Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
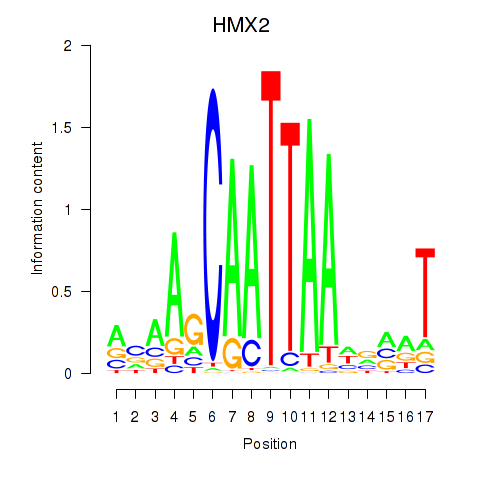
Results for HMX2
Z-value: 0.66
Transcription factors associated with HMX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX2
|
ENSG00000188816.3 | HMX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HMX2 | hg19_v2_chr10_+_124907638_124907660 | 0.56 | 1.5e-01 | Click! |
Activity profile of HMX2 motif
Sorted Z-values of HMX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_+_95066823 | 0.62 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr4_-_138453606 | 0.55 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr6_-_46293378 | 0.52 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr5_+_156696362 | 0.49 |
ENST00000377576.3 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr5_+_119867159 | 0.47 |
ENST00000505123.1 |
PRR16 |
proline rich 16 |
| chr4_-_159080806 | 0.47 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr12_-_15374343 | 0.45 |
ENST00000256953.2 ENST00000546331.1 |
RERG |
RAS-like, estrogen-regulated, growth inhibitor |
| chr15_+_41056255 | 0.44 |
ENST00000561160.1 ENST00000559445.1 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
| chr15_+_41056218 | 0.40 |
ENST00000260447.4 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
| chr2_+_152214098 | 0.40 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr11_+_5710919 | 0.38 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr4_-_138453559 | 0.38 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr9_+_99690592 | 0.37 |
ENST00000354649.3 |
NUTM2G |
NUT family member 2G |
| chr2_-_224467093 | 0.36 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chr13_-_38172863 | 0.34 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr8_-_13134045 | 0.31 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr2_-_145277569 | 0.29 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr6_+_26124373 | 0.24 |
ENST00000377791.2 ENST00000602637.1 |
HIST1H2AC |
histone cluster 1, H2ac |
| chr2_-_190044480 | 0.23 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr2_+_158114051 | 0.23 |
ENST00000259056.4 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr19_-_44384291 | 0.23 |
ENST00000324394.6 |
ZNF404 |
zinc finger protein 404 |
| chr6_+_112408768 | 0.22 |
ENST00000368656.2 ENST00000604268.1 |
FAM229B |
family with sequence similarity 229, member B |
| chr5_-_146781153 | 0.22 |
ENST00000520473.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr1_-_110933611 | 0.21 |
ENST00000472422.2 ENST00000437429.2 |
SLC16A4 |
solute carrier family 16, member 4 |
| chr19_+_15852203 | 0.20 |
ENST00000305892.1 |
OR10H3 |
olfactory receptor, family 10, subfamily H, member 3 |
| chr15_-_55700457 | 0.19 |
ENST00000442196.3 ENST00000563171.1 ENST00000425574.3 |
CCPG1 |
cell cycle progression 1 |
| chrX_-_57164058 | 0.19 |
ENST00000374906.3 |
SPIN2A |
spindlin family, member 2A |
| chr4_+_70894130 | 0.18 |
ENST00000526767.1 ENST00000530128.1 ENST00000381057.3 |
HTN3 |
histatin 3 |
| chr8_-_93978357 | 0.18 |
ENST00000522925.1 ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK |
triple QxxK/R motif containing |
| chrX_-_154493791 | 0.18 |
ENST00000369454.3 |
RAB39B |
RAB39B, member RAS oncogene family |
| chr12_+_133614119 | 0.18 |
ENST00000327668.7 |
ZNF84 |
zinc finger protein 84 |
| chr15_-_55700522 | 0.17 |
ENST00000564092.1 ENST00000310958.6 |
CCPG1 |
cell cycle progression 1 |
| chr20_+_5986727 | 0.17 |
ENST00000378863.4 |
CRLS1 |
cardiolipin synthase 1 |
| chr8_-_93978309 | 0.17 |
ENST00000517858.1 ENST00000378861.5 |
TRIQK |
triple QxxK/R motif containing |
| chr8_-_93978333 | 0.16 |
ENST00000524037.1 ENST00000520430.1 ENST00000521617.1 |
TRIQK |
triple QxxK/R motif containing |
| chr8_-_93978346 | 0.16 |
ENST00000523580.1 |
TRIQK |
triple QxxK/R motif containing |
| chr3_-_121379739 | 0.16 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr13_+_21714711 | 0.16 |
ENST00000607003.1 ENST00000492245.1 |
SAP18 |
Sin3A-associated protein, 18kDa |
| chr3_-_114343039 | 0.15 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr2_+_109237717 | 0.15 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr15_+_55700741 | 0.15 |
ENST00000569691.1 |
C15orf65 |
chromosome 15 open reading frame 65 |
| chr2_-_160654745 | 0.15 |
ENST00000259053.4 ENST00000429078.2 |
CD302 |
CD302 molecule |
| chr2_+_24272543 | 0.14 |
ENST00000380991.4 |
FKBP1B |
FK506 binding protein 1B, 12.6 kDa |
| chr1_+_70820451 | 0.14 |
ENST00000361764.4 ENST00000359875.5 ENST00000370940.5 ENST00000531950.1 ENST00000432224.1 |
HHLA3 |
HERV-H LTR-associating 3 |
| chr15_-_50558223 | 0.14 |
ENST00000267845.3 |
HDC |
histidine decarboxylase |
| chr1_-_110933663 | 0.13 |
ENST00000369781.4 ENST00000541986.1 ENST00000369779.4 |
SLC16A4 |
solute carrier family 16, member 4 |
| chr17_-_2966901 | 0.13 |
ENST00000575751.1 |
OR1D5 |
olfactory receptor, family 1, subfamily D, member 5 |
| chr1_+_99127225 | 0.13 |
ENST00000370189.5 ENST00000529992.1 |
SNX7 |
sorting nexin 7 |
| chr2_-_152590982 | 0.13 |
ENST00000409198.1 ENST00000397345.3 ENST00000427231.2 |
NEB |
nebulin |
| chr1_-_202858227 | 0.13 |
ENST00000367262.3 |
RABIF |
RAB interacting factor |
| chr8_+_77318769 | 0.12 |
ENST00000518732.1 |
RP11-706J10.1 |
long intergenic non-protein coding RNA 1111 |
| chrX_+_79675965 | 0.12 |
ENST00000308293.5 |
FAM46D |
family with sequence similarity 46, member D |
| chr3_+_38537960 | 0.12 |
ENST00000453767.1 |
EXOG |
endo/exonuclease (5'-3'), endonuclease G-like |
| chr4_+_86749045 | 0.12 |
ENST00000514229.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr10_-_49813090 | 0.11 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr1_+_144151520 | 0.11 |
ENST00000369372.4 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
| chr12_-_13248562 | 0.11 |
ENST00000457134.2 ENST00000537302.1 |
GSG1 |
germ cell associated 1 |
| chr4_-_69817481 | 0.11 |
ENST00000251566.4 |
UGT2A3 |
UDP glucuronosyltransferase 2 family, polypeptide A3 |
| chr1_-_108231101 | 0.11 |
ENST00000544443.1 ENST00000415432.2 |
VAV3 |
vav 3 guanine nucleotide exchange factor |
| chr9_+_5890802 | 0.10 |
ENST00000381477.3 ENST00000381476.1 ENST00000381471.1 |
MLANA |
melan-A |
| chr10_-_22292613 | 0.10 |
ENST00000376980.3 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr10_+_81462983 | 0.10 |
ENST00000448135.1 ENST00000429828.1 ENST00000372321.1 |
NUTM2B |
NUT family member 2B |
| chr11_-_327537 | 0.10 |
ENST00000602735.1 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr1_+_196946680 | 0.10 |
ENST00000256785.4 |
CFHR5 |
complement factor H-related 5 |
| chr13_+_24144509 | 0.10 |
ENST00000248484.4 |
TNFRSF19 |
tumor necrosis factor receptor superfamily, member 19 |
| chr1_+_196946664 | 0.09 |
ENST00000367414.5 |
CFHR5 |
complement factor H-related 5 |
| chr4_-_120243545 | 0.09 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr14_+_55595762 | 0.09 |
ENST00000254301.9 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
| chr12_+_25205568 | 0.09 |
ENST00000548766.1 ENST00000556887.1 |
LRMP |
lymphoid-restricted membrane protein |
| chr10_-_121296045 | 0.09 |
ENST00000392865.1 |
RGS10 |
regulator of G-protein signaling 10 |
| chr12_-_10541575 | 0.09 |
ENST00000540818.1 |
KLRK1 |
killer cell lectin-like receptor subfamily K, member 1 |
| chr5_+_140868717 | 0.09 |
ENST00000252087.1 |
PCDHGC5 |
protocadherin gamma subfamily C, 5 |
| chr4_+_86748898 | 0.09 |
ENST00000509300.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr4_-_100212132 | 0.08 |
ENST00000209668.2 |
ADH1A |
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr6_+_25652432 | 0.08 |
ENST00000377961.2 |
SCGN |
secretagogin, EF-hand calcium binding protein |
| chr2_-_201374781 | 0.08 |
ENST00000359878.3 ENST00000409157.1 |
KCTD18 |
potassium channel tetramerization domain containing 18 |
| chr6_-_26199471 | 0.08 |
ENST00000341023.1 |
HIST1H2AD |
histone cluster 1, H2ad |
| chr6_+_114178512 | 0.08 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr20_+_5986756 | 0.08 |
ENST00000452938.1 |
CRLS1 |
cardiolipin synthase 1 |
| chr19_+_58144529 | 0.08 |
ENST00000347302.3 ENST00000254182.7 ENST00000391703.3 ENST00000541801.1 ENST00000299871.5 ENST00000544273.1 |
ZNF211 |
zinc finger protein 211 |
| chr14_+_55595960 | 0.07 |
ENST00000554715.1 |
LGALS3 |
lectin, galactoside-binding, soluble, 3 |
| chr19_+_40877583 | 0.07 |
ENST00000596470.1 |
PLD3 |
phospholipase D family, member 3 |
| chr19_+_18668672 | 0.07 |
ENST00000601630.1 ENST00000599000.1 ENST00000595073.1 ENST00000596785.1 |
KXD1 |
KxDL motif containing 1 |
| chr10_-_22292675 | 0.07 |
ENST00000376946.1 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr20_+_56964169 | 0.07 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr21_+_31768348 | 0.07 |
ENST00000355459.2 |
KRTAP13-1 |
keratin associated protein 13-1 |
| chr19_+_18668572 | 0.07 |
ENST00000540691.1 ENST00000539106.1 ENST00000222307.4 |
KXD1 |
KxDL motif containing 1 |
| chr4_+_25915822 | 0.07 |
ENST00000506197.2 |
SMIM20 |
small integral membrane protein 20 |
| chr17_-_29641084 | 0.07 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr7_-_14880892 | 0.07 |
ENST00000406247.3 ENST00000399322.3 ENST00000258767.5 |
DGKB |
diacylglycerol kinase, beta 90kDa |
| chr4_+_25915896 | 0.06 |
ENST00000514384.1 |
SMIM20 |
small integral membrane protein 20 |
| chr5_-_43397184 | 0.06 |
ENST00000513525.1 |
CCL28 |
chemokine (C-C motif) ligand 28 |
| chr17_-_79105734 | 0.06 |
ENST00000417379.1 |
AATK |
apoptosis-associated tyrosine kinase |
| chr20_+_33104199 | 0.06 |
ENST00000357156.2 ENST00000417166.2 ENST00000300469.9 ENST00000374846.3 |
DYNLRB1 |
dynein, light chain, roadblock-type 1 |
| chr5_+_150404904 | 0.06 |
ENST00000521632.1 |
GPX3 |
glutathione peroxidase 3 (plasma) |
| chr15_+_67835517 | 0.06 |
ENST00000395476.2 |
MAP2K5 |
mitogen-activated protein kinase kinase 5 |
| chr19_-_51568324 | 0.06 |
ENST00000595547.1 ENST00000335422.3 ENST00000595793.1 ENST00000596955.1 |
KLK13 |
kallikrein-related peptidase 13 |
| chr2_-_111334678 | 0.06 |
ENST00000329516.3 ENST00000330331.5 ENST00000446930.1 |
RGPD6 |
RANBP2-like and GRIP domain containing 6 |
| chr14_+_65381079 | 0.06 |
ENST00000549115.1 ENST00000607599.1 ENST00000548752.2 ENST00000359118.2 ENST00000552002.2 ENST00000551947.1 ENST00000551093.1 ENST00000542227.1 ENST00000447296.2 ENST00000549987.1 |
CHURC1 FNTB CHURC1-FNTB |
churchill domain containing 1 farnesyltransferase, CAAX box, beta CHURC1-FNTB readthrough |
| chr21_+_44073860 | 0.06 |
ENST00000335512.4 ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A |
phosphodiesterase 9A |
| chr17_-_295730 | 0.06 |
ENST00000329099.4 |
FAM101B |
family with sequence similarity 101, member B |
| chr6_+_151561085 | 0.06 |
ENST00000402676.2 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr6_-_2751146 | 0.06 |
ENST00000268446.5 ENST00000274643.7 |
MYLK4 |
myosin light chain kinase family, member 4 |
| chr13_+_27998681 | 0.06 |
ENST00000381140.4 |
GTF3A |
general transcription factor IIIA |
| chr13_+_50570019 | 0.06 |
ENST00000442421.1 |
TRIM13 |
tripartite motif containing 13 |
| chrX_+_134478706 | 0.06 |
ENST00000370761.3 ENST00000339249.4 ENST00000370760.3 |
ZNF449 |
zinc finger protein 449 |
| chr1_-_151804314 | 0.06 |
ENST00000318247.6 |
RORC |
RAR-related orphan receptor C |
| chr21_+_44073916 | 0.06 |
ENST00000349112.3 ENST00000398224.3 |
PDE9A |
phosphodiesterase 9A |
| chr7_-_7680601 | 0.05 |
ENST00000396682.2 |
RPA3 |
replication protein A3, 14kDa |
| chr3_+_119013185 | 0.05 |
ENST00000264245.4 |
ARHGAP31 |
Rho GTPase activating protein 31 |
| chr3_+_16216137 | 0.05 |
ENST00000339732.5 |
GALNT15 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 15 |
| chr20_+_48429356 | 0.05 |
ENST00000361573.2 ENST00000541138.1 ENST00000539601.1 |
SLC9A8 |
solute carrier family 9, subfamily A (NHE8, cation proton antiporter 8), member 8 |
| chr17_+_67498538 | 0.05 |
ENST00000589647.1 |
MAP2K6 |
mitogen-activated protein kinase kinase 6 |
| chr19_+_9361606 | 0.05 |
ENST00000456448.1 |
OR7E24 |
olfactory receptor, family 7, subfamily E, member 24 |
| chr12_-_10151773 | 0.05 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr8_-_67974552 | 0.05 |
ENST00000357849.4 |
COPS5 |
COP9 signalosome subunit 5 |
| chr1_-_11918988 | 0.05 |
ENST00000376468.3 |
NPPB |
natriuretic peptide B |
| chr12_-_71182695 | 0.05 |
ENST00000342084.4 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr19_-_51340469 | 0.05 |
ENST00000326856.4 |
KLK15 |
kallikrein-related peptidase 15 |
| chr3_+_39424828 | 0.05 |
ENST00000273158.4 ENST00000431510.1 |
SLC25A38 |
solute carrier family 25, member 38 |
| chr7_-_32529973 | 0.04 |
ENST00000410044.1 ENST00000409987.1 ENST00000409782.1 ENST00000450169.2 |
LSM5 |
LSM5 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr1_+_24019099 | 0.04 |
ENST00000443624.1 ENST00000458455.1 |
RPL11 |
ribosomal protein L11 |
| chr1_+_109102652 | 0.04 |
ENST00000370035.3 ENST00000405454.1 |
FAM102B |
family with sequence similarity 102, member B |
| chr14_-_106494587 | 0.04 |
ENST00000390597.2 |
IGHV2-5 |
immunoglobulin heavy variable 2-5 |
| chr18_+_76829385 | 0.04 |
ENST00000426216.2 ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B |
ATPase, class II, type 9B |
| chr2_+_28618532 | 0.04 |
ENST00000545753.1 |
FOSL2 |
FOS-like antigen 2 |
| chr6_-_9977801 | 0.04 |
ENST00000316020.6 ENST00000491508.1 |
OFCC1 |
orofacial cleft 1 candidate 1 |
| chr5_+_3596168 | 0.04 |
ENST00000302006.3 |
IRX1 |
iroquois homeobox 1 |
| chr7_-_77325545 | 0.04 |
ENST00000447009.1 ENST00000416650.1 ENST00000440088.1 ENST00000430801.1 ENST00000398043.2 |
RSBN1L-AS1 |
RSBN1L antisense RNA 1 |
| chr11_-_26588634 | 0.04 |
ENST00000436318.2 ENST00000281268.8 |
MUC15 |
mucin 15, cell surface associated |
| chr10_-_12238071 | 0.04 |
ENST00000491614.1 ENST00000537776.1 |
NUDT5 |
nudix (nucleoside diphosphate linked moiety X)-type motif 5 |
| chr5_-_58295712 | 0.04 |
ENST00000317118.8 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr4_+_83956237 | 0.04 |
ENST00000264389.2 |
COPS4 |
COP9 signalosome subunit 4 |
| chr4_+_76649797 | 0.04 |
ENST00000538159.1 ENST00000514213.2 |
USO1 |
USO1 vesicle transport factor |
| chr5_-_142814241 | 0.04 |
ENST00000504572.1 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr14_-_101295407 | 0.04 |
ENST00000596284.1 |
AL117190.2 |
AL117190.2 |
| chrX_+_119737806 | 0.04 |
ENST00000371317.5 |
MCTS1 |
malignant T cell amplified sequence 1 |
| chr7_+_75024903 | 0.04 |
ENST00000323819.3 ENST00000430211.1 |
TRIM73 |
tripartite motif containing 73 |
| chr10_-_7513904 | 0.04 |
ENST00000420395.1 |
RP5-1031D4.2 |
RP5-1031D4.2 |
| chr14_-_20801427 | 0.04 |
ENST00000557665.1 ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1 |
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr18_+_45778672 | 0.04 |
ENST00000600091.1 |
AC091150.1 |
HCG1818186; Uncharacterized protein |
| chr2_+_45168875 | 0.03 |
ENST00000260653.3 |
SIX3 |
SIX homeobox 3 |
| chr5_+_167913450 | 0.03 |
ENST00000231572.3 ENST00000538719.1 |
RARS |
arginyl-tRNA synthetase |
| chr6_-_106773491 | 0.03 |
ENST00000360666.4 |
ATG5 |
autophagy related 5 |
| chr19_+_12175504 | 0.03 |
ENST00000439326.3 |
ZNF844 |
zinc finger protein 844 |
| chr17_-_72772462 | 0.03 |
ENST00000582870.1 ENST00000581136.1 ENST00000357814.3 ENST00000579218.1 ENST00000583476.1 ENST00000580301.1 ENST00000583757.1 ENST00000582524.1 |
NAT9 |
N-acetyltransferase 9 (GCN5-related, putative) |
| chrX_+_133594168 | 0.03 |
ENST00000298556.7 |
HPRT1 |
hypoxanthine phosphoribosyltransferase 1 |
| chr13_+_21714913 | 0.03 |
ENST00000450573.1 ENST00000467636.1 |
SAP18 |
Sin3A-associated protein, 18kDa |
| chr11_-_101000445 | 0.03 |
ENST00000534013.1 |
PGR |
progesterone receptor |
| chr2_-_99279928 | 0.03 |
ENST00000414521.2 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr21_+_17909594 | 0.03 |
ENST00000441820.1 ENST00000602280.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr19_+_52772821 | 0.03 |
ENST00000439461.1 |
ZNF766 |
zinc finger protein 766 |
| chr13_+_49551020 | 0.03 |
ENST00000541916.1 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr4_+_83956312 | 0.03 |
ENST00000509317.1 ENST00000503682.1 ENST00000511653.1 |
COPS4 |
COP9 signalosome subunit 4 |
| chr4_-_70518941 | 0.03 |
ENST00000286604.4 ENST00000505512.1 ENST00000514019.1 |
UGT2A1 UGT2A1 |
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr20_-_47804894 | 0.03 |
ENST00000371828.3 ENST00000371856.2 ENST00000360426.4 ENST00000347458.5 ENST00000340954.7 ENST00000371802.1 ENST00000371792.1 ENST00000437404.2 |
STAU1 |
staufen double-stranded RNA binding protein 1 |
| chr1_-_70820357 | 0.03 |
ENST00000370944.4 ENST00000262346.6 |
ANKRD13C |
ankyrin repeat domain 13C |
| chr3_+_75955817 | 0.03 |
ENST00000487694.3 ENST00000602589.1 |
ROBO2 |
roundabout, axon guidance receptor, homolog 2 (Drosophila) |
| chr12_+_25205446 | 0.03 |
ENST00000557489.1 ENST00000354454.3 ENST00000536173.1 |
LRMP |
lymphoid-restricted membrane protein |
| chrX_+_36254051 | 0.03 |
ENST00000378657.4 |
CXorf30 |
chromosome X open reading frame 30 |
| chr14_+_31091511 | 0.03 |
ENST00000544052.2 ENST00000421551.3 ENST00000541123.1 ENST00000557076.1 ENST00000553693.1 ENST00000396629.2 |
SCFD1 |
sec1 family domain containing 1 |
| chr15_+_54305101 | 0.02 |
ENST00000260323.11 ENST00000545554.1 ENST00000537900.1 |
UNC13C |
unc-13 homolog C (C. elegans) |
| chr17_+_45973516 | 0.02 |
ENST00000376741.4 |
SP2 |
Sp2 transcription factor |
| chr14_+_23564700 | 0.02 |
ENST00000554203.1 |
C14orf119 |
chromosome 14 open reading frame 119 |
| chr13_-_103426081 | 0.02 |
ENST00000376022.1 ENST00000376021.4 |
TEX30 |
testis expressed 30 |
| chr12_+_26164645 | 0.02 |
ENST00000542004.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr13_-_103426112 | 0.02 |
ENST00000376032.4 ENST00000376029.3 |
TEX30 |
testis expressed 30 |
| chr1_-_111174054 | 0.02 |
ENST00000369770.3 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr17_-_47755436 | 0.02 |
ENST00000505581.1 ENST00000514121.1 ENST00000393328.2 ENST00000509079.1 ENST00000393331.3 ENST00000347630.2 ENST00000504102.1 |
SPOP |
speckle-type POZ protein |
| chr17_+_8243154 | 0.02 |
ENST00000328248.2 ENST00000584943.1 |
ODF4 |
outer dense fiber of sperm tails 4 |
| chr18_+_5748793 | 0.02 |
ENST00000566533.1 ENST00000562452.2 |
RP11-945C19.1 |
RP11-945C19.1 |
| chr6_-_151773232 | 0.02 |
ENST00000444024.1 ENST00000367303.4 |
RMND1 |
required for meiotic nuclear division 1 homolog (S. cerevisiae) |
| chrX_+_36053908 | 0.02 |
ENST00000378660.2 |
CHDC2 |
calponin homology domain containing 2 |
| chr8_-_37797621 | 0.02 |
ENST00000524298.1 ENST00000307599.4 |
GOT1L1 |
glutamic-oxaloacetic transaminase 1-like 1 |
| chr9_-_116102530 | 0.02 |
ENST00000374195.3 ENST00000341761.4 |
WDR31 |
WD repeat domain 31 |
| chr1_+_64669294 | 0.02 |
ENST00000371077.5 |
UBE2U |
ubiquitin-conjugating enzyme E2U (putative) |
| chr20_+_42136308 | 0.02 |
ENST00000434666.1 ENST00000427442.2 ENST00000439769.1 ENST00000418998.1 |
L3MBTL1 |
l(3)mbt-like 1 (Drosophila) |
| chr10_+_99205959 | 0.02 |
ENST00000352634.4 ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16 |
zinc finger, DHHC-type containing 16 |
| chr14_+_73706308 | 0.02 |
ENST00000554301.1 ENST00000555445.1 |
PAPLN |
papilin, proteoglycan-like sulfated glycoprotein |
| chr9_-_98784042 | 0.02 |
ENST00000412122.2 |
LINC00092 |
long intergenic non-protein coding RNA 92 |
| chr9_+_130159409 | 0.02 |
ENST00000373371.3 |
SLC2A8 |
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr5_-_114961673 | 0.02 |
ENST00000333314.3 |
TMED7-TICAM2 |
TMED7-TICAM2 readthrough |
| chr2_+_166152283 | 0.02 |
ENST00000375427.2 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
| chr6_-_116833500 | 0.02 |
ENST00000356128.4 |
TRAPPC3L |
trafficking protein particle complex 3-like |
| chr12_+_128399965 | 0.02 |
ENST00000540882.1 ENST00000542089.1 |
LINC00507 |
long intergenic non-protein coding RNA 507 |
| chr17_+_1936687 | 0.01 |
ENST00000570477.1 |
DPH1 |
diphthamide biosynthesis 1 |
| chr10_+_99205894 | 0.01 |
ENST00000370854.3 ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16 |
zinc finger, DHHC-type containing 16 |
| chr3_-_187009798 | 0.01 |
ENST00000337774.5 |
MASP1 |
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr12_+_128399917 | 0.01 |
ENST00000544645.1 |
LINC00507 |
long intergenic non-protein coding RNA 507 |
| chr14_+_57857262 | 0.01 |
ENST00000555166.1 ENST00000556492.1 ENST00000554703.1 |
NAA30 |
N(alpha)-acetyltransferase 30, NatC catalytic subunit |
| chr10_-_99205607 | 0.01 |
ENST00000477692.2 ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1 |
exosome component 1 |
| chr1_-_213189168 | 0.01 |
ENST00000366962.3 ENST00000360506.2 |
ANGEL2 |
angel homolog 2 (Drosophila) |
| chr1_+_115397424 | 0.01 |
ENST00000369522.3 ENST00000455987.1 |
SYCP1 |
synaptonemal complex protein 1 |
| chr1_+_192544857 | 0.01 |
ENST00000367459.3 ENST00000469578.2 |
RGS1 |
regulator of G-protein signaling 1 |
| chr18_-_24765248 | 0.01 |
ENST00000580774.1 ENST00000284224.8 |
CHST9 |
carbohydrate (N-acetylgalactosamine 4-0) sulfotransferase 9 |
| chr19_-_52674696 | 0.01 |
ENST00000597252.1 |
ZNF836 |
zinc finger protein 836 |
| chr11_-_40315640 | 0.01 |
ENST00000278198.2 |
LRRC4C |
leucine rich repeat containing 4C |
| chr7_+_92158083 | 0.01 |
ENST00000265732.5 ENST00000481551.1 ENST00000496410.1 |
RBM48 |
RNA binding motif protein 48 |
| chr4_-_46126093 | 0.01 |
ENST00000295452.4 |
GABRG1 |
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr1_+_33546714 | 0.01 |
ENST00000294517.6 ENST00000358680.3 ENST00000373443.3 ENST00000398167.1 |
ADC |
arginine decarboxylase |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.1 | 0.2 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.4 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.3 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.0 | 0.5 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.2 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.3 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.1 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.0 | 0.1 | GO:2000342 | negative regulation of chemokine (C-X-C motif) ligand 2 production(GO:2000342) |
| 0.0 | 0.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) positive regulation of axon regeneration(GO:0048680) action potential propagation(GO:0098870) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.0 | GO:0002188 | translation reinitiation(GO:0002188) |
| 0.0 | 0.0 | GO:0009946 | proximal/distal axis specification(GO:0009946) neuroblast differentiation(GO:0014016) |
| 0.0 | 0.0 | GO:2000435 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
| 0.0 | 0.0 | GO:1901069 | guanosine-containing compound catabolic process(GO:1901069) |
| 0.0 | 0.1 | GO:0006982 | response to lipid hydroperoxide(GO:0006982) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.0 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.1 | 0.5 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.2 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0005219 | ryanodine-sensitive calcium-release channel activity(GO:0005219) |
| 0.0 | 0.2 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.2 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.0 | 0.4 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.1 | GO:0019784 | NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |