Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for HMX3
Z-value: 0.66
Transcription factors associated with HMX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HMX3
|
ENSG00000188620.9 | HMX3 |
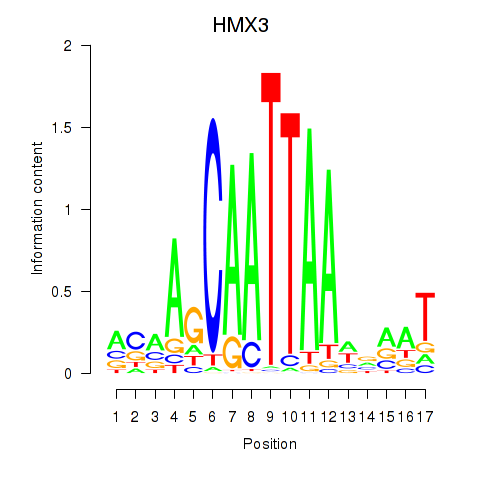
Activity profile of HMX3 motif
Sorted Z-values of HMX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HMX3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_13134045 | 0.47 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr6_+_73076432 | 0.46 |
ENST00000414192.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr11_+_5410607 | 0.42 |
ENST00000328611.3 |
OR51M1 |
olfactory receptor, family 51, subfamily M, member 1 |
| chr6_+_72922590 | 0.40 |
ENST00000523963.1 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr4_-_138453606 | 0.38 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr6_-_46293378 | 0.36 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr12_-_91576429 | 0.35 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr5_-_20575959 | 0.34 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr6_+_72922505 | 0.34 |
ENST00000401910.3 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr8_+_97506033 | 0.32 |
ENST00000518385.1 |
SDC2 |
syndecan 2 |
| chr1_+_87012753 | 0.31 |
ENST00000370563.3 |
CLCA4 |
chloride channel accessory 4 |
| chr4_+_70894130 | 0.30 |
ENST00000526767.1 ENST00000530128.1 ENST00000381057.3 |
HTN3 |
histatin 3 |
| chr7_+_29603394 | 0.30 |
ENST00000319694.2 |
PRR15 |
proline rich 15 |
| chr2_+_152214098 | 0.29 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr12_-_91573132 | 0.29 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr2_-_225811747 | 0.29 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr5_+_95066823 | 0.28 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr2_-_145277569 | 0.27 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr2_+_158114051 | 0.26 |
ENST00000259056.4 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr17_-_46507567 | 0.26 |
ENST00000584924.1 |
SKAP1 |
src kinase associated phosphoprotein 1 |
| chr1_-_86861660 | 0.26 |
ENST00000486215.1 |
ODF2L |
outer dense fiber of sperm tails 2-like |
| chr3_+_157154578 | 0.25 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr7_-_138363824 | 0.23 |
ENST00000419765.3 |
SVOPL |
SVOP-like |
| chr16_+_33204156 | 0.23 |
ENST00000398667.4 |
TP53TG3C |
TP53 target 3C |
| chr6_+_117198400 | 0.23 |
ENST00000332958.2 |
RFX6 |
regulatory factor X, 6 |
| chr17_-_2966901 | 0.22 |
ENST00000575751.1 |
OR1D5 |
olfactory receptor, family 1, subfamily D, member 5 |
| chr21_+_25801041 | 0.22 |
ENST00000453784.2 ENST00000423581.1 |
AP000476.1 |
AP000476.1 |
| chr8_+_134125727 | 0.22 |
ENST00000521107.1 |
TG |
thyroglobulin |
| chr2_+_108994466 | 0.22 |
ENST00000272452.2 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
| chr12_-_91546926 | 0.22 |
ENST00000550758.1 |
DCN |
decorin |
| chr6_+_72926145 | 0.21 |
ENST00000425662.2 ENST00000453976.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr2_+_108994633 | 0.21 |
ENST00000409309.3 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
| chr3_+_186560462 | 0.21 |
ENST00000412955.2 |
ADIPOQ |
adiponectin, C1Q and collagen domain containing |
| chr1_+_64239657 | 0.21 |
ENST00000371080.1 ENST00000371079.1 |
ROR1 |
receptor tyrosine kinase-like orphan receptor 1 |
| chr3_+_186560476 | 0.21 |
ENST00000320741.2 ENST00000444204.2 |
ADIPOQ |
adiponectin, C1Q and collagen domain containing |
| chr12_+_12202785 | 0.21 |
ENST00000586576.1 ENST00000464885.2 |
BCL2L14 |
BCL2-like 14 (apoptosis facilitator) |
| chr4_-_138453559 | 0.20 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr5_+_156696362 | 0.20 |
ENST00000377576.3 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr15_-_55700522 | 0.20 |
ENST00000564092.1 ENST00000310958.6 |
CCPG1 |
cell cycle progression 1 |
| chr16_-_90096309 | 0.19 |
ENST00000408886.2 |
C16orf3 |
chromosome 16 open reading frame 3 |
| chr9_-_130541017 | 0.19 |
ENST00000314830.8 |
SH2D3C |
SH2 domain containing 3C |
| chr2_-_166060552 | 0.19 |
ENST00000283254.7 ENST00000453007.1 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr7_+_80275953 | 0.19 |
ENST00000538969.1 ENST00000544133.1 ENST00000433696.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr15_-_78913628 | 0.19 |
ENST00000348639.3 |
CHRNA3 |
cholinergic receptor, nicotinic, alpha 3 (neuronal) |
| chr5_+_119867159 | 0.19 |
ENST00000505123.1 |
PRR16 |
proline rich 16 |
| chr18_+_76829385 | 0.19 |
ENST00000426216.2 ENST00000307671.7 ENST00000586672.1 ENST00000586722.1 |
ATP9B |
ATPase, class II, type 9B |
| chr1_-_72566613 | 0.19 |
ENST00000306821.3 |
NEGR1 |
neuronal growth regulator 1 |
| chr2_-_166060571 | 0.19 |
ENST00000360093.3 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr7_+_138943265 | 0.18 |
ENST00000483726.1 |
UBN2 |
ubinuclein 2 |
| chr6_-_132272504 | 0.17 |
ENST00000367976.3 |
CTGF |
connective tissue growth factor |
| chr19_-_44809121 | 0.17 |
ENST00000591609.1 ENST00000589799.1 ENST00000291182.4 ENST00000589248.1 |
ZNF235 |
zinc finger protein 235 |
| chr4_+_55095264 | 0.17 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr16_-_32688053 | 0.17 |
ENST00000398682.4 |
TP53TG3 |
TP53 target 3 |
| chr6_+_151561085 | 0.17 |
ENST00000402676.2 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr8_-_23712312 | 0.17 |
ENST00000290271.2 |
STC1 |
stanniocalcin 1 |
| chr16_+_14844670 | 0.16 |
ENST00000553201.1 |
NPIPA2 |
nuclear pore complex interacting protein family, member A2 |
| chr6_+_132891461 | 0.16 |
ENST00000275198.1 |
TAAR6 |
trace amine associated receptor 6 |
| chr11_-_124670550 | 0.16 |
ENST00000239614.4 |
MSANTD2 |
Myb/SANT-like DNA-binding domain containing 2 |
| chr12_+_51318513 | 0.16 |
ENST00000332160.4 |
METTL7A |
methyltransferase like 7A |
| chr15_-_50558223 | 0.16 |
ENST00000267845.3 |
HDC |
histidine decarboxylase |
| chr1_+_171060018 | 0.16 |
ENST00000367755.4 ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3 |
flavin containing monooxygenase 3 |
| chr17_-_29641104 | 0.15 |
ENST00000577894.1 ENST00000330927.4 |
EVI2B |
ecotropic viral integration site 2B |
| chr9_-_21482312 | 0.15 |
ENST00000448696.3 |
IFNE |
interferon, epsilon |
| chr7_+_91570165 | 0.15 |
ENST00000356239.3 ENST00000359028.2 ENST00000358100.2 |
AKAP9 |
A kinase (PRKA) anchor protein 9 |
| chr17_-_64225508 | 0.15 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr3_-_114343039 | 0.15 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr17_-_29641084 | 0.15 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr7_+_138915102 | 0.15 |
ENST00000486663.1 |
UBN2 |
ubinuclein 2 |
| chr9_-_104357277 | 0.15 |
ENST00000374806.1 |
PPP3R2 |
protein phosphatase 3, regulatory subunit B, beta |
| chr1_+_101361626 | 0.15 |
ENST00000370112.4 |
SLC30A7 |
solute carrier family 30 (zinc transporter), member 7 |
| chr13_+_50570019 | 0.15 |
ENST00000442421.1 |
TRIM13 |
tripartite motif containing 13 |
| chr12_-_11214893 | 0.15 |
ENST00000533467.1 |
TAS2R46 |
taste receptor, type 2, member 46 |
| chr12_-_10151773 | 0.15 |
ENST00000298527.6 ENST00000348658.4 |
CLEC1B |
C-type lectin domain family 1, member B |
| chr2_+_238395803 | 0.15 |
ENST00000264605.3 |
MLPH |
melanophilin |
| chr6_+_88757507 | 0.15 |
ENST00000237201.1 |
SPACA1 |
sperm acrosome associated 1 |
| chr16_+_32264040 | 0.14 |
ENST00000398664.3 |
TP53TG3D |
TP53 target 3D |
| chr1_+_144151520 | 0.14 |
ENST00000369372.4 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
| chr4_+_88754113 | 0.14 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr11_+_62104897 | 0.14 |
ENST00000415229.2 ENST00000535727.1 ENST00000301776.5 |
ASRGL1 |
asparaginase like 1 |
| chr19_+_58144529 | 0.14 |
ENST00000347302.3 ENST00000254182.7 ENST00000391703.3 ENST00000541801.1 ENST00000299871.5 ENST00000544273.1 |
ZNF211 |
zinc finger protein 211 |
| chr18_+_22040593 | 0.14 |
ENST00000256906.4 |
HRH4 |
histamine receptor H4 |
| chr8_+_38585704 | 0.14 |
ENST00000519416.1 ENST00000520615.1 |
TACC1 |
transforming, acidic coiled-coil containing protein 1 |
| chr3_+_141103634 | 0.14 |
ENST00000507722.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr18_+_22040620 | 0.14 |
ENST00000426880.2 |
HRH4 |
histamine receptor H4 |
| chr2_-_224467093 | 0.14 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chr12_-_122985067 | 0.14 |
ENST00000540586.1 ENST00000543897.1 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
| chr15_-_55700457 | 0.14 |
ENST00000442196.3 ENST00000563171.1 ENST00000425574.3 |
CCPG1 |
cell cycle progression 1 |
| chr8_-_93029865 | 0.14 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr3_+_133502877 | 0.13 |
ENST00000466490.2 |
SRPRB |
signal recognition particle receptor, B subunit |
| chrX_-_107682702 | 0.13 |
ENST00000372216.4 |
COL4A6 |
collagen, type IV, alpha 6 |
| chr11_+_94277017 | 0.13 |
ENST00000358752.2 |
FUT4 |
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr9_-_97090926 | 0.13 |
ENST00000335456.7 ENST00000253262.4 ENST00000341207.4 |
NUTM2F |
NUT family member 2F |
| chr6_+_116691001 | 0.13 |
ENST00000537543.1 |
DSE |
dermatan sulfate epimerase |
| chr19_-_52255107 | 0.13 |
ENST00000595042.1 ENST00000304748.4 |
FPR1 |
formyl peptide receptor 1 |
| chr4_-_159080806 | 0.13 |
ENST00000590648.1 |
FAM198B |
family with sequence similarity 198, member B |
| chr1_+_170501270 | 0.13 |
ENST00000367763.3 ENST00000367762.1 |
GORAB |
golgin, RAB6-interacting |
| chr7_+_75024903 | 0.13 |
ENST00000323819.3 ENST00000430211.1 |
TRIM73 |
tripartite motif containing 73 |
| chr2_+_223536428 | 0.13 |
ENST00000446656.3 |
MOGAT1 |
monoacylglycerol O-acyltransferase 1 |
| chr19_-_43690642 | 0.13 |
ENST00000407356.1 ENST00000407568.1 ENST00000404580.1 ENST00000599812.1 |
PSG5 |
pregnancy specific beta-1-glycoprotein 5 |
| chr5_-_94417339 | 0.13 |
ENST00000429576.2 ENST00000508509.1 ENST00000510732.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr3_-_105588231 | 0.13 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr7_+_107224364 | 0.12 |
ENST00000491150.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr17_-_42992856 | 0.12 |
ENST00000588316.1 ENST00000435360.2 ENST00000586793.1 ENST00000588735.1 ENST00000588037.1 ENST00000592320.1 ENST00000253408.5 |
GFAP |
glial fibrillary acidic protein |
| chr1_+_196946664 | 0.12 |
ENST00000367414.5 |
CFHR5 |
complement factor H-related 5 |
| chr10_-_22292613 | 0.12 |
ENST00000376980.3 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr19_+_58193337 | 0.12 |
ENST00000601064.1 ENST00000282296.5 ENST00000356715.4 |
ZNF551 |
zinc finger protein 551 |
| chr2_-_111334678 | 0.12 |
ENST00000329516.3 ENST00000330331.5 ENST00000446930.1 |
RGPD6 |
RANBP2-like and GRIP domain containing 6 |
| chr5_-_43397184 | 0.12 |
ENST00000513525.1 |
CCL28 |
chemokine (C-C motif) ligand 28 |
| chr12_+_51818749 | 0.12 |
ENST00000514353.3 |
SLC4A8 |
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr17_-_8702667 | 0.12 |
ENST00000329805.4 |
MFSD6L |
major facilitator superfamily domain containing 6-like |
| chr8_+_95565947 | 0.12 |
ENST00000523011.1 |
RP11-267M23.4 |
RP11-267M23.4 |
| chr21_+_44073860 | 0.12 |
ENST00000335512.4 ENST00000539837.1 ENST00000291539.6 ENST00000380328.2 ENST00000398232.3 ENST00000398234.3 ENST00000398236.3 ENST00000328862.6 ENST00000335440.6 ENST00000398225.3 ENST00000398229.3 ENST00000398227.3 |
PDE9A |
phosphodiesterase 9A |
| chr14_-_80697396 | 0.12 |
ENST00000557010.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr3_+_38537960 | 0.12 |
ENST00000453767.1 |
EXOG |
endo/exonuclease (5'-3'), endonuclease G-like |
| chr2_-_190044480 | 0.12 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr6_-_26108355 | 0.12 |
ENST00000338379.4 |
HIST1H1T |
histone cluster 1, H1t |
| chrX_+_55744166 | 0.12 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chr3_+_169629354 | 0.12 |
ENST00000428432.2 ENST00000335556.3 |
SAMD7 |
sterile alpha motif domain containing 7 |
| chr3_-_79816965 | 0.12 |
ENST00000464233.1 |
ROBO1 |
roundabout, axon guidance receptor, homolog 1 (Drosophila) |
| chr5_+_129083772 | 0.11 |
ENST00000564719.1 |
KIAA1024L |
KIAA1024-like |
| chr21_+_44073916 | 0.11 |
ENST00000349112.3 ENST00000398224.3 |
PDE9A |
phosphodiesterase 9A |
| chr12_-_89920030 | 0.11 |
ENST00000413530.1 ENST00000547474.1 |
GALNT4 POC1B-GALNT4 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr3_+_195413160 | 0.11 |
ENST00000599448.1 |
LINC00969 |
long intergenic non-protein coding RNA 969 |
| chr7_+_149571045 | 0.11 |
ENST00000479613.1 ENST00000606024.1 ENST00000464662.1 ENST00000425642.2 |
ATP6V0E2 |
ATPase, H+ transporting V0 subunit e2 |
| chr12_-_90024360 | 0.11 |
ENST00000393164.2 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr16_+_14802801 | 0.11 |
ENST00000526520.1 ENST00000531598.2 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr6_-_135271219 | 0.11 |
ENST00000367847.2 ENST00000367845.2 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
| chr5_+_42756903 | 0.11 |
ENST00000361970.5 ENST00000388827.4 |
CCDC152 |
coiled-coil domain containing 152 |
| chr1_+_196946680 | 0.11 |
ENST00000256785.4 |
CFHR5 |
complement factor H-related 5 |
| chr5_+_140227357 | 0.11 |
ENST00000378122.3 |
PCDHA9 |
protocadherin alpha 9 |
| chr6_-_29343068 | 0.11 |
ENST00000396806.3 |
OR12D3 |
olfactory receptor, family 12, subfamily D, member 3 |
| chrX_+_79675965 | 0.11 |
ENST00000308293.5 |
FAM46D |
family with sequence similarity 46, member D |
| chr16_-_50913188 | 0.11 |
ENST00000569986.1 |
CTD-2034I21.1 |
CTD-2034I21.1 |
| chr1_+_43291220 | 0.11 |
ENST00000372514.3 |
ERMAP |
erythroblast membrane-associated protein (Scianna blood group) |
| chr16_+_16429787 | 0.11 |
ENST00000331436.4 ENST00000541593.1 |
AC138969.4 |
Protein PKD1P1 |
| chr7_+_39773160 | 0.11 |
ENST00000340510.4 |
LINC00265 |
long intergenic non-protein coding RNA 265 |
| chr9_-_4666421 | 0.11 |
ENST00000381895.5 |
SPATA6L |
spermatogenesis associated 6-like |
| chr12_-_11150474 | 0.10 |
ENST00000538986.1 |
TAS2R20 |
taste receptor, type 2, member 20 |
| chr4_-_70518941 | 0.10 |
ENST00000286604.4 ENST00000505512.1 ENST00000514019.1 |
UGT2A1 UGT2A1 |
UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus UDP glucuronosyltransferase 2 family, polypeptide A1, complex locus |
| chr18_+_61144160 | 0.10 |
ENST00000489441.1 ENST00000424602.1 |
SERPINB5 |
serpin peptidase inhibitor, clade B (ovalbumin), member 5 |
| chr2_+_102953608 | 0.10 |
ENST00000311734.2 ENST00000409584.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chrX_+_55744228 | 0.10 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr6_+_29624758 | 0.10 |
ENST00000376917.3 ENST00000376902.3 ENST00000533330.2 ENST00000376888.2 |
MOG |
myelin oligodendrocyte glycoprotein |
| chr10_-_91011548 | 0.10 |
ENST00000456827.1 |
LIPA |
lipase A, lysosomal acid, cholesterol esterase |
| chr16_-_11922665 | 0.10 |
ENST00000573319.1 ENST00000577041.1 ENST00000574028.1 ENST00000571259.1 ENST00000573037.1 ENST00000571158.1 |
BCAR4 |
breast cancer anti-estrogen resistance 4 (non-protein coding) |
| chr16_+_14805546 | 0.10 |
ENST00000552140.1 |
NPIPA3 |
nuclear pore complex interacting protein family, member A3 |
| chr1_+_160709076 | 0.10 |
ENST00000359331.4 ENST00000495334.1 |
SLAMF7 |
SLAM family member 7 |
| chr5_+_15500280 | 0.10 |
ENST00000504595.1 |
FBXL7 |
F-box and leucine-rich repeat protein 7 |
| chr5_+_140868717 | 0.10 |
ENST00000252087.1 |
PCDHGC5 |
protocadherin gamma subfamily C, 5 |
| chr1_-_202858227 | 0.10 |
ENST00000367262.3 |
RABIF |
RAB interacting factor |
| chr14_-_105420241 | 0.10 |
ENST00000557457.1 |
AHNAK2 |
AHNAK nucleoprotein 2 |
| chr11_+_93517393 | 0.10 |
ENST00000251871.3 ENST00000530819.1 |
MED17 |
mediator complex subunit 17 |
| chr19_-_10341948 | 0.10 |
ENST00000590320.1 ENST00000592342.1 ENST00000588952.1 |
S1PR2 DNMT1 |
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr7_-_150020578 | 0.10 |
ENST00000478393.1 |
ACTR3C |
ARP3 actin-related protein 3 homolog C (yeast) |
| chr14_+_22615942 | 0.10 |
ENST00000390457.2 |
TRAV27 |
T cell receptor alpha variable 27 |
| chr6_+_12290586 | 0.10 |
ENST00000379375.5 |
EDN1 |
endothelin 1 |
| chr11_+_43380459 | 0.10 |
ENST00000299240.6 ENST00000039989.4 |
TTC17 |
tetratricopeptide repeat domain 17 |
| chrX_+_71996972 | 0.10 |
ENST00000334036.5 |
DMRTC1B |
DMRT-like family C1B |
| chr19_+_42212526 | 0.10 |
ENST00000598976.1 ENST00000435837.2 ENST00000221992.6 ENST00000405816.1 |
CEA CEACAM5 |
Uncharacterized protein carcinoembryonic antigen-related cell adhesion molecule 5 |
| chrX_+_71353499 | 0.10 |
ENST00000373677.1 |
NHSL2 |
NHS-like 2 |
| chr17_+_1665253 | 0.10 |
ENST00000254722.4 |
SERPINF1 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 1 |
| chr3_+_130569592 | 0.09 |
ENST00000533801.2 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr18_+_43304137 | 0.09 |
ENST00000502059.2 ENST00000586951.1 ENST00000589322.2 ENST00000415427.3 ENST00000535474.1 ENST00000402943.2 |
SLC14A1 |
solute carrier family 14 (urea transporter), member 1 (Kidd blood group) |
| chr4_+_86749045 | 0.09 |
ENST00000514229.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr16_-_15472151 | 0.09 |
ENST00000360151.4 ENST00000543801.1 |
NPIPA5 |
nuclear pore complex interacting protein family, member A5 |
| chr12_-_89919965 | 0.09 |
ENST00000548729.1 |
POC1B-GALNT4 |
POC1B-GALNT4 readthrough |
| chr19_+_10123925 | 0.09 |
ENST00000591589.1 ENST00000171214.1 |
RDH8 |
retinol dehydrogenase 8 (all-trans) |
| chr16_+_16472912 | 0.09 |
ENST00000530217.2 |
NPIPA7 |
nuclear pore complex interacting protein family, member A7 |
| chr6_+_28249299 | 0.09 |
ENST00000405948.2 |
PGBD1 |
piggyBac transposable element derived 1 |
| chr1_+_11333245 | 0.09 |
ENST00000376810.5 |
UBIAD1 |
UbiA prenyltransferase domain containing 1 |
| chr6_-_116575226 | 0.09 |
ENST00000420283.1 |
TSPYL4 |
TSPY-like 4 |
| chr9_+_21409146 | 0.09 |
ENST00000380205.1 |
IFNA8 |
interferon, alpha 8 |
| chr2_+_170683942 | 0.09 |
ENST00000272793.5 |
UBR3 |
ubiquitin protein ligase E3 component n-recognin 3 (putative) |
| chr5_+_175490540 | 0.09 |
ENST00000515817.1 |
FAM153B |
family with sequence similarity 153, member B |
| chr15_-_56757329 | 0.09 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chr2_+_90458201 | 0.09 |
ENST00000603238.1 |
CH17-132F21.1 |
Uncharacterized protein |
| chr10_-_22292675 | 0.09 |
ENST00000376946.1 |
DNAJC1 |
DnaJ (Hsp40) homolog, subfamily C, member 1 |
| chr8_+_7801144 | 0.09 |
ENST00000443676.1 |
ZNF705B |
zinc finger protein 705B |
| chr19_+_54466179 | 0.09 |
ENST00000270458.2 |
CACNG8 |
calcium channel, voltage-dependent, gamma subunit 8 |
| chr12_-_91505608 | 0.09 |
ENST00000266718.4 |
LUM |
lumican |
| chr6_+_36839616 | 0.09 |
ENST00000359359.2 ENST00000510325.2 |
C6orf89 |
chromosome 6 open reading frame 89 |
| chr8_+_39442097 | 0.09 |
ENST00000265707.5 ENST00000379866.1 ENST00000520772.1 ENST00000541111.1 |
ADAM18 |
ADAM metallopeptidase domain 18 |
| chr6_+_28249332 | 0.09 |
ENST00000259883.3 |
PGBD1 |
piggyBac transposable element derived 1 |
| chr19_+_45449301 | 0.09 |
ENST00000591597.1 |
APOC2 |
apolipoprotein C-II |
| chr16_+_22517166 | 0.09 |
ENST00000356156.3 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr3_-_194119995 | 0.09 |
ENST00000323007.3 |
GP5 |
glycoprotein V (platelet) |
| chr2_-_158345462 | 0.09 |
ENST00000439355.1 ENST00000540637.1 |
CYTIP |
cytohesin 1 interacting protein |
| chr1_-_145826450 | 0.09 |
ENST00000462900.2 |
GPR89A |
G protein-coupled receptor 89A |
| chr2_+_73612858 | 0.09 |
ENST00000409009.1 ENST00000264448.6 ENST00000377715.1 |
ALMS1 |
Alstrom syndrome 1 |
| chr6_+_158589374 | 0.09 |
ENST00000607778.1 |
GTF2H5 |
general transcription factor IIH, polypeptide 5 |
| chr19_+_52772832 | 0.09 |
ENST00000593703.1 ENST00000601711.1 ENST00000599581.1 |
ZNF766 |
zinc finger protein 766 |
| chr20_+_45338126 | 0.09 |
ENST00000359271.2 |
SLC2A10 |
solute carrier family 2 (facilitated glucose transporter), member 10 |
| chr10_+_89420706 | 0.09 |
ENST00000427144.2 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr2_-_70475701 | 0.09 |
ENST00000282574.4 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr14_+_21423611 | 0.09 |
ENST00000304625.2 |
RNASE2 |
ribonuclease, RNase A family, 2 (liver, eosinophil-derived neurotoxin) |
| chr10_+_43633914 | 0.09 |
ENST00000374466.3 ENST00000374464.1 |
CSGALNACT2 |
chondroitin sulfate N-acetylgalactosaminyltransferase 2 |
| chr16_+_15031300 | 0.09 |
ENST00000328085.6 |
NPIPA1 |
nuclear pore complex interacting protein family, member A1 |
| chrX_-_154493791 | 0.08 |
ENST00000369454.3 |
RAB39B |
RAB39B, member RAS oncogene family |
| chr8_-_13372253 | 0.08 |
ENST00000316609.5 |
DLC1 |
deleted in liver cancer 1 |
| chr6_-_135271260 | 0.08 |
ENST00000265605.2 |
ALDH8A1 |
aldehyde dehydrogenase 8 family, member A1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097018 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.2 | GO:0035790 | platelet-derived growth factor receptor-alpha signaling pathway(GO:0035790) |
| 0.1 | 1.4 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.3 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.1 | 0.2 | GO:0034059 | response to anoxia(GO:0034059) |
| 0.1 | 0.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.2 | GO:0060084 | synaptic transmission involved in micturition(GO:0060084) |
| 0.1 | 0.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.2 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.3 | GO:0070543 | response to linoleic acid(GO:0070543) |
| 0.0 | 0.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.3 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.2 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.1 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.0 | 0.1 | GO:0021827 | cerebral cortex tangential migration using cell-cell interactions(GO:0021823) postnatal olfactory bulb interneuron migration(GO:0021827) chemorepulsion involved in postnatal olfactory bulb interneuron migration(GO:0021836) |
| 0.0 | 0.0 | GO:0042149 | cellular response to glucose starvation(GO:0042149) |
| 0.0 | 0.6 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0060585 | regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) |
| 0.0 | 0.2 | GO:0015688 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:1904714 | chaperone-mediated autophagy(GO:0061684) regulation of chaperone-mediated autophagy(GO:1904714) |
| 0.0 | 0.2 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.0 | 0.2 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.0 | 0.3 | GO:0003310 | pancreatic A cell differentiation(GO:0003310) |
| 0.0 | 0.2 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:0000023 | maltose metabolic process(GO:0000023) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.0 | 0.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.1 | GO:0071918 | urea transmembrane transport(GO:0071918) |
| 0.0 | 0.1 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.0 | GO:0071480 | cellular response to gamma radiation(GO:0071480) |
| 0.0 | 0.1 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.2 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.3 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0071279 | cellular response to cobalt ion(GO:0071279) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.3 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.0 | 0.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.1 | GO:1903371 | establishment of protein localization to endoplasmic reticulum membrane(GO:0097051) regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.2 | GO:0042904 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.0 | 0.1 | GO:0071934 | thiamine transmembrane transport(GO:0071934) |
| 0.0 | 0.1 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.1 | GO:0006532 | aspartate biosynthetic process(GO:0006532) |
| 0.0 | 0.1 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.0 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.2 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.1 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.0 | GO:0050894 | determination of affect(GO:0050894) |
| 0.0 | 0.0 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.0 | 0.1 | GO:0051169 | nuclear export(GO:0051168) nuclear transport(GO:0051169) |
| 0.0 | 0.1 | GO:2001166 | regulation of histone H2B ubiquitination(GO:2001166) positive regulation of histone H2B ubiquitination(GO:2001168) |
| 0.0 | 0.0 | GO:1902623 | negative regulation of granulocyte chemotaxis(GO:0071623) negative regulation of neutrophil chemotaxis(GO:0090024) negative regulation of neutrophil migration(GO:1902623) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0044026 | DNA hypermethylation(GO:0044026) |
| 0.0 | 0.1 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.0 | GO:0060369 | positive regulation of Fc receptor mediated stimulatory signaling pathway(GO:0060369) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.0 | GO:0014063 | negative regulation of serotonin secretion(GO:0014063) |
| 0.0 | 0.0 | GO:0046373 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.1 | GO:0035672 | oligopeptide transmembrane transport(GO:0035672) |
| 0.0 | 0.1 | GO:1902109 | negative regulation of mitochondrial membrane permeability involved in apoptotic process(GO:1902109) |
| 0.0 | 0.1 | GO:0006420 | arginyl-tRNA aminoacylation(GO:0006420) |
| 0.0 | 0.1 | GO:0019317 | fucose catabolic process(GO:0019317) L-fucose metabolic process(GO:0042354) L-fucose catabolic process(GO:0042355) |
| 0.0 | 0.1 | GO:0002430 | complement receptor mediated signaling pathway(GO:0002430) |
| 0.0 | 0.0 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.1 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0070935 | 3'-UTR-mediated mRNA stabilization(GO:0070935) |
| 0.0 | 0.0 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.1 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.0 | 1.4 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.1 | GO:0048237 | rough endoplasmic reticulum lumen(GO:0048237) |
| 0.0 | 0.4 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0005587 | collagen type IV trimer(GO:0005587) |
| 0.0 | 0.1 | GO:0045293 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.0 | 0.0 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.0 | 0.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 0.1 | GO:0005785 | signal recognition particle receptor complex(GO:0005785) |
| 0.0 | 0.1 | GO:0031166 | integral component of vacuolar membrane(GO:0031166) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.1 | GO:0034363 | intermediate-density lipoprotein particle(GO:0034363) |
| 0.0 | 0.0 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.0 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.1 | 0.2 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.0 | 0.1 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.0 | 0.3 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.2 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.2 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.2 | GO:0047237 | glucuronylgalactosylproteoglycan 4-beta-N-acetylgalactosaminyltransferase activity(GO:0047237) |
| 0.0 | 0.1 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.2 | GO:1904929 | coreceptor activity involved in Wnt signaling pathway, planar cell polarity pathway(GO:1904929) |
| 0.0 | 0.4 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0004020 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 0.3 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.1 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.0 | 0.1 | GO:0015265 | urea channel activity(GO:0015265) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0004144 | diacylglycerol O-acyltransferase activity(GO:0004144) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.2 | GO:0001758 | retinal dehydrogenase activity(GO:0001758) |
| 0.0 | 0.1 | GO:0008046 | axon guidance receptor activity(GO:0008046) |
| 0.0 | 0.1 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) |
| 0.0 | 0.0 | GO:0033823 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0034235 | GPI anchor binding(GO:0034235) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.0 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.0 | 0.4 | GO:1905030 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.0 | GO:0031877 | somatostatin receptor binding(GO:0031877) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.0 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.0 | 0.0 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.0 | GO:0070336 | flap-structured DNA binding(GO:0070336) |
| 0.0 | 0.4 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
| 0.0 | 0.0 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.2 | GO:0033038 | bitter taste receptor activity(GO:0033038) |
| 0.0 | 0.1 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.0 | 0.0 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.8 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.1 | GO:0004814 | arginine-tRNA ligase activity(GO:0004814) |
| 0.0 | 0.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.0 | GO:0016433 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) rRNA (adenine) methyltransferase activity(GO:0016433) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0055056 | D-glucose transmembrane transporter activity(GO:0055056) |
| 0.0 | 0.3 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.0 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0045134 | uridine-diphosphatase activity(GO:0045134) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.0 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |
| 0.0 | 0.2 | REACTOME CHONDROITIN SULFATE BIOSYNTHESIS | Genes involved in Chondroitin sulfate biosynthesis |
| 0.0 | 0.1 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |