Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
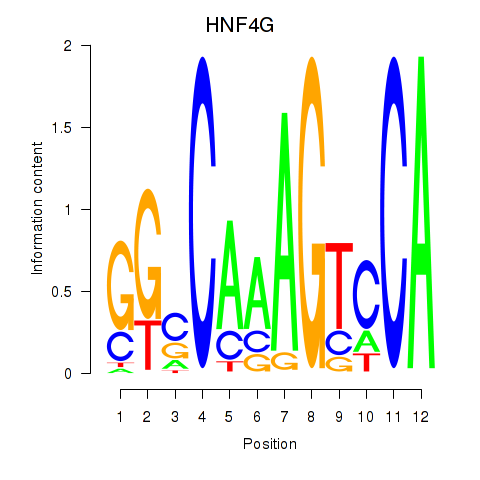
Results for HNF4G
Z-value: 0.75
Transcription factors associated with HNF4G
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HNF4G
|
ENSG00000164749.7 | HNF4G |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HNF4G | hg19_v2_chr8_+_76452097_76452126 | 0.67 | 6.7e-02 | Click! |
Activity profile of HNF4G motif
Sorted Z-values of HNF4G motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HNF4G
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr17_-_39677971 | 1.25 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr19_-_6720686 | 0.89 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chr12_-_52845910 | 0.76 |
ENST00000252252.3 |
KRT6B |
keratin 6B |
| chr12_-_52887034 | 0.71 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr10_+_118187379 | 0.67 |
ENST00000369230.3 |
PNLIPRP3 |
pancreatic lipase-related protein 3 |
| chr1_-_201368707 | 0.64 |
ENST00000391967.2 |
LAD1 |
ladinin 1 |
| chr20_+_58251716 | 0.63 |
ENST00000355648.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr18_+_34124507 | 0.62 |
ENST00000591635.1 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr11_-_118134997 | 0.62 |
ENST00000278937.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr11_+_1860200 | 0.57 |
ENST00000381911.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr8_-_57232656 | 0.56 |
ENST00000396721.2 |
SDR16C5 |
short chain dehydrogenase/reductase family 16C, member 5 |
| chr11_-_118135160 | 0.56 |
ENST00000438295.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr8_-_57233103 | 0.56 |
ENST00000303749.3 ENST00000522671.1 |
SDR16C5 |
short chain dehydrogenase/reductase family 16C, member 5 |
| chr1_-_201368653 | 0.54 |
ENST00000367313.3 |
LAD1 |
ladinin 1 |
| chr2_-_238499303 | 0.51 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr8_-_145642267 | 0.48 |
ENST00000301305.3 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
| chr14_-_94854926 | 0.46 |
ENST00000402629.1 ENST00000556091.1 ENST00000554720.1 |
SERPINA1 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 1 |
| chr1_+_152956549 | 0.44 |
ENST00000307122.2 |
SPRR1A |
small proline-rich protein 1A |
| chr20_-_43883197 | 0.44 |
ENST00000338380.2 |
SLPI |
secretory leukocyte peptidase inhibitor |
| chr1_+_35246775 | 0.43 |
ENST00000373366.2 |
GJB3 |
gap junction protein, beta 3, 31kDa |
| chr8_-_145641864 | 0.42 |
ENST00000276833.5 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
| chr1_+_155099927 | 0.42 |
ENST00000368407.3 |
EFNA1 |
ephrin-A1 |
| chr16_+_3115378 | 0.41 |
ENST00000529550.1 ENST00000551122.1 ENST00000525643.2 ENST00000548807.1 ENST00000528163.2 |
IL32 |
interleukin 32 |
| chr11_-_119999611 | 0.41 |
ENST00000529044.1 |
TRIM29 |
tripartite motif containing 29 |
| chr17_-_76123101 | 0.39 |
ENST00000392467.3 |
TMC6 |
transmembrane channel-like 6 |
| chr8_-_144952631 | 0.38 |
ENST00000525985.1 |
EPPK1 |
epiplakin 1 |
| chr11_-_119999539 | 0.38 |
ENST00000541857.1 |
TRIM29 |
tripartite motif containing 29 |
| chr1_+_160370344 | 0.38 |
ENST00000368061.2 |
VANGL2 |
VANGL planar cell polarity protein 2 |
| chr2_-_238499725 | 0.37 |
ENST00000264601.3 |
RAB17 |
RAB17, member RAS oncogene family |
| chr1_+_35247859 | 0.37 |
ENST00000373362.3 |
GJB3 |
gap junction protein, beta 3, 31kDa |
| chr17_+_54671047 | 0.37 |
ENST00000332822.4 |
NOG |
noggin |
| chr2_-_238499131 | 0.36 |
ENST00000538644.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr11_+_1856034 | 0.36 |
ENST00000341958.3 |
SYT8 |
synaptotagmin VIII |
| chr7_-_143105941 | 0.35 |
ENST00000275815.3 |
EPHA1 |
EPH receptor A1 |
| chr12_-_56882136 | 0.33 |
ENST00000311966.4 |
GLS2 |
glutaminase 2 (liver, mitochondrial) |
| chr2_-_238499337 | 0.33 |
ENST00000411462.1 ENST00000409822.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr12_-_95009837 | 0.33 |
ENST00000551457.1 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
| chr7_+_145813453 | 0.31 |
ENST00000361727.3 |
CNTNAP2 |
contactin associated protein-like 2 |
| chr16_+_3115611 | 0.31 |
ENST00000530890.1 ENST00000444393.3 ENST00000533097.2 ENST00000008180.9 ENST00000396890.2 ENST00000525228.1 ENST00000548652.1 ENST00000525377.2 ENST00000530538.2 ENST00000549213.1 ENST00000552936.1 ENST00000548476.1 ENST00000552664.1 ENST00000552356.1 ENST00000551513.1 ENST00000382213.3 ENST00000548246.1 |
IL32 |
interleukin 32 |
| chr1_+_156252708 | 0.31 |
ENST00000295694.5 ENST00000357501.2 |
TMEM79 |
transmembrane protein 79 |
| chr11_+_67071050 | 0.31 |
ENST00000376757.5 |
SSH3 |
slingshot protein phosphatase 3 |
| chr1_+_27668505 | 0.31 |
ENST00000318074.5 |
SYTL1 |
synaptotagmin-like 1 |
| chr11_+_1860682 | 0.30 |
ENST00000381906.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr2_-_235405168 | 0.30 |
ENST00000339728.3 |
ARL4C |
ADP-ribosylation factor-like 4C |
| chr14_-_92333873 | 0.29 |
ENST00000435962.2 |
TC2N |
tandem C2 domains, nuclear |
| chr11_+_1860832 | 0.29 |
ENST00000252898.7 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr16_+_3115298 | 0.29 |
ENST00000325568.5 ENST00000534507.1 |
IL32 |
interleukin 32 |
| chr12_+_6949964 | 0.29 |
ENST00000541978.1 ENST00000435982.2 |
GNB3 |
guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| chr4_+_100737954 | 0.29 |
ENST00000296414.7 ENST00000512369.1 |
DAPP1 |
dual adaptor of phosphotyrosine and 3-phosphoinositides |
| chrX_+_138612889 | 0.28 |
ENST00000218099.2 ENST00000394090.2 |
F9 |
coagulation factor IX |
| chr11_-_118023490 | 0.28 |
ENST00000324727.4 |
SCN4B |
sodium channel, voltage-gated, type IV, beta subunit |
| chr11_+_65779283 | 0.28 |
ENST00000312134.2 |
CST6 |
cystatin E/M |
| chr11_+_67070919 | 0.27 |
ENST00000308127.4 ENST00000308298.7 |
SSH3 |
slingshot protein phosphatase 3 |
| chr2_+_233925064 | 0.27 |
ENST00000359570.5 ENST00000538935.1 |
INPP5D |
inositol polyphosphate-5-phosphatase, 145kDa |
| chr4_-_84256024 | 0.27 |
ENST00000311412.5 |
HPSE |
heparanase |
| chr11_-_66496430 | 0.27 |
ENST00000533211.1 |
SPTBN2 |
spectrin, beta, non-erythrocytic 2 |
| chr22_+_45072925 | 0.26 |
ENST00000006251.7 |
PRR5 |
proline rich 5 (renal) |
| chr22_+_45072958 | 0.26 |
ENST00000403581.1 |
PRR5 |
proline rich 5 (renal) |
| chr2_-_31637560 | 0.25 |
ENST00000379416.3 |
XDH |
xanthine dehydrogenase |
| chr8_+_27348649 | 0.25 |
ENST00000521780.1 ENST00000380476.3 ENST00000518379.1 ENST00000521684.1 |
EPHX2 |
epoxide hydrolase 2, cytoplasmic |
| chr12_+_10366016 | 0.24 |
ENST00000546017.1 ENST00000535576.1 ENST00000539170.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr8_+_27348626 | 0.24 |
ENST00000517536.1 |
EPHX2 |
epoxide hydrolase 2, cytoplasmic |
| chr9_-_117568365 | 0.23 |
ENST00000374045.4 |
TNFSF15 |
tumor necrosis factor (ligand) superfamily, member 15 |
| chr1_-_209824643 | 0.23 |
ENST00000391911.1 ENST00000415782.1 |
LAMB3 |
laminin, beta 3 |
| chr16_+_3115323 | 0.23 |
ENST00000531965.1 ENST00000396887.3 ENST00000529699.1 ENST00000526464.2 ENST00000440815.3 |
IL32 |
interleukin 32 |
| chr11_+_1855645 | 0.23 |
ENST00000381968.3 ENST00000381978.3 |
SYT8 |
synaptotagmin VIII |
| chr1_-_41131326 | 0.22 |
ENST00000372684.3 |
RIMS3 |
regulating synaptic membrane exocytosis 3 |
| chr4_+_3465027 | 0.22 |
ENST00000389653.2 ENST00000507039.1 ENST00000340083.5 |
DOK7 |
docking protein 7 |
| chr18_+_19749386 | 0.22 |
ENST00000269216.3 |
GATA6 |
GATA binding protein 6 |
| chr3_-_116163830 | 0.22 |
ENST00000333617.4 |
LSAMP |
limbic system-associated membrane protein |
| chr16_+_57406368 | 0.22 |
ENST00000006053.6 ENST00000563383.1 |
CX3CL1 |
chemokine (C-X3-C motif) ligand 1 |
| chr14_+_22966642 | 0.22 |
ENST00000390496.1 |
TRAJ41 |
T cell receptor alpha joining 41 |
| chr18_-_53253323 | 0.21 |
ENST00000540999.1 ENST00000563888.2 |
TCF4 |
transcription factor 4 |
| chrX_-_48827976 | 0.21 |
ENST00000218176.3 |
KCND1 |
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr2_-_85108164 | 0.21 |
ENST00000409520.2 |
TRABD2A |
TraB domain containing 2A |
| chr5_-_179636153 | 0.20 |
ENST00000361132.4 |
RASGEF1C |
RasGEF domain family, member 1C |
| chr18_-_53253112 | 0.20 |
ENST00000568673.1 ENST00000562847.1 ENST00000568147.1 |
TCF4 |
transcription factor 4 |
| chr16_+_4838412 | 0.20 |
ENST00000589327.1 |
SMIM22 |
small integral membrane protein 22 |
| chr12_+_113354341 | 0.20 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_+_26869597 | 0.19 |
ENST00000530003.1 |
RPS6KA1 |
ribosomal protein S6 kinase, 90kDa, polypeptide 1 |
| chr14_-_23623577 | 0.19 |
ENST00000422941.2 ENST00000453702.1 |
SLC7A8 |
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr2_-_237416071 | 0.19 |
ENST00000309507.5 ENST00000431676.2 |
IQCA1 |
IQ motif containing with AAA domain 1 |
| chr17_+_7608511 | 0.19 |
ENST00000226091.2 |
EFNB3 |
ephrin-B3 |
| chr16_+_3014217 | 0.19 |
ENST00000572045.1 |
KREMEN2 |
kringle containing transmembrane protein 2 |
| chr16_+_4838393 | 0.19 |
ENST00000589721.1 |
SMIM22 |
small integral membrane protein 22 |
| chr7_+_143701022 | 0.19 |
ENST00000408922.2 |
OR6B1 |
olfactory receptor, family 6, subfamily B, member 1 |
| chr7_-_71868354 | 0.19 |
ENST00000412588.1 |
CALN1 |
calneuron 1 |
| chr16_+_2867164 | 0.19 |
ENST00000455114.1 ENST00000450020.3 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr16_+_2867228 | 0.19 |
ENST00000005995.3 ENST00000574813.1 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr8_+_22436635 | 0.18 |
ENST00000452226.1 ENST00000397760.4 ENST00000339162.7 ENST00000397761.2 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr1_+_167599330 | 0.18 |
ENST00000367854.3 ENST00000361496.3 |
RCSD1 |
RCSD domain containing 1 |
| chr12_-_48963829 | 0.18 |
ENST00000301046.2 ENST00000549817.1 |
LALBA |
lactalbumin, alpha- |
| chr1_+_24120143 | 0.18 |
ENST00000374501.1 |
LYPLA2 |
lysophospholipase II |
| chr14_-_81893734 | 0.17 |
ENST00000555447.1 |
STON2 |
stonin 2 |
| chr11_-_1780261 | 0.17 |
ENST00000427721.1 |
RP11-295K3.1 |
RP11-295K3.1 |
| chr17_-_17740325 | 0.17 |
ENST00000338854.5 |
SREBF1 |
sterol regulatory element binding transcription factor 1 |
| chr18_+_29598335 | 0.17 |
ENST00000217740.3 |
RNF125 |
ring finger protein 125, E3 ubiquitin protein ligase |
| chr1_-_85462623 | 0.17 |
ENST00000370608.3 |
MCOLN2 |
mucolipin 2 |
| chr8_+_32405728 | 0.17 |
ENST00000523079.1 ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1 |
neuregulin 1 |
| chr8_+_32405785 | 0.16 |
ENST00000287842.3 |
NRG1 |
neuregulin 1 |
| chr6_+_155585147 | 0.16 |
ENST00000367165.3 |
CLDN20 |
claudin 20 |
| chr3_+_122785895 | 0.16 |
ENST00000316218.7 |
PDIA5 |
protein disulfide isomerase family A, member 5 |
| chr19_+_6464243 | 0.16 |
ENST00000600229.1 ENST00000356762.3 |
CRB3 |
crumbs homolog 3 (Drosophila) |
| chr19_+_7660716 | 0.16 |
ENST00000160298.4 ENST00000446248.2 |
CAMSAP3 |
calmodulin regulated spectrin-associated protein family, member 3 |
| chr12_-_58220078 | 0.16 |
ENST00000549039.1 |
CTDSP2 |
CTD (carboxy-terminal domain, RNA polymerase II, polypeptide A) small phosphatase 2 |
| chr11_-_111781554 | 0.16 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chr9_-_117853297 | 0.15 |
ENST00000542877.1 ENST00000537320.1 ENST00000341037.4 |
TNC |
tenascin C |
| chr11_+_63137251 | 0.15 |
ENST00000310969.4 ENST00000279178.3 |
SLC22A9 |
solute carrier family 22 (organic anion transporter), member 9 |
| chr20_-_44144249 | 0.15 |
ENST00000217428.6 |
SPINT3 |
serine peptidase inhibitor, Kunitz type, 3 |
| chr19_+_6464502 | 0.15 |
ENST00000308243.7 |
CRB3 |
crumbs homolog 3 (Drosophila) |
| chr19_+_18496957 | 0.15 |
ENST00000252809.3 |
GDF15 |
growth differentiation factor 15 |
| chr1_-_43751230 | 0.15 |
ENST00000523677.1 |
C1orf210 |
chromosome 1 open reading frame 210 |
| chr3_-_52488048 | 0.15 |
ENST00000232975.3 |
TNNC1 |
troponin C type 1 (slow) |
| chr2_+_196521458 | 0.15 |
ENST00000409086.3 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
| chr3_-_48471454 | 0.14 |
ENST00000296440.6 ENST00000448774.2 |
PLXNB1 |
plexin B1 |
| chr8_-_145159083 | 0.14 |
ENST00000398712.2 |
SHARPIN |
SHANK-associated RH domain interactor |
| chr17_+_7531281 | 0.14 |
ENST00000575729.1 ENST00000340624.5 |
SHBG |
sex hormone-binding globulin |
| chr4_-_40517984 | 0.14 |
ENST00000381795.6 |
RBM47 |
RNA binding motif protein 47 |
| chr8_+_21911054 | 0.13 |
ENST00000519850.1 ENST00000381470.3 |
DMTN |
dematin actin binding protein |
| chr2_+_196521845 | 0.13 |
ENST00000359634.5 ENST00000412905.1 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
| chr2_+_85811525 | 0.13 |
ENST00000306384.4 |
VAMP5 |
vesicle-associated membrane protein 5 |
| chr12_+_109273806 | 0.13 |
ENST00000228476.3 ENST00000547768.1 |
DAO |
D-amino-acid oxidase |
| chr11_-_111781610 | 0.13 |
ENST00000525823.1 |
CRYAB |
crystallin, alpha B |
| chr20_-_50419055 | 0.13 |
ENST00000217086.4 |
SALL4 |
spalt-like transcription factor 4 |
| chr1_-_1356628 | 0.13 |
ENST00000442470.1 ENST00000537107.1 |
ANKRD65 |
ankyrin repeat domain 65 |
| chr17_-_17494972 | 0.12 |
ENST00000435340.2 ENST00000255389.5 ENST00000395781.2 |
PEMT |
phosphatidylethanolamine N-methyltransferase |
| chr11_-_66725837 | 0.12 |
ENST00000393958.2 ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC |
pyruvate carboxylase |
| chr12_+_119772502 | 0.12 |
ENST00000536742.1 ENST00000327554.2 |
CCDC60 |
coiled-coil domain containing 60 |
| chr14_-_105531759 | 0.12 |
ENST00000329797.3 ENST00000539291.2 ENST00000392585.2 |
GPR132 |
G protein-coupled receptor 132 |
| chr1_-_44482979 | 0.12 |
ENST00000360584.2 ENST00000357730.2 ENST00000528803.1 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr19_-_41356347 | 0.12 |
ENST00000301141.5 |
CYP2A6 |
cytochrome P450, family 2, subfamily A, polypeptide 6 |
| chr4_+_144257915 | 0.12 |
ENST00000262995.4 |
GAB1 |
GRB2-associated binding protein 1 |
| chr16_+_84209738 | 0.12 |
ENST00000564928.1 |
DNAAF1 |
dynein, axonemal, assembly factor 1 |
| chr8_-_102218292 | 0.12 |
ENST00000518336.1 ENST00000520454.1 |
ZNF706 |
zinc finger protein 706 |
| chrX_-_130423386 | 0.12 |
ENST00000370903.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr22_-_42526802 | 0.12 |
ENST00000359033.4 ENST00000389970.3 ENST00000360608.5 |
CYP2D6 |
cytochrome P450, family 2, subfamily D, polypeptide 6 |
| chr4_+_71200681 | 0.11 |
ENST00000273936.5 |
CABS1 |
calcium-binding protein, spermatid-specific 1 |
| chr12_-_11175219 | 0.11 |
ENST00000390673.2 |
TAS2R19 |
taste receptor, type 2, member 19 |
| chr17_-_38256973 | 0.11 |
ENST00000246672.3 |
NR1D1 |
nuclear receptor subfamily 1, group D, member 1 |
| chr1_-_22263790 | 0.11 |
ENST00000374695.3 |
HSPG2 |
heparan sulfate proteoglycan 2 |
| chr14_+_105046094 | 0.11 |
ENST00000331952.2 |
C14orf180 |
chromosome 14 open reading frame 180 |
| chr4_-_120243545 | 0.11 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr5_-_115872142 | 0.11 |
ENST00000510263.1 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr4_+_89378261 | 0.11 |
ENST00000264350.3 |
HERC5 |
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr4_-_46126093 | 0.11 |
ENST00000295452.4 |
GABRG1 |
gamma-aminobutyric acid (GABA) A receptor, gamma 1 |
| chr7_-_22539771 | 0.11 |
ENST00000406890.2 ENST00000424363.1 |
STEAP1B |
STEAP family member 1B |
| chr15_-_68497657 | 0.10 |
ENST00000448060.2 ENST00000467889.1 |
CALML4 |
calmodulin-like 4 |
| chr11_-_111781454 | 0.10 |
ENST00000533280.1 |
CRYAB |
crystallin, alpha B |
| chr9_+_133320339 | 0.10 |
ENST00000372394.1 ENST00000372393.3 ENST00000422569.1 |
ASS1 |
argininosuccinate synthase 1 |
| chr11_-_74442430 | 0.10 |
ENST00000376332.3 |
CHRDL2 |
chordin-like 2 |
| chr2_+_230787201 | 0.10 |
ENST00000283946.3 |
FBXO36 |
F-box protein 36 |
| chr1_+_2407754 | 0.10 |
ENST00000419816.2 ENST00000378486.3 ENST00000378488.3 ENST00000288766.5 |
PLCH2 |
phospholipase C, eta 2 |
| chr3_+_38495333 | 0.10 |
ENST00000352511.4 |
ACVR2B |
activin A receptor, type IIB |
| chr11_-_60719213 | 0.10 |
ENST00000227880.3 |
SLC15A3 |
solute carrier family 15 (oligopeptide transporter), member 3 |
| chr1_-_1356719 | 0.10 |
ENST00000520296.1 |
ANKRD65 |
ankyrin repeat domain 65 |
| chr21_+_42688657 | 0.10 |
ENST00000357985.2 |
FAM3B |
family with sequence similarity 3, member B |
| chr6_+_135502501 | 0.10 |
ENST00000527615.1 ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr9_+_116298778 | 0.10 |
ENST00000462143.1 |
RGS3 |
regulator of G-protein signaling 3 |
| chr6_-_83775489 | 0.09 |
ENST00000369747.3 |
UBE3D |
ubiquitin protein ligase E3D |
| chr4_-_151936416 | 0.09 |
ENST00000510413.1 ENST00000507224.1 |
LRBA |
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr1_-_6550625 | 0.09 |
ENST00000377725.1 ENST00000340850.5 |
PLEKHG5 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr2_+_230787213 | 0.09 |
ENST00000409992.1 |
FBXO36 |
F-box protein 36 |
| chr19_+_46003056 | 0.09 |
ENST00000401593.1 ENST00000396736.2 |
PPM1N |
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr3_+_52812523 | 0.09 |
ENST00000540715.1 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr2_+_242750274 | 0.09 |
ENST00000405370.1 |
NEU4 |
sialidase 4 |
| chr3_+_184080387 | 0.09 |
ENST00000455712.1 |
POLR2H |
polymerase (RNA) II (DNA directed) polypeptide H |
| chr14_+_70346125 | 0.09 |
ENST00000361956.3 ENST00000381280.4 |
SMOC1 |
SPARC related modular calcium binding 1 |
| chr3_-_141747439 | 0.09 |
ENST00000467667.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr17_-_73840415 | 0.09 |
ENST00000592386.1 ENST00000412096.2 ENST00000586147.1 |
UNC13D |
unc-13 homolog D (C. elegans) |
| chr6_-_70506963 | 0.09 |
ENST00000370577.3 |
LMBRD1 |
LMBR1 domain containing 1 |
| chr22_+_18111594 | 0.09 |
ENST00000399782.1 |
BCL2L13 |
BCL2-like 13 (apoptosis facilitator) |
| chr14_-_21562648 | 0.09 |
ENST00000555270.1 |
ZNF219 |
zinc finger protein 219 |
| chr3_+_118905564 | 0.09 |
ENST00000460625.1 |
UPK1B |
uroplakin 1B |
| chr17_+_48423453 | 0.09 |
ENST00000017003.2 ENST00000509778.1 ENST00000507602.1 |
XYLT2 |
xylosyltransferase II |
| chr6_-_112408661 | 0.09 |
ENST00000368662.5 |
TUBE1 |
tubulin, epsilon 1 |
| chr1_-_177939041 | 0.09 |
ENST00000308284.6 |
SEC16B |
SEC16 homolog B (S. cerevisiae) |
| chr7_+_29874341 | 0.09 |
ENST00000409290.1 ENST00000242140.5 |
WIPF3 |
WAS/WASL interacting protein family, member 3 |
| chr2_-_75426826 | 0.09 |
ENST00000305249.5 |
TACR1 |
tachykinin receptor 1 |
| chr16_+_21244986 | 0.09 |
ENST00000311620.5 |
ANKS4B |
ankyrin repeat and sterile alpha motif domain containing 4B |
| chr19_+_1041212 | 0.09 |
ENST00000433129.1 |
ABCA7 |
ATP-binding cassette, sub-family A (ABC1), member 7 |
| chr19_-_15490568 | 0.09 |
ENST00000269701.2 |
AKAP8 |
A kinase (PRKA) anchor protein 8 |
| chr9_-_104145795 | 0.09 |
ENST00000259407.2 |
BAAT |
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr22_-_29711645 | 0.09 |
ENST00000401450.3 |
RASL10A |
RAS-like, family 10, member A |
| chr16_-_11367452 | 0.09 |
ENST00000327157.2 |
PRM3 |
protamine 3 |
| chr14_+_103589789 | 0.09 |
ENST00000558056.1 ENST00000560869.1 |
TNFAIP2 |
tumor necrosis factor, alpha-induced protein 2 |
| chr8_+_133931648 | 0.09 |
ENST00000519178.1 ENST00000542445.1 |
TG |
thyroglobulin |
| chr13_+_25670268 | 0.08 |
ENST00000281589.3 |
PABPC3 |
poly(A) binding protein, cytoplasmic 3 |
| chr17_-_73840774 | 0.08 |
ENST00000207549.4 |
UNC13D |
unc-13 homolog D (C. elegans) |
| chr2_-_85829496 | 0.08 |
ENST00000409668.1 |
TMEM150A |
transmembrane protein 150A |
| chr1_-_201390846 | 0.08 |
ENST00000367312.1 ENST00000555340.2 ENST00000361379.4 |
TNNI1 |
troponin I type 1 (skeletal, slow) |
| chr19_-_633576 | 0.08 |
ENST00000588649.2 |
POLRMT |
polymerase (RNA) mitochondrial (DNA directed) |
| chrX_-_15288154 | 0.08 |
ENST00000380483.3 ENST00000380485.3 ENST00000380488.4 |
ASB9 |
ankyrin repeat and SOCS box containing 9 |
| chr20_+_13202418 | 0.08 |
ENST00000262487.4 |
ISM1 |
isthmin 1, angiogenesis inhibitor |
| chr7_+_47834880 | 0.08 |
ENST00000258776.4 |
C7orf69 |
chromosome 7 open reading frame 69 |
| chr19_+_1065922 | 0.08 |
ENST00000539243.2 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr13_+_113812956 | 0.08 |
ENST00000375547.2 ENST00000342783.4 |
PROZ |
protein Z, vitamin K-dependent plasma glycoprotein |
| chr14_-_23388338 | 0.08 |
ENST00000555209.1 ENST00000554256.1 ENST00000557403.1 ENST00000557549.1 ENST00000555676.1 ENST00000557571.1 ENST00000557464.1 ENST00000554618.1 ENST00000556862.1 ENST00000555722.1 ENST00000346528.5 ENST00000542016.2 ENST00000399922.2 ENST00000557227.1 ENST00000359890.3 |
RBM23 |
RNA binding motif protein 23 |
| chr19_-_14629224 | 0.08 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr20_+_42574317 | 0.08 |
ENST00000358131.5 |
TOX2 |
TOX high mobility group box family member 2 |
| chr17_+_36452989 | 0.08 |
ENST00000312513.5 ENST00000582535.1 |
MRPL45 |
mitochondrial ribosomal protein L45 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.3 | 0.9 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.2 | 0.9 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.2 | 0.7 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.2 | 0.6 | GO:0046271 | phenylpropanoid catabolic process(GO:0046271) |
| 0.1 | 0.4 | GO:0060490 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.4 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.3 | GO:0071109 | superior temporal gyrus development(GO:0071109) |
| 0.1 | 0.3 | GO:0045658 | regulation of neutrophil differentiation(GO:0045658) negative regulation of neutrophil differentiation(GO:0045659) |
| 0.1 | 0.2 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.1 | 0.4 | GO:0048570 | notochord morphogenesis(GO:0048570) fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.2 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 0.2 | GO:1904806 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.1 | 0.3 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.3 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 1.4 | GO:0031065 | positive regulation of histone deacetylation(GO:0031065) |
| 0.1 | 0.2 | GO:0005989 | lactose metabolic process(GO:0005988) lactose biosynthetic process(GO:0005989) |
| 0.1 | 0.3 | GO:0030200 | heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.1 | 0.3 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.1 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.0 | 0.3 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0046416 | D-amino acid metabolic process(GO:0046416) |
| 0.0 | 0.3 | GO:2000491 | positive regulation of hepatic stellate cell activation(GO:2000491) |
| 0.0 | 0.1 | GO:0019074 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.5 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.2 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0070859 | circadian temperature homeostasis(GO:0060086) positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.1 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.2 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.0 | 0.4 | GO:0051547 | regulation of keratinocyte migration(GO:0051547) |
| 0.0 | 0.2 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.2 | GO:0009698 | phenylpropanoid metabolic process(GO:0009698) |
| 0.0 | 0.0 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:1904106 | protein localization to microvillus(GO:1904106) |
| 0.0 | 0.1 | GO:1902994 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) |
| 0.0 | 0.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.2 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.1 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.2 | GO:0002084 | protein depalmitoylation(GO:0002084) |
| 0.0 | 0.1 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 1.4 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.1 | GO:0060061 | Spemann organizer formation(GO:0060061) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.8 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.1 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.0 | 0.0 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.1 | GO:1902460 | regulation of mesenchymal stem cell proliferation(GO:1902460) positive regulation of mesenchymal stem cell proliferation(GO:1902462) |
| 0.0 | 0.0 | GO:0051289 | protein homotetramerization(GO:0051289) |
| 0.0 | 0.2 | GO:0039532 | negative regulation of viral-induced cytoplasmic pattern recognition receptor signaling pathway(GO:0039532) |
| 0.0 | 0.0 | GO:0036343 | psychomotor behavior(GO:0036343) |
| 0.0 | 0.2 | GO:0016198 | axon choice point recognition(GO:0016198) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.3 | GO:0021692 | cerebellar Purkinje cell layer morphogenesis(GO:0021692) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.0 | GO:0019364 | NADP catabolic process(GO:0006742) pyridine nucleotide catabolic process(GO:0019364) |
| 0.0 | 0.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.0 | GO:0006424 | glutamyl-tRNA aminoacylation(GO:0006424) |
| 0.0 | 0.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) |
| 0.0 | 0.3 | GO:0086016 | AV node cell action potential(GO:0086016) AV node cell to bundle of His cell signaling(GO:0086027) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.0 | GO:1901526 | positive regulation of macromitophagy(GO:1901526) positive regulation of mitophagy in response to mitochondrial depolarization(GO:1904925) |
| 0.0 | 0.0 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0045217 | cell-cell junction maintenance(GO:0045217) |
| 0.0 | 0.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.0 | GO:0014707 | branchiomeric skeletal muscle development(GO:0014707) |
| 0.0 | 0.9 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.6 | GO:0010591 | regulation of lamellipodium assembly(GO:0010591) |
| 0.0 | 0.2 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.1 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.0 | 1.6 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.0 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:0035637 | multicellular organismal signaling(GO:0035637) |
| 0.0 | 0.0 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.0 | GO:0061181 | regulation of chondrocyte development(GO:0061181) |
| 0.0 | 0.1 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.2 | GO:0043320 | natural killer cell degranulation(GO:0043320) |
| 0.0 | 0.0 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.0 | 0.1 | GO:1902455 | negative regulation of stem cell population maintenance(GO:1902455) |
| 0.0 | 0.1 | GO:0015705 | iodide transport(GO:0015705) |
| 0.0 | 0.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.1 | GO:2000051 | Wnt receptor catabolic process(GO:0038018) negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.0 | GO:0003330 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 1.4 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.6 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.8 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 2.5 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.1 | GO:0071797 | LUBAC complex(GO:0071797) |
| 0.0 | 0.5 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 1.8 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0033010 | paranodal junction(GO:0033010) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.3 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.2 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.2 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 1.5 | GO:0005604 | basement membrane(GO:0005604) |
| 0.0 | 0.1 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.3 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.5 | GO:0042577 | lipid phosphatase activity(GO:0042577) |
| 0.1 | 0.6 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.1 | 0.3 | GO:0030305 | beta-glucuronidase activity(GO:0004566) heparanase activity(GO:0030305) |
| 0.1 | 0.3 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.3 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.3 | GO:0016314 | inositol-polyphosphate 5-phosphatase activity(GO:0004445) phosphatidylinositol-3,4,5-trisphosphate 3-phosphatase activity(GO:0016314) phosphatidylinositol-3,4,5-trisphosphate 5-phosphatase activity(GO:0034485) |
| 0.0 | 0.2 | GO:0004461 | lactose synthase activity(GO:0004461) |
| 0.0 | 1.1 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.1 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.8 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.4 | GO:0019215 | intermediate filament binding(GO:0019215) |
| 0.0 | 0.3 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 1.2 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0032810 | sterol response element binding(GO:0032810) |
| 0.0 | 0.2 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.4 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.3 | GO:0043546 | molybdopterin cofactor binding(GO:0043546) |
| 0.0 | 0.8 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 1.6 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0052794 | exo-alpha-(2->3)-sialidase activity(GO:0052794) exo-alpha-(2->6)-sialidase activity(GO:0052795) exo-alpha-(2->8)-sialidase activity(GO:0052796) |
| 0.0 | 0.5 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.2 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0008474 | palmitoyl-(protein) hydrolase activity(GO:0008474) palmitoyl hydrolase activity(GO:0098599) |
| 0.0 | 0.2 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.6 | GO:0008157 | protein phosphatase 1 binding(GO:0008157) |
| 0.0 | 0.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.3 | GO:0001054 | RNA polymerase I activity(GO:0001054) |
| 0.0 | 0.2 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.4 | GO:0016712 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced flavin or flavoprotein as one donor, and incorporation of one atom of oxygen(GO:0016712) |
| 0.0 | 0.1 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 1.2 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.0 | GO:0004818 | glutamate-tRNA ligase activity(GO:0004818) |
| 0.0 | 0.4 | GO:0030507 | spectrin binding(GO:0030507) |
| 0.0 | 0.0 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.0 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.0 | 0.1 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.0 | 0.1 | GO:0004075 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.0 | GO:0008177 | succinate dehydrogenase (ubiquinone) activity(GO:0008177) |
| 0.0 | 0.0 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.1 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.0 | 2.2 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0000285 | 1-phosphatidylinositol-3-phosphate 5-kinase activity(GO:0000285) |
| 0.0 | 0.0 | GO:0004609 | phosphatidylserine decarboxylase activity(GO:0004609) |
| 0.0 | 0.1 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.8 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 0.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.9 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.8 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 1.2 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.3 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |
| 0.0 | 0.2 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.4 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.4 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.3 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME VIRAL MESSENGER RNA SYNTHESIS | Genes involved in Viral Messenger RNA Synthesis |
| 0.0 | 0.3 | REACTOME GLUTAMATE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Glutamate Neurotransmitter Release Cycle |
| 0.0 | 0.5 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |