Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for HNF4A
Z-value: 0.33
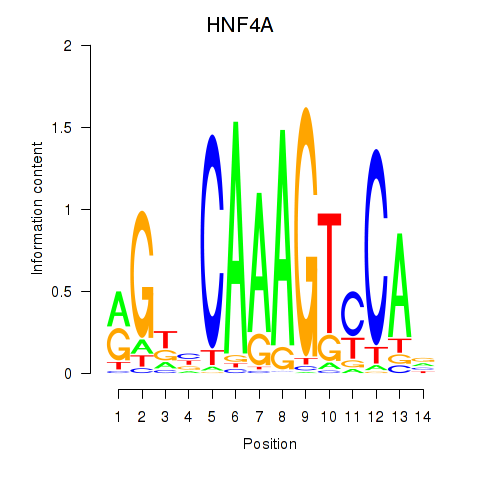
Transcription factors associated with HNF4A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF4A
|
ENSG00000101076.12 | HNF4A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HNF4A | hg19_v2_chr20_+_42984330_42984445 | -0.72 | 4.5e-02 | Click! |
Activity profile of HNF4A motif
Sorted Z-values of HNF4A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HNF4A
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_56075330 | 0.41 |
ENST00000394252.3 |
METTL7B |
methyltransferase like 7B |
| chr13_+_76334498 | 0.21 |
ENST00000534657.1 |
LMO7 |
LIM domain 7 |
| chr11_-_118550346 | 0.20 |
ENST00000530256.1 |
TREH |
trehalase (brush-border membrane glycoprotein) |
| chr16_-_3350614 | 0.20 |
ENST00000268674.2 |
TIGD7 |
tigger transposable element derived 7 |
| chr1_-_72566613 | 0.19 |
ENST00000306821.3 |
NEGR1 |
neuronal growth regulator 1 |
| chr8_+_70404996 | 0.16 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr1_+_145727681 | 0.16 |
ENST00000417171.1 ENST00000451928.2 |
PDZK1 |
PDZ domain containing 1 |
| chrX_+_66764375 | 0.15 |
ENST00000374690.3 |
AR |
androgen receptor |
| chr13_+_76334795 | 0.14 |
ENST00000526202.1 ENST00000465261.2 |
LMO7 |
LIM domain 7 |
| chr1_-_177939348 | 0.13 |
ENST00000464631.2 |
SEC16B |
SEC16 homolog B (S. cerevisiae) |
| chr3_+_37284824 | 0.13 |
ENST00000431105.1 |
GOLGA4 |
golgin A4 |
| chr12_+_56114189 | 0.12 |
ENST00000548082.1 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr19_+_35629702 | 0.12 |
ENST00000351325.4 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr2_-_28113217 | 0.12 |
ENST00000444339.2 |
RBKS |
ribokinase |
| chr12_+_56114151 | 0.12 |
ENST00000547072.1 ENST00000552930.1 ENST00000257895.5 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr19_+_35630022 | 0.12 |
ENST00000589209.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr13_+_76334567 | 0.12 |
ENST00000321797.8 |
LMO7 |
LIM domain 7 |
| chr17_-_7082668 | 0.11 |
ENST00000573083.1 ENST00000574388.1 |
ASGR1 |
asialoglycoprotein receptor 1 |
| chr6_-_29343068 | 0.11 |
ENST00000396806.3 |
OR12D3 |
olfactory receptor, family 12, subfamily D, member 3 |
| chr7_+_120628731 | 0.10 |
ENST00000310396.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr2_-_219433014 | 0.10 |
ENST00000418019.1 ENST00000454775.1 ENST00000338465.5 ENST00000415516.1 ENST00000258399.3 |
USP37 |
ubiquitin specific peptidase 37 |
| chr15_+_63340775 | 0.10 |
ENST00000559281.1 ENST00000317516.7 |
TPM1 |
tropomyosin 1 (alpha) |
| chr9_-_35685452 | 0.09 |
ENST00000607559.1 |
TPM2 |
tropomyosin 2 (beta) |
| chr1_-_167059830 | 0.09 |
ENST00000367868.3 |
GPA33 |
glycoprotein A33 (transmembrane) |
| chr14_-_39572279 | 0.09 |
ENST00000536508.1 |
SEC23A |
Sec23 homolog A (S. cerevisiae) |
| chr20_-_44485835 | 0.09 |
ENST00000457981.1 ENST00000426915.1 ENST00000217455.4 |
ACOT8 |
acyl-CoA thioesterase 8 |
| chr15_+_63340858 | 0.08 |
ENST00000560615.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr20_+_57875457 | 0.08 |
ENST00000337938.2 ENST00000311585.7 ENST00000371028.2 |
EDN3 |
endothelin 3 |
| chr15_+_63340734 | 0.08 |
ENST00000560959.1 |
TPM1 |
tropomyosin 1 (alpha) |
| chr11_+_118868830 | 0.08 |
ENST00000334418.1 |
CCDC84 |
coiled-coil domain containing 84 |
| chr3_-_49851313 | 0.08 |
ENST00000333486.3 |
UBA7 |
ubiquitin-like modifier activating enzyme 7 |
| chr1_+_23695680 | 0.07 |
ENST00000454117.1 ENST00000335648.3 ENST00000518821.1 ENST00000437367.2 |
C1orf213 |
chromosome 1 open reading frame 213 |
| chr17_-_28618867 | 0.07 |
ENST00000394819.3 ENST00000577623.1 |
BLMH |
bleomycin hydrolase |
| chr9_+_139839686 | 0.07 |
ENST00000371634.2 |
C8G |
complement component 8, gamma polypeptide |
| chr4_-_76928641 | 0.07 |
ENST00000264888.5 |
CXCL9 |
chemokine (C-X-C motif) ligand 9 |
| chr1_-_155271213 | 0.07 |
ENST00000342741.4 |
PKLR |
pyruvate kinase, liver and RBC |
| chr14_-_39572345 | 0.07 |
ENST00000548032.2 ENST00000556092.1 ENST00000557280.1 ENST00000545328.2 ENST00000553970.1 |
SEC23A |
Sec23 homolog A (S. cerevisiae) |
| chr12_-_6665200 | 0.07 |
ENST00000336604.4 ENST00000396840.2 ENST00000356896.4 |
IFFO1 |
intermediate filament family orphan 1 |
| chr1_+_78511586 | 0.07 |
ENST00000370759.3 |
GIPC2 |
GIPC PDZ domain containing family, member 2 |
| chr9_+_139839711 | 0.07 |
ENST00000224181.3 |
C8G |
complement component 8, gamma polypeptide |
| chr20_+_62694461 | 0.07 |
ENST00000343484.5 ENST00000395053.3 |
TCEA2 |
transcription elongation factor A (SII), 2 |
| chr16_+_8768422 | 0.07 |
ENST00000268251.8 ENST00000564714.1 |
ABAT |
4-aminobutyrate aminotransferase |
| chr15_-_63448973 | 0.06 |
ENST00000462430.1 |
RPS27L |
ribosomal protein S27-like |
| chr2_-_222436988 | 0.06 |
ENST00000409854.1 ENST00000281821.2 ENST00000392071.4 ENST00000443796.1 |
EPHA4 |
EPH receptor A4 |
| chr2_+_28113583 | 0.06 |
ENST00000344773.2 ENST00000379624.1 ENST00000342045.2 ENST00000379632.2 ENST00000361704.2 |
BRE |
brain and reproductive organ-expressed (TNFRSF1A modulator) |
| chrX_+_135388147 | 0.06 |
ENST00000394141.1 |
GPR112 |
G protein-coupled receptor 112 |
| chr3_+_191046810 | 0.06 |
ENST00000392455.3 ENST00000392456.3 |
CCDC50 |
coiled-coil domain containing 50 |
| chr10_-_17171817 | 0.06 |
ENST00000377833.4 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
| chr7_+_100547156 | 0.06 |
ENST00000379458.4 |
MUC3A |
Protein LOC100131514 |
| chrX_+_65382381 | 0.06 |
ENST00000519389.1 |
HEPH |
hephaestin |
| chr19_+_10197463 | 0.05 |
ENST00000590378.1 ENST00000397881.3 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr15_-_101142362 | 0.05 |
ENST00000559577.1 ENST00000561308.1 ENST00000560133.1 ENST00000560941.1 ENST00000559736.1 ENST00000560272.1 |
LINS |
lines homolog (Drosophila) |
| chr15_-_100273544 | 0.05 |
ENST00000409796.1 ENST00000545021.1 ENST00000344791.2 ENST00000332728.4 ENST00000450512.1 |
LYSMD4 |
LysM, putative peptidoglycan-binding, domain containing 4 |
| chr19_+_45449301 | 0.05 |
ENST00000591597.1 |
APOC2 |
apolipoprotein C-II |
| chr2_+_74648848 | 0.05 |
ENST00000409791.1 ENST00000426787.1 ENST00000348227.4 |
WDR54 |
WD repeat domain 54 |
| chr6_-_30080876 | 0.05 |
ENST00000376734.3 |
TRIM31 |
tripartite motif containing 31 |
| chr6_+_31982539 | 0.05 |
ENST00000435363.2 ENST00000425700.2 |
C4B |
complement component 4B (Chido blood group) |
| chr6_+_46761118 | 0.05 |
ENST00000230588.4 |
MEP1A |
meprin A, alpha (PABA peptide hydrolase) |
| chr7_+_100663353 | 0.05 |
ENST00000306151.4 |
MUC17 |
mucin 17, cell surface associated |
| chr10_+_70320413 | 0.05 |
ENST00000373644.4 |
TET1 |
tet methylcytosine dioxygenase 1 |
| chr8_+_96037205 | 0.05 |
ENST00000396124.4 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr22_-_31063782 | 0.05 |
ENST00000404885.1 ENST00000403268.1 ENST00000407308.1 ENST00000342474.4 ENST00000334679.3 |
DUSP18 |
dual specificity phosphatase 18 |
| chr4_-_120225600 | 0.05 |
ENST00000399075.4 |
C4orf3 |
chromosome 4 open reading frame 3 |
| chr22_+_19950060 | 0.04 |
ENST00000449653.1 |
COMT |
catechol-O-methyltransferase |
| chr2_+_10442993 | 0.04 |
ENST00000423674.1 ENST00000307845.3 |
HPCAL1 |
hippocalcin-like 1 |
| chr14_+_100485712 | 0.04 |
ENST00000544450.2 |
EVL |
Enah/Vasp-like |
| chr2_+_44502597 | 0.04 |
ENST00000260649.6 ENST00000409387.1 |
SLC3A1 |
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr16_-_87970122 | 0.04 |
ENST00000309893.2 |
CA5A |
carbonic anhydrase VA, mitochondrial |
| chr6_+_30131318 | 0.04 |
ENST00000376688.1 |
TRIM15 |
tripartite motif containing 15 |
| chr12_-_54121212 | 0.04 |
ENST00000548263.1 ENST00000430117.2 ENST00000550804.1 ENST00000549173.1 ENST00000551900.1 ENST00000546619.1 ENST00000548177.1 ENST00000549349.1 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
| chr12_-_54121261 | 0.04 |
ENST00000549784.1 ENST00000262059.4 |
CALCOCO1 |
calcium binding and coiled-coil domain 1 |
| chr6_+_31949801 | 0.04 |
ENST00000428956.2 ENST00000498271.1 |
C4A |
complement component 4A (Rodgers blood group) |
| chr12_-_53594227 | 0.04 |
ENST00000550743.2 |
ITGB7 |
integrin, beta 7 |
| chr20_+_36661910 | 0.04 |
ENST00000373433.4 |
RPRD1B |
regulation of nuclear pre-mRNA domain containing 1B |
| chr19_+_41594377 | 0.04 |
ENST00000330436.3 |
CYP2A13 |
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr14_+_38677123 | 0.04 |
ENST00000267377.2 |
SSTR1 |
somatostatin receptor 1 |
| chrX_-_77041685 | 0.04 |
ENST00000373344.5 ENST00000395603.3 |
ATRX |
alpha thalassemia/mental retardation syndrome X-linked |
| chr12_-_46384334 | 0.04 |
ENST00000369367.3 ENST00000266589.6 ENST00000395453.2 ENST00000395454.2 |
SCAF11 |
SR-related CTD-associated factor 11 |
| chr17_+_4853442 | 0.04 |
ENST00000522301.1 |
ENO3 |
enolase 3 (beta, muscle) |
| chr18_+_29769978 | 0.04 |
ENST00000269202.6 ENST00000581447.1 |
MEP1B |
meprin A, beta |
| chr6_-_30080863 | 0.04 |
ENST00000540829.1 |
TRIM31 |
tripartite motif containing 31 |
| chr5_+_133842243 | 0.04 |
ENST00000515627.2 |
AC005355.2 |
AC005355.2 |
| chr5_-_32444828 | 0.03 |
ENST00000265069.8 |
ZFR |
zinc finger RNA binding protein |
| chr16_+_24857552 | 0.03 |
ENST00000568579.1 ENST00000567758.1 ENST00000569071.1 ENST00000539472.1 |
SLC5A11 |
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr12_+_121416340 | 0.03 |
ENST00000257555.6 ENST00000400024.2 |
HNF1A |
HNF1 homeobox A |
| chrX_-_152939133 | 0.03 |
ENST00000370150.1 |
PNCK |
pregnancy up-regulated nonubiquitous CaM kinase |
| chr20_+_57875758 | 0.03 |
ENST00000395654.3 |
EDN3 |
endothelin 3 |
| chr6_+_43028182 | 0.03 |
ENST00000394058.1 |
KLC4 |
kinesin light chain 4 |
| chrX_+_118108601 | 0.03 |
ENST00000371628.3 |
LONRF3 |
LON peptidase N-terminal domain and ring finger 3 |
| chrX_-_152939252 | 0.03 |
ENST00000340888.3 |
PNCK |
pregnancy up-regulated nonubiquitous CaM kinase |
| chr19_-_15235906 | 0.03 |
ENST00000600984.1 |
ILVBL |
ilvB (bacterial acetolactate synthase)-like |
| chr20_-_36661826 | 0.03 |
ENST00000373448.2 ENST00000373447.3 |
TTI1 |
TELO2 interacting protein 1 |
| chr1_-_43855479 | 0.03 |
ENST00000290663.6 ENST00000372457.4 |
MED8 |
mediator complex subunit 8 |
| chr5_-_42812143 | 0.03 |
ENST00000514985.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr1_+_43855545 | 0.03 |
ENST00000372450.4 ENST00000310739.4 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr12_+_121416489 | 0.03 |
ENST00000541395.1 ENST00000544413.1 |
HNF1A |
HNF1 homeobox A |
| chr8_-_98290087 | 0.03 |
ENST00000322128.3 |
TSPYL5 |
TSPY-like 5 |
| chr10_+_18689637 | 0.03 |
ENST00000377315.4 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr4_+_109541772 | 0.03 |
ENST00000506397.1 ENST00000394668.2 |
RPL34 |
ribosomal protein L34 |
| chr11_-_63536113 | 0.03 |
ENST00000433688.1 ENST00000546282.2 |
C11orf95 RP11-466C23.4 |
chromosome 11 open reading frame 95 RP11-466C23.4 |
| chr4_+_159593271 | 0.03 |
ENST00000512251.1 ENST00000511912.1 |
ETFDH |
electron-transferring-flavoprotein dehydrogenase |
| chr3_+_63897605 | 0.03 |
ENST00000487717.1 |
ATXN7 |
ataxin 7 |
| chrX_+_118108571 | 0.03 |
ENST00000304778.7 |
LONRF3 |
LON peptidase N-terminal domain and ring finger 3 |
| chr1_+_43855560 | 0.02 |
ENST00000562955.1 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chr19_+_45449228 | 0.02 |
ENST00000252490.4 |
APOC2 |
apolipoprotein C-II |
| chr9_-_27005686 | 0.02 |
ENST00000380055.5 |
LRRC19 |
leucine rich repeat containing 19 |
| chr5_-_42811986 | 0.02 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr1_-_23694794 | 0.02 |
ENST00000374608.3 |
ZNF436 |
zinc finger protein 436 |
| chr7_-_75452673 | 0.02 |
ENST00000416943.1 |
CCL24 |
chemokine (C-C motif) ligand 24 |
| chr10_+_52152766 | 0.02 |
ENST00000596442.1 |
AC069547.2 |
Uncharacterized protein |
| chr20_+_44486246 | 0.02 |
ENST00000255152.2 ENST00000454862.2 |
ZSWIM3 |
zinc finger, SWIM-type containing 3 |
| chr6_-_33385854 | 0.02 |
ENST00000488478.1 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr8_+_96037255 | 0.02 |
ENST00000286687.4 |
NDUFAF6 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 6 |
| chr12_+_121416437 | 0.02 |
ENST00000402929.1 ENST00000535955.1 ENST00000538626.1 ENST00000543427.1 |
HNF1A |
HNF1 homeobox A |
| chr6_-_33385823 | 0.02 |
ENST00000494751.1 ENST00000374496.3 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr17_+_4675175 | 0.02 |
ENST00000270560.3 |
TM4SF5 |
transmembrane 4 L six family member 5 |
| chr4_+_109541740 | 0.02 |
ENST00000394665.1 |
RPL34 |
ribosomal protein L34 |
| chr1_+_199996702 | 0.02 |
ENST00000367362.3 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
| chr6_-_31926629 | 0.02 |
ENST00000375425.5 ENST00000426722.1 ENST00000441998.1 ENST00000444811.2 ENST00000375429.3 |
NELFE |
negative elongation factor complex member E |
| chr20_+_57875658 | 0.02 |
ENST00000371025.3 |
EDN3 |
endothelin 3 |
| chr10_-_96829246 | 0.02 |
ENST00000371270.3 ENST00000535898.1 ENST00000539050.1 |
CYP2C8 |
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chr22_+_30163340 | 0.02 |
ENST00000330029.6 ENST00000401406.3 |
UQCR10 |
ubiquinol-cytochrome c reductase, complex III subunit X |
| chr5_-_173043591 | 0.02 |
ENST00000285908.5 ENST00000480951.1 ENST00000311086.4 |
BOD1 |
biorientation of chromosomes in cell division 1 |
| chr9_-_15307200 | 0.02 |
ENST00000506891.1 ENST00000541445.1 ENST00000512701.2 ENST00000380850.4 ENST00000297615.5 ENST00000355694.2 |
TTC39B |
tetratricopeptide repeat domain 39B |
| chr16_+_24857309 | 0.02 |
ENST00000565769.1 ENST00000449109.2 ENST00000424767.2 ENST00000545376.1 ENST00000569520.1 |
SLC5A11 |
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr6_-_33385655 | 0.02 |
ENST00000440279.3 ENST00000607266.1 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr14_+_64680854 | 0.02 |
ENST00000458046.2 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
| chr7_+_192969 | 0.02 |
ENST00000313766.5 |
FAM20C |
family with sequence similarity 20, member C |
| chr13_+_52586517 | 0.02 |
ENST00000523764.1 ENST00000521508.1 |
ALG11 |
ALG11, alpha-1,2-mannosyltransferase |
| chr18_-_61034743 | 0.02 |
ENST00000406396.3 |
KDSR |
3-ketodihydrosphingosine reductase |
| chr11_-_10315741 | 0.02 |
ENST00000256190.8 |
SBF2 |
SET binding factor 2 |
| chr7_+_45928079 | 0.02 |
ENST00000468955.1 |
IGFBP1 |
insulin-like growth factor binding protein 1 |
| chr7_+_45927956 | 0.02 |
ENST00000275525.3 ENST00000457280.1 |
IGFBP1 |
insulin-like growth factor binding protein 1 |
| chr14_-_101295407 | 0.02 |
ENST00000596284.1 |
AL117190.2 |
AL117190.2 |
| chr19_-_7698599 | 0.02 |
ENST00000311069.5 |
PCP2 |
Purkinje cell protein 2 |
| chrX_+_65382433 | 0.02 |
ENST00000374727.3 |
HEPH |
hephaestin |
| chr6_+_30594619 | 0.02 |
ENST00000318999.7 ENST00000376485.4 ENST00000376478.2 ENST00000319027.5 ENST00000376483.4 ENST00000329992.8 ENST00000330083.5 |
ATAT1 |
alpha tubulin acetyltransferase 1 |
| chr3_-_141868357 | 0.02 |
ENST00000489671.1 ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr7_-_15601595 | 0.02 |
ENST00000342526.3 |
AGMO |
alkylglycerol monooxygenase |
| chr6_+_147527103 | 0.02 |
ENST00000179882.6 |
STXBP5 |
syntaxin binding protein 5 (tomosyn) |
| chr6_+_31691121 | 0.02 |
ENST00000480039.1 ENST00000375810.4 ENST00000375805.2 ENST00000375809.3 ENST00000375804.2 ENST00000375814.3 ENST00000375806.2 |
C6orf25 |
chromosome 6 open reading frame 25 |
| chr16_+_24857162 | 0.02 |
ENST00000347898.3 |
SLC5A11 |
solute carrier family 5 (sodium/inositol cotransporter), member 11 |
| chr3_-_125313934 | 0.02 |
ENST00000296220.5 |
OSBPL11 |
oxysterol binding protein-like 11 |
| chr1_-_43855444 | 0.02 |
ENST00000372455.4 |
MED8 |
mediator complex subunit 8 |
| chr19_+_4639514 | 0.02 |
ENST00000327473.4 |
TNFAIP8L1 |
tumor necrosis factor, alpha-induced protein 8-like 1 |
| chr2_+_241544834 | 0.02 |
ENST00000319838.5 ENST00000403859.1 ENST00000438013.2 |
GPR35 |
G protein-coupled receptor 35 |
| chr4_+_109541722 | 0.02 |
ENST00000394667.3 ENST00000502534.1 |
RPL34 |
ribosomal protein L34 |
| chr19_+_45449266 | 0.02 |
ENST00000592257.1 |
APOC2 |
apolipoprotein C-II |
| chr19_-_14201776 | 0.02 |
ENST00000269724.5 |
SAMD1 |
sterile alpha motif domain containing 1 |
| chr18_+_54318566 | 0.02 |
ENST00000589935.1 ENST00000357574.3 |
WDR7 |
WD repeat domain 7 |
| chr11_-_32456891 | 0.02 |
ENST00000452863.3 |
WT1 |
Wilms tumor 1 |
| chr12_-_109219937 | 0.02 |
ENST00000546697.1 |
SSH1 |
slingshot protein phosphatase 1 |
| chr6_+_116937636 | 0.01 |
ENST00000368581.4 ENST00000229554.5 ENST00000368580.4 |
RSPH4A |
radial spoke head 4 homolog A (Chlamydomonas) |
| chr15_+_50716576 | 0.01 |
ENST00000560297.1 ENST00000307179.4 ENST00000396444.3 ENST00000433963.1 ENST00000425032.3 |
USP8 |
ubiquitin specific peptidase 8 |
| chr7_+_33765593 | 0.01 |
ENST00000311067.3 |
RP11-89N17.1 |
HCG1643653; Uncharacterized protein |
| chr12_+_121163538 | 0.01 |
ENST00000242592.4 |
ACADS |
acyl-CoA dehydrogenase, C-2 to C-3 short chain |
| chr19_-_39881777 | 0.01 |
ENST00000595564.1 ENST00000221265.3 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr14_+_39735411 | 0.01 |
ENST00000603904.1 |
RP11-407N17.3 |
cTAGE family member 5 isoform 4 |
| chr12_-_21757774 | 0.01 |
ENST00000261195.2 |
GYS2 |
glycogen synthase 2 (liver) |
| chr3_+_63898275 | 0.01 |
ENST00000538065.1 |
ATXN7 |
ataxin 7 |
| chr3_-_158390282 | 0.01 |
ENST00000264265.3 |
LXN |
latexin |
| chr2_-_219134343 | 0.01 |
ENST00000447885.1 ENST00000420660.1 |
AAMP |
angio-associated, migratory cell protein |
| chr19_+_10196981 | 0.01 |
ENST00000591813.1 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr5_-_102455801 | 0.01 |
ENST00000508629.1 ENST00000399004.2 |
GIN1 |
gypsy retrotransposon integrase 1 |
| chr5_-_79950371 | 0.01 |
ENST00000511032.1 ENST00000504396.1 ENST00000505337.1 |
DHFR |
dihydrofolate reductase |
| chr19_+_46806856 | 0.01 |
ENST00000300862.3 |
HIF3A |
hypoxia inducible factor 3, alpha subunit |
| chr20_-_3644046 | 0.01 |
ENST00000290417.2 ENST00000319242.3 |
GFRA4 |
GDNF family receptor alpha 4 |
| chr1_-_155270770 | 0.01 |
ENST00000392414.3 |
PKLR |
pyruvate kinase, liver and RBC |
| chr5_+_1801503 | 0.01 |
ENST00000274137.5 ENST00000469176.1 |
NDUFS6 |
NADH dehydrogenase (ubiquinone) Fe-S protein 6, 13kDa (NADH-coenzyme Q reductase) |
| chr1_+_113217345 | 0.01 |
ENST00000357443.2 |
MOV10 |
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr17_+_4692230 | 0.01 |
ENST00000331264.7 |
GLTPD2 |
glycolipid transfer protein domain containing 2 |
| chr3_-_125775629 | 0.01 |
ENST00000383598.2 |
SLC41A3 |
solute carrier family 41, member 3 |
| chr1_-_161193349 | 0.01 |
ENST00000469730.2 ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2 |
apolipoprotein A-II |
| chr1_+_199996733 | 0.01 |
ENST00000236914.3 |
NR5A2 |
nuclear receptor subfamily 5, group A, member 2 |
| chr14_+_23352374 | 0.01 |
ENST00000267396.4 ENST00000536884.1 |
REM2 |
RAS (RAD and GEM)-like GTP binding 2 |
| chr3_+_46538981 | 0.01 |
ENST00000296142.3 |
RTP3 |
receptor (chemosensory) transporter protein 3 |
| chr19_+_39881951 | 0.01 |
ENST00000315588.5 ENST00000594368.1 ENST00000599213.2 ENST00000596297.1 |
MED29 |
mediator complex subunit 29 |
| chr3_-_187009798 | 0.01 |
ENST00000337774.5 |
MASP1 |
mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) |
| chr17_+_61554413 | 0.01 |
ENST00000538928.1 ENST00000290866.4 ENST00000428043.1 |
ACE |
angiotensin I converting enzyme |
| chr16_+_81272287 | 0.01 |
ENST00000425577.2 ENST00000564552.1 |
BCMO1 |
beta-carotene 15,15'-monooxygenase 1 |
| chr11_+_119056178 | 0.01 |
ENST00000525131.1 ENST00000531114.1 ENST00000355547.5 ENST00000322712.4 |
PDZD3 |
PDZ domain containing 3 |
| chr8_-_16859690 | 0.01 |
ENST00000180166.5 |
FGF20 |
fibroblast growth factor 20 |
| chr2_-_11810284 | 0.01 |
ENST00000306928.5 |
NTSR2 |
neurotensin receptor 2 |
| chr19_-_12886327 | 0.01 |
ENST00000397668.3 ENST00000587178.1 ENST00000264827.5 |
HOOK2 |
hook microtubule-tethering protein 2 |
| chr1_+_113217309 | 0.01 |
ENST00000544796.1 ENST00000369644.1 |
MOV10 |
Mov10, Moloney leukemia virus 10, homolog (mouse) |
| chr1_-_151138323 | 0.01 |
ENST00000368908.5 |
LYSMD1 |
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr14_-_76447336 | 0.01 |
ENST00000556285.1 |
TGFB3 |
transforming growth factor, beta 3 |
| chr15_-_41120896 | 0.01 |
ENST00000299174.5 ENST00000427255.2 |
PPP1R14D |
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr5_+_79950463 | 0.01 |
ENST00000265081.6 |
MSH3 |
mutS homolog 3 |
| chr1_+_151138500 | 0.01 |
ENST00000368905.4 |
SCNM1 |
sodium channel modifier 1 |
| chr3_+_46919235 | 0.01 |
ENST00000449590.1 |
PTH1R |
parathyroid hormone 1 receptor |
| chr22_+_25003606 | 0.01 |
ENST00000432867.1 |
GGT1 |
gamma-glutamyltransferase 1 |
| chr11_-_69590101 | 0.01 |
ENST00000168712.1 |
FGF4 |
fibroblast growth factor 4 |
| chr16_+_2255841 | 0.00 |
ENST00000301725.7 |
MLST8 |
MTOR associated protein, LST8 homolog (S. cerevisiae) |
| chr1_+_110163202 | 0.00 |
ENST00000531203.1 ENST00000256578.3 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr11_-_64660916 | 0.00 |
ENST00000413053.1 |
MIR194-2 |
microRNA 194-2 |
| chr14_-_76447494 | 0.00 |
ENST00000238682.3 |
TGFB3 |
transforming growth factor, beta 3 |
| chr4_-_109541539 | 0.00 |
ENST00000509984.1 ENST00000507248.1 ENST00000506795.1 |
RPL34-AS1 |
RPL34 antisense RNA 1 (head to head) |
| chr11_-_45939565 | 0.00 |
ENST00000525192.1 ENST00000378750.5 |
PEX16 |
peroxisomal biogenesis factor 16 |
| chr1_+_94884023 | 0.00 |
ENST00000315713.5 |
ABCD3 |
ATP-binding cassette, sub-family D (ALD), member 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.0 | 0.2 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.2 | GO:0010731 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.0 | 0.3 | GO:0003065 | positive regulation of heart rate by epinephrine(GO:0003065) |
| 0.0 | 0.1 | GO:0035623 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.0 | 0.1 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.1 | GO:0060697 | positive regulation of phospholipid catabolic process(GO:0060697) |
| 0.0 | 0.1 | GO:0014826 | cellular magnesium ion homeostasis(GO:0010961) vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.1 | GO:1904448 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.0 | GO:0090310 | negative regulation of methylation-dependent chromatin silencing(GO:0090310) |
| 0.0 | 0.2 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.1 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.0 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.0 | 0.2 | GO:0016559 | peroxisome fission(GO:0016559) |
| 0.0 | 0.1 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0005862 | muscle thin filament tropomyosin(GO:0005862) |
| 0.0 | 0.2 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.0 | 0.0 | GO:0031933 | telomeric heterochromatin(GO:0031933) |
| 0.0 | 0.0 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.0 | 0.2 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0005124 | scavenger receptor binding(GO:0005124) |
| 0.0 | 0.2 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.1 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.0 | 0.2 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.0 | GO:0070579 | methylcytosine dioxygenase activity(GO:0070579) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.1 | GO:0060230 | lipoprotein lipase activator activity(GO:0060230) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |