccharide-induced TNF factor |
| chr19_-_46272462 | 0.27 |
ENST00000317578.6 |
SIX5 |
SIX homeobox 5 |
| chr19_+_39390587 | 0.27 |
ENST00000572515.1 ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr2_+_202047843 | 0.27 |
ENST00000272879.5 ENST00000374650.3 ENST00000346817.5 ENST00000313728.7 ENST00000448480.1 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr10_+_74653330 | 0.26 |
ENST00000334011.5 |
OIT3 |
oncoprotein induced transcript 3 |
| chr9_-_71629010 | 0.26 |
ENST00000377276.2 |
PRKACG |
protein kinase, cAMP-dependent, catalytic, gamma |
| chr14_+_23842018 | 0.26 |
ENST00000397242.2 ENST00000329715.2 |
IL25 |
interleukin 25 |
| chr3_+_52821841 | 0.26 |
ENST00000405128.3 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr3_+_97887544 | 0.26 |
ENST00000356526.2 |
OR5H15 |
olfactory receptor, family 5, subfamily H, member 15 |
| chr1_+_46713357 | 0.26 |
ENST00000442598.1 |
RAD54L |
RAD54-like (S. cerevisiae) |
| chrX_-_139015153 | 0.25 |
ENST00000370557.1 |
ATP11C |
ATPase, class VI, type 11C |
| chr17_-_48943706 | 0.25 |
ENST00000499247.2 |
TOB1 |
transducer of ERBB2, 1 |
| chr19_-_2042065 | 0.25 |
ENST00000591588.1 ENST00000591142.1 |
MKNK2 |
MAP kinase interacting serine/threonine kinase 2 |
| chr5_+_147443534 | 0.25 |
ENST00000398454.1 ENST00000359874.3 ENST00000508733.1 ENST00000256084.7 |
SPINK5 |
serine peptidase inhibitor, Kazal type 5 |
| chr19_-_4831701 | 0.25 |
ENST00000248244.5 |
TICAM1 |
toll-like receptor adaptor molecule 1 |
| chr1_+_46713404 | 0.25 |
ENST00000371975.4 ENST00000469835.1 |
RAD54L |
RAD54-like (S. cerevisiae) |
| chr17_-_41050716 | 0.25 |
ENST00000417193.1 ENST00000301683.3 ENST00000436546.1 ENST00000431109.2 |
LINC00671 |
long intergenic non-protein coding RNA 671 |
| chr14_+_22771851 | 0.25 |
ENST00000390466.1 |
TRAV39 |
T cell receptor alpha variable 39 |
| chr14_+_103243813 | 0.25 |
ENST00000560371.1 ENST00000347662.4 ENST00000392745.2 ENST00000539721.1 ENST00000560463.1 |
TRAF3 |
TNF receptor-associated factor 3 |
| chr7_-_8302298 | 0.25 |
ENST00000446305.1 |
ICA1 |
islet cell autoantigen 1, 69kDa |
| chr1_+_84873913 | 0.24 |
ENST00000370662.3 |
DNASE2B |
deoxyribonuclease II beta |
| chr3_-_9291063 | 0.24 |
ENST00000383836.3 |
SRGAP3 |
SLIT-ROBO Rho GTPase activating protein 3 |
| chr11_-_75062730 | 0.24 |
ENST00000420843.2 ENST00000360025.3 |
ARRB1 |
arrestin, beta 1 |
| chr3_-_119813264 | 0.24 |
ENST00000264235.8 |
GSK3B |
glycogen synthase kinase 3 beta |
| chr4_-_80993717 | 0.24 |
ENST00000307333.7 |
ANTXR2 |
anthrax toxin receptor 2 |
| chr1_-_10532531 | 0.24 |
ENST00000377036.2 ENST00000377038.3 |
DFFA |
DNA fragmentation factor, 45kDa, alpha polypeptide |
| chr12_-_55375622 | 0.24 |
ENST00000316577.8 |
TESPA1 |
thymocyte expressed, positive selection associated 1 |
| chr16_-_20556492 | 0.24 |
ENST00000568098.1 |
ACSM2B |
acyl-CoA synthetase medium-chain family member 2B |
| chr14_+_22675388 | 0.24 |
ENST00000390461.2 |
TRAV34 |
T cell receptor alpha variable 34 |
| chr17_-_26876350 | 0.23 |
ENST00000470125.1 |
UNC119 |
unc-119 homolog (C. elegans) |
| chr14_+_103589789 | 0.23 |
ENST00000558056.1 ENST00000560869.1 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
| chr16_+_56970567 | 0.23 |
ENST00000563911.1 |
HERPUD1 |
homocysteine-inducible, endoplasmic reticulum stress-inducible, ubiquitin-like domain member 1 |
| chr12_+_10365404 | 0.23 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chrX_-_15353629 | 0.23 |
ENST00000333590.4 ENST00000428964.1 ENST00000542278.1 |
PIGA |
phosphatidylinositol glycan anchor biosynthesis, class A |
| chr11_-_104893863 | 0.23 |
ENST00000260315.3 ENST00000526056.1 ENST00000531367.1 ENST00000456094.1 ENST00000444749.2 ENST00000393141.2 ENST00000418434.1 ENST00000393139.2 |
CASP5 |
caspase 5, apoptosis-related cysteine peptidase |
| chr1_+_156611704 | 0.23 |
ENST00000329117.5 |
BCAN |
brevican |
| chr9_-_14722715 | 0.23 |
ENST00000380911.3 |
CER1 |
cerberus 1, DAN family BMP antagonist |
| chr2_+_102314161 | 0.23 |
ENST00000425019.1 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr3_-_121264848 | 0.23 |
ENST00000264233.5 |
POLQ |
polymerase (DNA directed), theta |
| chr19_-_44324750 | 0.22 |
ENST00000594049.1 ENST00000414615.2 |
LYPD5 |
LY6/PLAUR domain containing 5 |
| chr7_+_107110488 | 0.22 |
ENST00000304402.4 |
GPR22 |
G protein-coupled receptor 22 |
| chr15_-_41408409 | 0.22 |
ENST00000361937.3 |
INO80 |
INO80 complex subunit |
| chr2_+_202047596 | 0.22 |
ENST00000286186.6 ENST00000360132.3 |
CASP10 |
caspase 10, apoptosis-related cysteine peptidase |
| chr10_-_125851961 | 0.22 |
ENST00000346248.5 |
CHST15 |
carbohydrate (N-acetylgalactosamine 4-sulfate 6-O) sulfotransferase 15 |
| chr2_-_74753332 | 0.22 |
ENST00000451518.1 ENST00000404568.3 |
DQX1 |
DEAQ box RNA-dependent ATPase 1 |
| chr6_-_32908792 | 0.22 |
ENST00000418107.2 |
HLA-DMB |
major histocompatibility complex, class II, DM beta |
| chr12_+_52445191 | 0.22 |
ENST00000243050.1 ENST00000394825.1 ENST00000550763.1 ENST00000394824.2 ENST00000548232.1 ENST00000562373.1 |
NR4A1 |
nuclear receptor subfamily 4, group A, member 1 |
| chr17_-_41277467 | 0.22 |
ENST00000494123.1 ENST00000346315.3 ENST00000309486.4 ENST00000468300.1 ENST00000354071.3 ENST00000352993.3 ENST00000471181.2 |
BRCA1 |
breast cancer 1, early onset |
| chr12_-_113909877 | 0.21 |
ENST00000261731.3 |
LHX5 |
LIM homeobox 5 |
| chr17_-_40333150 | 0.21 |
ENST00000264661.3 |
KCNH4 |
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr7_-_100034060 | 0.21 |
ENST00000292330.2 |
PPP1R35 |
protein phosphatase 1, regulatory subunit 35 |
| chr9_-_131486367 | 0.21 |
ENST00000372663.4 ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12 |
zinc finger, DHHC-type containing 12 |
| chr2_-_74753305 | 0.21 |
ENST00000393951.2 |
DQX1 |
DEAQ box RNA-dependent ATPase 1 |
| chr8_-_70747205 | 0.21 |
ENST00000260126.4 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr3_-_49142178 | 0.20 |
ENST00000452739.1 ENST00000414533.1 ENST00000417025.1 |
QARS |
glutaminyl-tRNA synthetase |
| chr12_+_53662073 | 0.20 |
ENST00000553219.1 ENST00000257934.4 |
ESPL1 |
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr14_-_24804269 | 0.20 |
ENST00000310677.4 ENST00000554068.2 ENST00000559167.1 ENST00000561138.1 |
ADCY4 |
adenylate cyclase 4 |
| chr6_-_29399744 | 0.20 |
ENST00000377154.1 |
OR5V1 |
olfactory receptor, family 5, subfamily V, member 1 |
| chr1_+_54359854 | 0.20 |
ENST00000361921.3 ENST00000322679.6 ENST00000532493.1 ENST00000525202.1 ENST00000524406.1 ENST00000388876.3 |
DIO1 |
deiodinase, iodothyronine, type I |
| chr5_+_142149955 | 0.20 |
ENST00000378004.3 |
ARHGAP26 |
Rho GTPase activating protein 26 |
| chr13_+_30002741 | 0.19 |
ENST00000380808.2 |
MTUS2 |
microtubule associated tumor suppressor candidate 2 |
| chr9_-_32526184 | 0.19 |
ENST00000545044.1 ENST00000379868.1 |
DDX58 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr6_-_31239846 | 0.19 |
ENST00000415537.1 ENST00000376228.5 ENST00000383329.3 |
HLA-C |
major histocompatibility complex, class I, C |
| chr14_-_69446034 | 0.19 |
ENST00000193403.6 |
ACTN1 |
actinin, alpha 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.0 | GO:0098582 | innate vocalization behavior(GO:0098582) |
| 0.4 | 1.1 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.3 | 2.6 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 1.1 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.2 | 0.9 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.2 | 0.6 | GO:1902769 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.2 | 0.6 | GO:1902534 | single-organism membrane invagination(GO:1902534) |
| 0.2 | 2.1 | GO:0032782 | bile acid secretion(GO:0032782) |
| 0.2 | 0.5 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.2 | 0.5 | GO:0071947 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.2 | 0.5 | GO:0018894 | dibenzo-p-dioxin metabolic process(GO:0018894) cellular alkene metabolic process(GO:0043449) |
| 0.2 | 0.5 | GO:0060058 | apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.1 | 0.4 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.1 | 0.6 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.1 | 0.4 | GO:0070427 | nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070427) |
| 0.1 | 1.3 | GO:0002480 | antigen processing and presentation of exogenous peptide antigen via MHC class I, TAP-independent(GO:0002480) |
| 0.1 | 0.4 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.1 | 0.4 | GO:2000620 | positive regulation of histone H4-K16 acetylation(GO:2000620) |
| 0.1 | 0.5 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.4 | GO:0032900 | negative regulation of low-density lipoprotein particle receptor catabolic process(GO:0032804) negative regulation of neurotrophin production(GO:0032900) negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.3 | GO:0060585 | detection of peptidoglycan(GO:0032499) activation of MAPK activity involved in innate immune response(GO:0035419) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 3.2 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.1 | 0.4 | GO:0010387 | COP9 signalosome assembly(GO:0010387) |
| 0.1 | 1.6 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.1 | 0.3 | GO:0042640 | anagen(GO:0042640) |
| 0.1 | 0.5 | GO:0036493 | positive regulation of translation in response to endoplasmic reticulum stress(GO:0036493) |
| 0.1 | 0.2 | GO:0071109 | superior temporal gyrus development(GO:0071109) negative regulation of dopaminergic neuron differentiation(GO:1904339) |
| 0.1 | 0.3 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 0.2 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.1 | 0.3 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.1 | 1.1 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.1 | 0.5 | GO:2000402 | negative regulation of lymphocyte migration(GO:2000402) |
| 0.1 | 0.2 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.1 | 0.1 | GO:0070100 | regulation of chemokine-mediated signaling pathway(GO:0070099) negative regulation of chemokine-mediated signaling pathway(GO:0070100) |
| 0.1 | 2.2 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.1 | 1.3 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 2.7 | GO:0050716 | positive regulation of interleukin-1 secretion(GO:0050716) |
| 0.1 | 0.5 | GO:0090240 | positive regulation of histone H4 acetylation(GO:0090240) |
| 0.1 | 0.2 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.1 | 2.2 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.1 | 0.3 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.1 | 0.3 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.1 | 0.7 | GO:2000252 | negative regulation of feeding behavior(GO:2000252) |
| 0.1 | 0.6 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.1 | 0.2 | GO:0048633 | positive regulation of skeletal muscle tissue growth(GO:0048633) |
| 0.1 | 0.3 | GO:0045357 | interferon-beta biosynthetic process(GO:0045350) regulation of interferon-beta biosynthetic process(GO:0045357) positive regulation of interferon-beta biosynthetic process(GO:0045359) |
| 0.1 | 0.6 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.8 | GO:0006776 | vitamin A metabolic process(GO:0006776) |
| 0.1 | 0.5 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.1 | 0.4 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.1 | 0.2 | GO:1900186 | negative regulation of clathrin-mediated endocytosis(GO:1900186) negative regulation of caveolin-mediated endocytosis(GO:2001287) |
| 0.1 | 0.9 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.1 | 0.2 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.2 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.4 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.6 | GO:0006032 | chitin metabolic process(GO:0006030) chitin catabolic process(GO:0006032) |
| 0.0 | 0.3 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.2 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.4 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.5 | GO:1905245 | regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902959) regulation of aspartic-type peptidase activity(GO:1905245) |
| 0.0 | 0.1 | GO:0021569 | rhombomere 3 development(GO:0021569) |
| 0.0 | 0.2 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.6 | GO:0015884 | folic acid transport(GO:0015884) |
| 0.0 | 2.2 | GO:0050919 | negative chemotaxis(GO:0050919) |
| 0.0 | 0.1 | GO:1901860 | positive regulation of mitochondrial DNA metabolic process(GO:1901860) |
| 0.0 | 0.1 | GO:0098886 | modification of dendritic spine(GO:0098886) |
| 0.0 | 0.1 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.3 | GO:0006701 | progesterone biosynthetic process(GO:0006701) |
| 0.0 | 0.1 | GO:0046832 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.3 | GO:0000160 | phosphorelay signal transduction system(GO:0000160) |
| 0.0 | 0.1 | GO:0031938 | regulation of chromatin silencing at telomere(GO:0031938) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.3 | GO:0032224 | positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.5 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.3 | GO:0060753 | regulation of mast cell chemotaxis(GO:0060753) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:0045229 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.2 | GO:0014010 | Schwann cell proliferation(GO:0014010) |
| 0.0 | 0.2 | GO:0036123 | histone H3-K9 dimethylation(GO:0036123) |
| 0.0 | 0.5 | GO:0008340 | determination of adult lifespan(GO:0008340) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:1990258 | box C/D snoRNA 3'-end processing(GO:0000494) box C/D snoRNA metabolic process(GO:0033967) box C/D snoRNA processing(GO:0034963) histone glutamine methylation(GO:1990258) |
| 0.0 | 0.8 | GO:0046716 | muscle cell cellular homeostasis(GO:0046716) |
| 0.0 | 0.3 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 2.2 | GO:0030049 | muscle filament sliding(GO:0030049) actin-myosin filament sliding(GO:0033275) |
| 0.0 | 0.4 | GO:0031441 | negative regulation of mRNA 3'-end processing(GO:0031441) |
| 0.0 | 0.2 | GO:0061469 | regulation of type B pancreatic cell proliferation(GO:0061469) |
| 0.0 | 0.2 | GO:0035897 | proteolysis in other organism(GO:0035897) |
| 0.0 | 0.3 | GO:0001675 | acrosome assembly(GO:0001675) |
| 0.0 | 0.2 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.0 | GO:2001183 | negative regulation of interleukin-12 secretion(GO:2001183) |
| 0.0 | 0.2 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
| 0.0 | 0.1 | GO:1900246 | positive regulation of RIG-I signaling pathway(GO:1900246) |
| 0.0 | 0.0 | GO:1903936 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.2 | GO:0016446 | somatic hypermutation of immunoglobulin genes(GO:0016446) |
| 0.0 | 0.1 | GO:0032929 | negative regulation of superoxide anion generation(GO:0032929) |
| 0.0 | 0.1 | GO:0071321 | cellular response to cGMP(GO:0071321) |
| 0.0 | 0.2 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 2.2 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.1 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.2 | GO:1903071 | positive regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903071) |
| 0.0 | 0.4 | GO:0010155 | regulation of proton transport(GO:0010155) |
| 0.0 | 2.8 | GO:0006024 | glycosaminoglycan biosynthetic process(GO:0006024) |
| 0.0 | 1.0 | GO:1901998 | toxin transport(GO:1901998) |
| 0.0 | 0.1 | GO:2000612 | thyroid-stimulating hormone secretion(GO:0070460) regulation of thyroid-stimulating hormone secretion(GO:2000612) |
| 0.0 | 0.1 | GO:0060056 | mammary gland involution(GO:0060056) |
| 0.0 | 0.4 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:0045082 | positive regulation of interleukin-10 biosynthetic process(GO:0045082) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.1 | GO:0060215 | primitive hemopoiesis(GO:0060215) |
| 0.0 | 0.4 | GO:0007617 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:1900363 | regulation of mRNA polyadenylation(GO:1900363) |
| 0.0 | 0.1 | GO:0070777 | D-aspartate transport(GO:0070777) D-aspartate import(GO:0070779) |
| 0.0 | 0.2 | GO:2001199 | negative regulation of dendritic cell differentiation(GO:2001199) |
| 0.0 | 0.7 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.0 | 0.4 | GO:0034199 | activation of protein kinase A activity(GO:0034199) |
| 0.0 | 0.1 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.1 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.0 | 0.3 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0050893 | sensory processing(GO:0050893) |
| 0.0 | 0.1 | GO:0019264 | glycine biosynthetic process from serine(GO:0019264) |
| 0.0 | 0.4 | GO:0010172 | embryonic body morphogenesis(GO:0010172) |
| 0.0 | 0.3 | GO:0071243 | cellular response to arsenic-containing substance(GO:0071243) |
| 0.0 | 0.1 | GO:0060010 | Sertoli cell fate commitment(GO:0060010) |
| 0.0 | 0.1 | GO:0035116 | embryonic hindlimb morphogenesis(GO:0035116) |
| 0.0 | 0.1 | GO:0042167 | porphyrin-containing compound catabolic process(GO:0006787) tetrapyrrole catabolic process(GO:0033015) heme catabolic process(GO:0042167) pigment catabolic process(GO:0046149) |
| 0.0 | 0.2 | GO:0051601 | exocyst localization(GO:0051601) |
| 0.0 | 0.1 | GO:0033590 | response to cobalamin(GO:0033590) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) positive regulation of guanylate cyclase activity(GO:0031284) |
| 0.0 | 0.1 | GO:0060437 | lung growth(GO:0060437) |
| 0.0 | 0.5 | GO:0045745 | positive regulation of G-protein coupled receptor protein signaling pathway(GO:0045745) |
| 0.0 | 0.3 | GO:0006309 | apoptotic DNA fragmentation(GO:0006309) |
| 0.0 | 1.5 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 1.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0050930 | induction of positive chemotaxis(GO:0050930) |
| 0.0 | 0.0 | GO:0035552 | oxidative single-stranded DNA demethylation(GO:0035552) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 3.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.2 | 2.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.2 | 0.5 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.2 | 2.1 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.1 | 1.1 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 1.3 | GO:0042612 | MHC class I protein complex(GO:0042612) |
| 0.1 | 0.4 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 0.4 | GO:0031436 | BRCA1-BARD1 complex(GO:0031436) |
| 0.1 | 2.2 | GO:0005861 | troponin complex(GO:0005861) |
| 0.1 | 0.8 | GO:0016012 | sarcoglycan complex(GO:0016012) |
| 0.1 | 2.4 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.1 | 0.2 | GO:0034271 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.1 | 0.4 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.1 | 0.6 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.1 | GO:0097444 | spine apparatus(GO:0097444) |
| 0.0 | 0.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.2 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.2 | GO:0072558 | NLRP1 inflammasome complex(GO:0072558) |
| 0.0 | 1.1 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.0 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.2 | GO:0033553 | rDNA heterochromatin(GO:0033553) |
| 0.0 | 0.7 | GO:0097342 | ripoptosome(GO:0097342) |
| 0.0 | 0.1 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.3 | GO:0098559 | cytoplasmic side of early endosome membrane(GO:0098559) cytoplasmic side of late endosome membrane(GO:0098560) |
| 0.0 | 1.1 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.1 | GO:0098855 | HCN channel complex(GO:0098855) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.3 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:0035032 | phosphatidylinositol 3-kinase complex, class III(GO:0035032) |
| 0.0 | 0.1 | GO:0030061 | mitochondrial crista(GO:0030061) |
| 0.0 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.3 | GO:0005605 | basal lamina(GO:0005605) |
| 0.0 | 0.3 | GO:0002199 | zona pellucida receptor complex(GO:0002199) |
| 0.0 | 1.3 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.2 | GO:0034715 | pICln-Sm protein complex(GO:0034715) |
| 0.0 | 0.2 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 0.1 | GO:0043196 | varicosity(GO:0043196) |
| 0.0 | 2.1 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.4 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.9 | GO:0008180 | COP9 signalosome(GO:0008180) |
| 0.0 | 0.3 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.0 | GO:0008537 | proteasome activator complex(GO:0008537) |
| 0.0 | 0.4 | GO:0001891 | phagocytic cup(GO:0001891) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.1 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.1 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 1.3 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
| 0.0 | 1.4 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 0.3 | GO:0044232 | organelle membrane contact site(GO:0044232) |
| 0.0 | 0.3 | GO:0031312 | extrinsic component of organelle membrane(GO:0031312) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0034464 | BBSome(GO:0034464) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 2.6 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 1.7 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.2 | 0.5 | GO:0031896 | V2 vasopressin receptor binding(GO:0031896) |
| 0.2 | 2.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.5 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.1 | 2.9 | GO:0038191 | neuropilin binding(GO:0038191) |
| 0.1 | 0.8 | GO:0016416 | O-palmitoyltransferase activity(GO:0016416) |
| 0.1 | 1.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.1 | 3.0 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.5 | GO:0070576 | vitamin D 24-hydroxylase activity(GO:0070576) |
| 0.1 | 0.5 | GO:0038181 | bile acid receptor activity(GO:0038181) vitamin D response element binding(GO:0070644) |
| 0.1 | 0.5 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.1 | 0.7 | GO:0046979 | TAP2 binding(GO:0046979) |
| 0.1 | 0.4 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 0.2 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.1 | 0.5 | GO:0097199 | cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097199) |
| 0.1 | 0.4 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.1 | 0.8 | GO:0015379 | potassium:chloride symporter activity(GO:0015379) potassium ion symporter activity(GO:0022820) |
| 0.1 | 0.2 | GO:0050659 | N-acetylgalactosamine 4-sulfate 6-O-sulfotransferase activity(GO:0050659) |
| 0.1 | 0.2 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.1 | 0.2 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.1 | 0.5 | GO:0004945 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.1 | 0.5 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 2.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.1 | 0.3 | GO:0016531 | copper chaperone activity(GO:0016531) |
| 0.0 | 0.4 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.5 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.8 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.6 | GO:0004568 | chitinase activity(GO:0004568) |
| 0.0 | 0.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.2 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.4 | GO:0098599 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.3 | GO:0000155 | phosphorelay sensor kinase activity(GO:0000155) |
| 0.0 | 0.6 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.6 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 1.0 | GO:0005001 | transmembrane receptor protein tyrosine phosphatase activity(GO:0005001) transmembrane receptor protein phosphatase activity(GO:0019198) |
| 0.0 | 0.5 | GO:0045159 | myosin II binding(GO:0045159) |
| 0.0 | 0.6 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.6 | GO:0048185 | activin binding(GO:0048185) |
| 0.0 | 0.2 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0036310 | annealing helicase activity(GO:0036310) |
| 0.0 | 0.6 | GO:0005542 | folic acid binding(GO:0005542) |
| 0.0 | 0.5 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0019862 | IgA binding(GO:0019862) |
| 0.0 | 0.1 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0004906 | interferon-gamma receptor activity(GO:0004906) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 1.1 | GO:0004198 | calcium-dependent cysteine-type endopeptidase activity(GO:0004198) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:1990259 | protein-glutamine N-methyltransferase activity(GO:0036009) histone-glutamine methyltransferase activity(GO:1990259) |
| 0.0 | 0.2 | GO:0036132 | 13-prostaglandin reductase activity(GO:0036132) 15-oxoprostaglandin 13-oxidase activity(GO:0047522) |
| 0.0 | 0.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.5 | GO:0035613 | RNA stem-loop binding(GO:0035613) |
| 0.0 | 0.1 | GO:0005222 | intracellular cAMP activated cation channel activity(GO:0005222) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.5 | GO:0017166 | vinculin binding(GO:0017166) |
| 0.0 | 0.2 | GO:0051575 | 5'-deoxyribose-5-phosphate lyase activity(GO:0051575) |
| 0.0 | 1.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.0 | 0.1 | GO:0010484 | H3 histone acetyltransferase activity(GO:0010484) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.2 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
| 0.0 | 0.5 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.3 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.3 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.3 | GO:0016641 | oxidoreductase activity, acting on the CH-NH2 group of donors, oxygen as acceptor(GO:0016641) |
| 0.0 | 0.5 | GO:0042605 | peptide antigen binding(GO:0042605) |
| 0.0 | 0.7 | GO:0051059 | NF-kappaB binding(GO:0051059) |
| 0.0 | 0.2 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 0.2 | GO:0031996 | thioesterase binding(GO:0031996) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0004691 | cAMP-dependent protein kinase activity(GO:0004691) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.1 | GO:1901612 | cardiolipin binding(GO:1901612) |
| 0.0 | 0.2 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.1 | GO:0008732 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.9 | GO:0097110 | scaffold protein binding(GO:0097110) |
| 0.0 | 3.7 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.0 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.1 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.3 | GO:0017127 | cholesterol transporter activity(GO:0017127) |
| 0.0 | 0.3 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.0 | 1.3 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.3 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 2.0 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.1 | 0.5 | PID IL6 7 PATHWAY | IL6-mediated signaling events |
| 0.0 | 1.5 | PID NFKAPPAB ATYPICAL PATHWAY | Atypical NF-kappaB pathway |
| 0.0 | 1.2 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.8 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.9 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 1.0 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
| 0.0 | 0.7 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 1.4 | PID AURORA B PATHWAY | Aurora B signaling |
| 0.0 | 0.8 | PID IL8 CXCR2 PATHWAY | IL8- and CXCR2-mediated signaling events |
| 0.0 | 0.6 | SA CASPASE CASCADE | Apoptosis is mediated by caspases, cysteine proteases arranged in a proteolytic cascade. |
| 0.0 | 1.2 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.7 | PID RXR VDR PATHWAY | RXR and RAR heterodimerization with other nuclear receptor |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 1.1 | PID CASPASE PATHWAY | Caspase cascade in apoptosis |
| 0.0 | 0.8 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.6 | PID ERBB1 RECEPTOR PROXIMAL PATHWAY | EGF receptor (ErbB1) signaling pathway |
| 0.0 | 0.2 | PID IL5 PATHWAY | IL5-mediated signaling events |
| 0.0 | 0.5 | PID CXCR3 PATHWAY | CXCR3-mediated signaling events |
| 0.0 | 0.2 | PID EPHB FWD PATHWAY | EPHB forward signaling |
| 0.0 | 0.4 | PID BARD1 PATHWAY | BARD1 signaling events |
| 0.0 | 0.3 | ST P38 MAPK PATHWAY | p38 MAPK Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | REACTOME ENDOSOMAL VACUOLAR PATHWAY | Genes involved in Endosomal/Vacuolar pathway |
| 0.1 | 3.4 | REACTOME TRAF6 MEDIATED NFKB ACTIVATION | Genes involved in TRAF6 mediated NF-kB activation |
| 0.1 | 2.0 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 0.5 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.3 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.6 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 1.7 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.9 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.2 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 2.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |
| 0.0 | 2.6 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.5 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.9 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 3.5 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.6 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.3 | REACTOME TAK1 ACTIVATES NFKB BY PHOSPHORYLATION AND ACTIVATION OF IKKS COMPLEX | Genes involved in TAK1 activates NFkB by phosphorylation and activation of IKKs complex |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.6 | REACTOME PROLACTIN RECEPTOR SIGNALING | Genes involved in Prolactin receptor signaling |
| 0.0 | 0.3 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.0 | 0.8 | REACTOME ACTIVATION OF GENES BY ATF4 | Genes involved in Activation of Genes by ATF4 |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.7 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.4 | REACTOME FANCONI ANEMIA PATHWAY | Genes involved in Fanconi Anemia pathway |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME APOPTOSIS INDUCED DNA FRAGMENTATION | Genes involved in Apoptosis induced DNA fragmentation |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.5 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.1 | REACTOME REGULATION OF IFNG SIGNALING | Genes involved in Regulation of IFNG signaling |
| 0.0 | 0.5 | REACTOME NEGATIVE REGULATORS OF RIG I MDA5 SIGNALING | Genes involved in Negative regulators of RIG-I/MDA5 signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.4 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |