Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
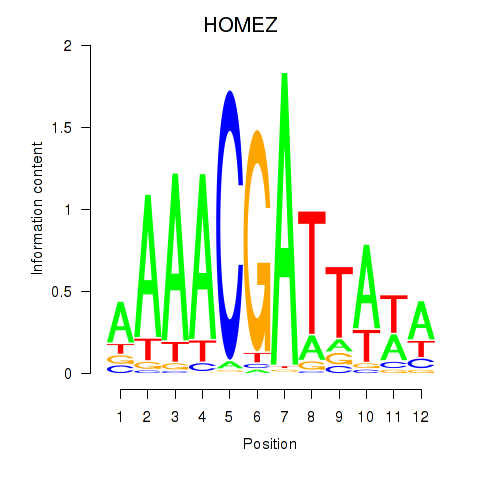
Results for HOMEZ
Z-value: 0.71
Transcription factors associated with HOMEZ
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOMEZ
|
ENSG00000215271.6 | HOMEZ |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOMEZ | hg19_v2_chr14_-_23755297_23755350 | 0.59 | 1.3e-01 | Click! |
Activity profile of HOMEZ motif
Sorted Z-values of HOMEZ motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOMEZ
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_16059818 | 0.79 |
ENST00000322107.1 |
OR10H4 |
olfactory receptor, family 10, subfamily H, member 4 |
| chr5_+_95066823 | 0.76 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr12_-_91539918 | 0.73 |
ENST00000548218.1 |
DCN |
decorin |
| chr1_+_163039143 | 0.60 |
ENST00000531057.1 ENST00000527809.1 ENST00000367908.4 |
RGS4 |
regulator of G-protein signaling 4 |
| chr1_+_163038565 | 0.59 |
ENST00000421743.2 |
RGS4 |
regulator of G-protein signaling 4 |
| chr9_+_99690592 | 0.57 |
ENST00000354649.3 |
NUTM2G |
NUT family member 2G |
| chrX_+_86772787 | 0.56 |
ENST00000373114.4 |
KLHL4 |
kelch-like family member 4 |
| chrX_+_12993202 | 0.52 |
ENST00000451311.2 ENST00000380636.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chrX_+_86772707 | 0.52 |
ENST00000373119.4 |
KLHL4 |
kelch-like family member 4 |
| chrX_-_154250989 | 0.49 |
ENST00000360256.4 |
F8 |
coagulation factor VIII, procoagulant component |
| chr5_+_92919043 | 0.48 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chrX_+_12993336 | 0.44 |
ENST00000380635.1 |
TMSB4X |
thymosin beta 4, X-linked |
| chr6_+_35996859 | 0.44 |
ENST00000472333.1 |
MAPK14 |
mitogen-activated protein kinase 14 |
| chr4_-_70626314 | 0.43 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chrX_-_92928557 | 0.42 |
ENST00000373079.3 ENST00000475430.2 |
NAP1L3 |
nucleosome assembly protein 1-like 3 |
| chr19_+_20959098 | 0.42 |
ENST00000360204.5 ENST00000594534.1 |
ZNF66 |
zinc finger protein 66 |
| chr4_+_70861647 | 0.41 |
ENST00000246895.4 ENST00000381060.2 |
STATH |
statherin |
| chr5_-_135290705 | 0.40 |
ENST00000274507.1 |
LECT2 |
leukocyte cell-derived chemotaxin 2 |
| chr8_-_13134045 | 0.37 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr6_-_11382478 | 0.36 |
ENST00000397378.3 ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
| chr1_+_109102652 | 0.36 |
ENST00000370035.3 ENST00000405454.1 |
FAM102B |
family with sequence similarity 102, member B |
| chr11_+_62104897 | 0.36 |
ENST00000415229.2 ENST00000535727.1 ENST00000301776.5 |
ASRGL1 |
asparaginase like 1 |
| chr2_+_201450591 | 0.34 |
ENST00000374700.2 |
AOX1 |
aldehyde oxidase 1 |
| chr17_+_66521936 | 0.34 |
ENST00000592800.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr2_+_109237717 | 0.34 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr5_-_35048047 | 0.33 |
ENST00000231420.6 |
AGXT2 |
alanine--glyoxylate aminotransferase 2 |
| chr1_-_72748417 | 0.33 |
ENST00000357731.5 |
NEGR1 |
neuronal growth regulator 1 |
| chrX_-_64196351 | 0.32 |
ENST00000374839.3 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr3_-_121379739 | 0.32 |
ENST00000428394.2 ENST00000314583.3 |
HCLS1 |
hematopoietic cell-specific Lyn substrate 1 |
| chr5_-_95158375 | 0.32 |
ENST00000512469.2 ENST00000379979.4 ENST00000505427.1 ENST00000508780.1 |
GLRX |
glutaredoxin (thioltransferase) |
| chrX_-_64196307 | 0.32 |
ENST00000545618.1 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr5_-_64920115 | 0.32 |
ENST00000381018.3 ENST00000274327.7 |
TRIM23 |
tripartite motif containing 23 |
| chr3_-_107941209 | 0.32 |
ENST00000492106.1 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr1_-_182360918 | 0.32 |
ENST00000339526.4 |
GLUL |
glutamate-ammonia ligase |
| chrX_-_119709637 | 0.32 |
ENST00000404115.3 |
CUL4B |
cullin 4B |
| chr1_-_153518270 | 0.31 |
ENST00000354332.4 ENST00000368716.4 |
S100A4 |
S100 calcium binding protein A4 |
| chr4_+_126237554 | 0.31 |
ENST00000394329.3 |
FAT4 |
FAT atypical cadherin 4 |
| chr3_-_169587621 | 0.30 |
ENST00000523069.1 ENST00000316428.5 ENST00000264676.5 |
LRRC31 |
leucine rich repeat containing 31 |
| chr6_+_111408698 | 0.30 |
ENST00000368851.5 |
SLC16A10 |
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr16_-_75467318 | 0.30 |
ENST00000283882.3 |
CFDP1 |
craniofacial development protein 1 |
| chr22_-_29075853 | 0.30 |
ENST00000397906.2 |
TTC28 |
tetratricopeptide repeat domain 28 |
| chr11_+_114168085 | 0.29 |
ENST00000541754.1 |
NNMT |
nicotinamide N-methyltransferase |
| chr15_+_63414760 | 0.29 |
ENST00000557972.1 |
LACTB |
lactamase, beta |
| chr10_-_38265517 | 0.28 |
ENST00000302609.7 |
ZNF25 |
zinc finger protein 25 |
| chrX_-_64196376 | 0.28 |
ENST00000447788.2 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr19_+_41119323 | 0.27 |
ENST00000599724.1 ENST00000597071.1 ENST00000243562.9 |
LTBP4 |
latent transforming growth factor beta binding protein 4 |
| chr12_+_26126681 | 0.27 |
ENST00000542865.1 |
RASSF8 |
Ras association (RalGDS/AF-6) domain family (N-terminal) member 8 |
| chr2_+_233497931 | 0.27 |
ENST00000264059.3 |
EFHD1 |
EF-hand domain family, member D1 |
| chr1_+_97188188 | 0.26 |
ENST00000541987.1 |
PTBP2 |
polypyrimidine tract binding protein 2 |
| chr10_-_72648541 | 0.26 |
ENST00000299299.3 |
PCBD1 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha |
| chr7_-_56119156 | 0.25 |
ENST00000421312.1 ENST00000416592.1 |
PSPH |
phosphoserine phosphatase |
| chr7_+_28725585 | 0.25 |
ENST00000396298.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr1_+_197170592 | 0.25 |
ENST00000535699.1 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chr18_-_19994830 | 0.25 |
ENST00000525417.1 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
| chr5_-_175388327 | 0.25 |
ENST00000432305.2 ENST00000505969.1 |
THOC3 |
THO complex 3 |
| chrX_+_9431324 | 0.25 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr17_+_9728828 | 0.25 |
ENST00000262441.5 |
GLP2R |
glucagon-like peptide 2 receptor |
| chr3_+_186358200 | 0.24 |
ENST00000382136.3 |
FETUB |
fetuin B |
| chr19_+_44617511 | 0.24 |
ENST00000262894.6 ENST00000588926.1 ENST00000592780.1 |
ZNF225 |
zinc finger protein 225 |
| chr3_-_126327398 | 0.24 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr19_-_39330818 | 0.23 |
ENST00000594769.1 ENST00000602021.1 |
AC104534.3 |
Delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial |
| chr12_+_60058458 | 0.23 |
ENST00000548610.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr15_-_99789736 | 0.23 |
ENST00000560235.1 ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23 |
tetratricopeptide repeat domain 23 |
| chr7_-_7679633 | 0.23 |
ENST00000401447.1 |
RPA3 |
replication protein A3, 14kDa |
| chr5_+_178450753 | 0.23 |
ENST00000444149.2 ENST00000519896.1 ENST00000522442.1 |
ZNF879 |
zinc finger protein 879 |
| chr12_+_12764773 | 0.23 |
ENST00000228865.2 |
CREBL2 |
cAMP responsive element binding protein-like 2 |
| chr1_+_12851545 | 0.23 |
ENST00000332296.7 |
PRAMEF1 |
PRAME family member 1 |
| chr9_-_95166884 | 0.22 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr17_+_37856214 | 0.22 |
ENST00000445658.2 |
ERBB2 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr14_+_74034310 | 0.21 |
ENST00000538782.1 |
ACOT2 |
acyl-CoA thioesterase 2 |
| chr12_+_10365404 | 0.21 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr3_-_112218378 | 0.21 |
ENST00000334529.5 |
BTLA |
B and T lymphocyte associated |
| chr7_-_14028488 | 0.21 |
ENST00000405358.4 |
ETV1 |
ets variant 1 |
| chr6_+_20534672 | 0.21 |
ENST00000274695.4 ENST00000378624.4 |
CDKAL1 |
CDK5 regulatory subunit associated protein 1-like 1 |
| chr9_-_21995249 | 0.21 |
ENST00000494262.1 |
CDKN2A |
cyclin-dependent kinase inhibitor 2A |
| chr2_+_61372226 | 0.21 |
ENST00000426997.1 |
C2orf74 |
chromosome 2 open reading frame 74 |
| chr4_-_100242549 | 0.21 |
ENST00000305046.8 ENST00000394887.3 |
ADH1B |
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr6_-_74019870 | 0.21 |
ENST00000370384.3 |
KHDC1 |
KH homology domain containing 1 |
| chr8_+_77593448 | 0.21 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr9_-_138391692 | 0.20 |
ENST00000429260.2 |
C9orf116 |
chromosome 9 open reading frame 116 |
| chr16_+_58283814 | 0.20 |
ENST00000443128.2 ENST00000219299.4 |
CCDC113 |
coiled-coil domain containing 113 |
| chr17_+_47074758 | 0.20 |
ENST00000290341.3 |
IGF2BP1 |
insulin-like growth factor 2 mRNA binding protein 1 |
| chr3_-_107941230 | 0.20 |
ENST00000264538.3 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr11_+_63304273 | 0.20 |
ENST00000439013.2 ENST00000255688.3 |
RARRES3 |
retinoic acid receptor responder (tazarotene induced) 3 |
| chr7_+_13141097 | 0.20 |
ENST00000411542.1 |
AC011288.2 |
AC011288.2 |
| chr9_-_37592561 | 0.20 |
ENST00000544379.1 ENST00000377773.5 ENST00000401811.3 ENST00000321301.6 |
TOMM5 |
translocase of outer mitochondrial membrane 5 homolog (yeast) |
| chr3_-_49466686 | 0.20 |
ENST00000273598.3 ENST00000436744.2 |
NICN1 |
nicolin 1 |
| chr1_-_54405773 | 0.20 |
ENST00000371376.1 |
HSPB11 |
heat shock protein family B (small), member 11 |
| chr19_+_13135386 | 0.19 |
ENST00000360105.4 ENST00000588228.1 ENST00000591028.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr10_-_32667660 | 0.19 |
ENST00000375110.2 |
EPC1 |
enhancer of polycomb homolog 1 (Drosophila) |
| chr12_+_69201923 | 0.19 |
ENST00000462284.1 ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2 |
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr1_+_74701062 | 0.19 |
ENST00000326637.3 |
TNNI3K |
TNNI3 interacting kinase |
| chr10_-_82049424 | 0.19 |
ENST00000372213.3 |
MAT1A |
methionine adenosyltransferase I, alpha |
| chr10_-_31288398 | 0.19 |
ENST00000538351.2 |
ZNF438 |
zinc finger protein 438 |
| chr8_-_99129384 | 0.19 |
ENST00000521560.1 ENST00000254878.3 |
HRSP12 |
heat-responsive protein 12 |
| chr1_+_202431859 | 0.18 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr16_-_4588822 | 0.18 |
ENST00000564828.1 |
CDIP1 |
cell death-inducing p53 target 1 |
| chr1_+_172628154 | 0.18 |
ENST00000340030.3 ENST00000367721.2 |
FASLG |
Fas ligand (TNF superfamily, member 6) |
| chr8_-_93978357 | 0.18 |
ENST00000522925.1 ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK |
triple QxxK/R motif containing |
| chr4_-_140222358 | 0.18 |
ENST00000505036.1 ENST00000544855.1 ENST00000539002.1 |
NDUFC1 |
NADH dehydrogenase (ubiquinone) 1, subcomplex unknown, 1, 6kDa |
| chr1_+_26758790 | 0.18 |
ENST00000427245.2 ENST00000525682.2 ENST00000236342.7 ENST00000526219.1 ENST00000374185.3 ENST00000360009.2 |
DHDDS |
dehydrodolichyl diphosphate synthase |
| chr3_-_112218205 | 0.18 |
ENST00000383680.4 |
BTLA |
B and T lymphocyte associated |
| chr12_-_92539614 | 0.17 |
ENST00000256015.3 |
BTG1 |
B-cell translocation gene 1, anti-proliferative |
| chr18_-_70532906 | 0.17 |
ENST00000299430.2 ENST00000397929.1 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr20_+_42219559 | 0.17 |
ENST00000373030.3 ENST00000373039.4 |
IFT52 |
intraflagellar transport 52 homolog (Chlamydomonas) |
| chr9_-_95166841 | 0.17 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr7_+_98923505 | 0.17 |
ENST00000432884.2 ENST00000262942.5 |
ARPC1A |
actin related protein 2/3 complex, subunit 1A, 41kDa |
| chr1_-_145076186 | 0.17 |
ENST00000369348.3 |
PDE4DIP |
phosphodiesterase 4D interacting protein |
| chr17_-_39183452 | 0.17 |
ENST00000361883.5 |
KRTAP1-5 |
keratin associated protein 1-5 |
| chr8_-_99129338 | 0.17 |
ENST00000520507.1 |
HRSP12 |
heat-responsive protein 12 |
| chr17_+_29664830 | 0.17 |
ENST00000444181.2 ENST00000417592.2 |
NF1 |
neurofibromin 1 |
| chr1_-_182642017 | 0.17 |
ENST00000367557.4 ENST00000258302.4 |
RGS8 |
regulator of G-protein signaling 8 |
| chr12_+_8995832 | 0.17 |
ENST00000541459.1 |
A2ML1 |
alpha-2-macroglobulin-like 1 |
| chr11_+_65266507 | 0.17 |
ENST00000544868.1 |
MALAT1 |
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr8_-_13372253 | 0.16 |
ENST00000316609.5 |
DLC1 |
deleted in liver cancer 1 |
| chr12_+_32655048 | 0.16 |
ENST00000427716.2 ENST00000266482.3 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
| chr7_-_73256838 | 0.16 |
ENST00000297873.4 |
WBSCR27 |
Williams Beuren syndrome chromosome region 27 |
| chr5_-_159827033 | 0.16 |
ENST00000523213.1 |
C5orf54 |
chromosome 5 open reading frame 54 |
| chrX_+_54834004 | 0.16 |
ENST00000375068.1 |
MAGED2 |
melanoma antigen family D, 2 |
| chrX_+_118708493 | 0.16 |
ENST00000371558.2 |
UBE2A |
ubiquitin-conjugating enzyme E2A |
| chr6_+_80816342 | 0.16 |
ENST00000369760.4 ENST00000356489.5 ENST00000320393.6 |
BCKDHB |
branched chain keto acid dehydrogenase E1, beta polypeptide |
| chr2_-_190627481 | 0.16 |
ENST00000264151.5 ENST00000520350.1 ENST00000521630.1 ENST00000517895.1 |
OSGEPL1 |
O-sialoglycoprotein endopeptidase-like 1 |
| chr8_+_77593474 | 0.16 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chrX_-_134429952 | 0.16 |
ENST00000370764.1 |
ZNF75D |
zinc finger protein 75D |
| chr1_+_32608566 | 0.16 |
ENST00000545542.1 |
KPNA6 |
karyopherin alpha 6 (importin alpha 7) |
| chr2_+_242289502 | 0.15 |
ENST00000451310.1 |
SEPT2 |
septin 2 |
| chr16_-_30032610 | 0.15 |
ENST00000574405.1 |
DOC2A |
double C2-like domains, alpha |
| chr12_+_12878829 | 0.15 |
ENST00000326765.6 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr3_-_138312971 | 0.15 |
ENST00000485115.1 ENST00000484888.1 ENST00000468900.1 ENST00000542237.1 ENST00000481834.1 |
CEP70 |
centrosomal protein 70kDa |
| chr6_+_7590413 | 0.15 |
ENST00000342415.5 |
SNRNP48 |
small nuclear ribonucleoprotein 48kDa (U11/U12) |
| chr6_-_34664612 | 0.15 |
ENST00000374023.3 ENST00000374026.3 |
C6orf106 |
chromosome 6 open reading frame 106 |
| chr5_-_159827073 | 0.15 |
ENST00000408953.3 |
C5orf54 |
chromosome 5 open reading frame 54 |
| chr8_-_93978309 | 0.15 |
ENST00000517858.1 ENST00000378861.5 |
TRIQK |
triple QxxK/R motif containing |
| chr9_-_13279406 | 0.15 |
ENST00000546205.1 |
MPDZ |
multiple PDZ domain protein |
| chr12_-_42631529 | 0.15 |
ENST00000548917.1 |
YAF2 |
YY1 associated factor 2 |
| chr6_-_31632962 | 0.14 |
ENST00000456540.1 ENST00000445768.1 |
GPANK1 |
G patch domain and ankyrin repeats 1 |
| chr4_+_108925787 | 0.14 |
ENST00000454409.2 |
HADH |
hydroxyacyl-CoA dehydrogenase |
| chr7_+_20687017 | 0.14 |
ENST00000258738.6 |
ABCB5 |
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr8_+_42552533 | 0.14 |
ENST00000289957.2 |
CHRNB3 |
cholinergic receptor, nicotinic, beta 3 (neuronal) |
| chr4_+_169842707 | 0.14 |
ENST00000503290.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chrX_-_80457385 | 0.14 |
ENST00000451455.1 ENST00000436386.1 ENST00000358130.2 |
HMGN5 |
high mobility group nucleosome binding domain 5 |
| chr1_-_180471947 | 0.14 |
ENST00000367595.3 |
ACBD6 |
acyl-CoA binding domain containing 6 |
| chr2_-_112237835 | 0.14 |
ENST00000442293.1 ENST00000439494.1 |
MIR4435-1HG |
MIR4435-1 host gene (non-protein coding) |
| chr1_+_24018269 | 0.14 |
ENST00000374550.3 |
RPL11 |
ribosomal protein L11 |
| chr8_+_107460147 | 0.14 |
ENST00000442977.2 |
OXR1 |
oxidation resistance 1 |
| chr12_-_91573249 | 0.14 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr18_+_11857439 | 0.14 |
ENST00000602628.1 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr11_-_36310958 | 0.13 |
ENST00000532705.1 ENST00000263401.5 ENST00000452374.2 |
COMMD9 |
COMM domain containing 9 |
| chr5_-_156569850 | 0.13 |
ENST00000524219.1 |
HAVCR2 |
hepatitis A virus cellular receptor 2 |
| chrX_-_114953669 | 0.13 |
ENST00000449327.1 |
RP1-241P17.4 |
Uncharacterized protein |
| chr5_+_79331164 | 0.13 |
ENST00000350881.2 |
THBS4 |
thrombospondin 4 |
| chr12_+_72148614 | 0.13 |
ENST00000261263.3 |
RAB21 |
RAB21, member RAS oncogene family |
| chr6_+_34725263 | 0.13 |
ENST00000374018.1 ENST00000374017.3 |
SNRPC |
small nuclear ribonucleoprotein polypeptide C |
| chr5_-_36242119 | 0.13 |
ENST00000511088.1 ENST00000282512.3 ENST00000506945.1 |
NADK2 |
NAD kinase 2, mitochondrial |
| chr19_+_24097706 | 0.13 |
ENST00000322487.7 ENST00000575986.1 |
ZNF726 |
zinc finger protein 726 |
| chr19_-_52643157 | 0.13 |
ENST00000597013.1 ENST00000600228.1 ENST00000596290.1 |
ZNF616 |
zinc finger protein 616 |
| chr17_+_30469473 | 0.13 |
ENST00000333942.6 ENST00000358365.3 ENST00000583994.1 ENST00000545287.2 |
RHOT1 |
ras homolog family member T1 |
| chr1_+_144220127 | 0.13 |
ENST00000369373.5 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
| chr2_-_99224915 | 0.13 |
ENST00000328709.3 ENST00000409997.1 |
COA5 |
cytochrome c oxidase assembly factor 5 |
| chr10_-_121296045 | 0.13 |
ENST00000392865.1 |
RGS10 |
regulator of G-protein signaling 10 |
| chr19_-_18995029 | 0.13 |
ENST00000596048.1 |
CERS1 |
ceramide synthase 1 |
| chr17_+_4853442 | 0.13 |
ENST00000522301.1 |
ENO3 |
enolase 3 (beta, muscle) |
| chr19_-_4124079 | 0.13 |
ENST00000394867.4 ENST00000262948.5 |
MAP2K2 |
mitogen-activated protein kinase kinase 2 |
| chr1_+_236557569 | 0.13 |
ENST00000334232.4 |
EDARADD |
EDAR-associated death domain |
| chr9_+_132962843 | 0.13 |
ENST00000458469.1 |
NCS1 |
neuronal calcium sensor 1 |
| chr11_+_120207787 | 0.13 |
ENST00000397843.2 ENST00000356641.3 |
ARHGEF12 |
Rho guanine nucleotide exchange factor (GEF) 12 |
| chr7_-_18067478 | 0.12 |
ENST00000506618.2 |
PRPS1L1 |
phosphoribosyl pyrophosphate synthetase 1-like 1 |
| chr4_-_159956333 | 0.12 |
ENST00000434826.2 |
C4orf45 |
chromosome 4 open reading frame 45 |
| chr19_+_12780512 | 0.12 |
ENST00000242796.4 |
WDR83 |
WD repeat domain 83 |
| chr10_+_53806501 | 0.12 |
ENST00000373975.2 |
PRKG1 |
protein kinase, cGMP-dependent, type I |
| chr19_+_44764031 | 0.12 |
ENST00000592581.1 ENST00000590668.1 ENST00000588489.1 ENST00000391958.2 |
ZNF233 |
zinc finger protein 233 |
| chr16_-_18801643 | 0.12 |
ENST00000322989.4 ENST00000563390.1 |
RPS15A |
ribosomal protein S15a |
| chr2_+_132160448 | 0.12 |
ENST00000437751.1 |
AC073869.19 |
long intergenic non-protein coding RNA 1120 |
| chr8_-_145652336 | 0.12 |
ENST00000529182.1 ENST00000526054.1 |
VPS28 |
vacuolar protein sorting 28 homolog (S. cerevisiae) |
| chr12_+_72233487 | 0.12 |
ENST00000482439.2 ENST00000550746.1 ENST00000491063.1 ENST00000319106.8 ENST00000485960.2 ENST00000393309.3 |
TBC1D15 |
TBC1 domain family, member 15 |
| chr17_-_77924627 | 0.12 |
ENST00000572862.1 ENST00000573782.1 ENST00000574427.1 ENST00000570373.1 ENST00000340848.7 ENST00000576768.1 |
TBC1D16 |
TBC1 domain family, member 16 |
| chr5_+_54398463 | 0.12 |
ENST00000274306.6 |
GZMA |
granzyme A (granzyme 1, cytotoxic T-lymphocyte-associated serine esterase 3) |
| chr6_+_31802364 | 0.12 |
ENST00000375640.3 ENST00000375641.2 |
C6orf48 |
chromosome 6 open reading frame 48 |
| chrX_-_48931648 | 0.12 |
ENST00000376386.3 ENST00000376390.4 |
PRAF2 |
PRA1 domain family, member 2 |
| chr2_+_87769459 | 0.12 |
ENST00000414030.1 ENST00000437561.1 |
LINC00152 |
long intergenic non-protein coding RNA 152 |
| chr5_+_118965244 | 0.12 |
ENST00000515256.1 ENST00000509264.1 |
FAM170A |
family with sequence similarity 170, member A |
| chr2_-_231860596 | 0.12 |
ENST00000441063.1 ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2 |
SPATA3 antisense RNA 1 (head to head) |
| chr4_-_170679024 | 0.12 |
ENST00000393381.2 |
C4orf27 |
chromosome 4 open reading frame 27 |
| chr1_-_153958805 | 0.12 |
ENST00000368575.3 |
RAB13 |
RAB13, member RAS oncogene family |
| chr16_-_20364030 | 0.12 |
ENST00000396134.2 ENST00000573567.1 ENST00000570757.1 ENST00000424589.1 ENST00000302509.4 ENST00000571174.1 ENST00000576688.1 |
UMOD |
uromodulin |
| chrX_-_151999269 | 0.12 |
ENST00000370277.3 |
CETN2 |
centrin, EF-hand protein, 2 |
| chr16_+_29674277 | 0.12 |
ENST00000395389.2 |
SPN |
sialophorin |
| chr1_-_6259641 | 0.12 |
ENST00000234875.4 |
RPL22 |
ribosomal protein L22 |
| chr17_+_6554971 | 0.12 |
ENST00000391428.2 |
C17orf100 |
chromosome 17 open reading frame 100 |
| chr10_-_79397391 | 0.12 |
ENST00000286628.8 ENST00000406533.3 ENST00000354353.5 ENST00000404857.1 |
KCNMA1 |
potassium large conductance calcium-activated channel, subfamily M, alpha member 1 |
| chr2_-_3523507 | 0.12 |
ENST00000327435.6 |
ADI1 |
acireductone dioxygenase 1 |
| chr16_+_33204156 | 0.12 |
ENST00000398667.4 |
TP53TG3C |
TP53 target 3C |
| chr4_-_99064387 | 0.11 |
ENST00000295268.3 |
STPG2 |
sperm-tail PG-rich repeat containing 2 |
| chrX_-_114468605 | 0.11 |
ENST00000538422.1 ENST00000317135.8 |
LRCH2 |
leucine-rich repeats and calponin homology (CH) domain containing 2 |
| chr6_+_34725181 | 0.11 |
ENST00000244520.5 |
SNRPC |
small nuclear ribonucleoprotein polypeptide C |
| chr6_+_31802685 | 0.11 |
ENST00000375639.2 ENST00000375638.3 ENST00000375635.2 ENST00000375642.2 ENST00000395789.1 |
C6orf48 |
chromosome 6 open reading frame 48 |
| chr8_-_93978346 | 0.11 |
ENST00000523580.1 |
TRIQK |
triple QxxK/R motif containing |
| chr16_-_20364122 | 0.11 |
ENST00000396138.4 ENST00000577168.1 |
UMOD |
uromodulin |
| chr19_-_22379753 | 0.11 |
ENST00000397121.2 |
ZNF676 |
zinc finger protein 676 |
| chr7_-_7680601 | 0.11 |
ENST00000396682.2 |
RPA3 |
replication protein A3, 14kDa |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:1903026 | negative regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903026) |
| 0.1 | 0.3 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.3 | GO:0006542 | glutamine biosynthetic process(GO:0006542) |
| 0.1 | 0.4 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.1 | 0.3 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.2 | GO:0044278 | cell wall disruption in other organism(GO:0044278) |
| 0.1 | 0.3 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.1 | 0.3 | GO:0035720 | intraciliary anterograde transport(GO:0035720) |
| 0.1 | 0.2 | GO:0036369 | transcription factor catabolic process(GO:0036369) cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.1 | 0.2 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.1 | 0.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.1 | 0.2 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.1 | 0.2 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.3 | GO:0034334 | adherens junction maintenance(GO:0034334) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.2 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.2 | GO:0072023 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.2 | GO:0030644 | cellular chloride ion homeostasis(GO:0030644) |
| 0.0 | 0.1 | GO:0002519 | natural killer cell tolerance induction(GO:0002519) regulation of tolerance induction dependent upon immune response(GO:0002652) negative regulation of response to tumor cell(GO:0002835) negative regulation of immune response to tumor cell(GO:0002838) negative regulation of natural killer cell mediated immune response to tumor cell(GO:0002856) negative regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002859) |
| 0.0 | 0.2 | GO:0097368 | establishment of Sertoli cell barrier(GO:0097368) |
| 0.0 | 0.1 | GO:2000397 | regulation of ubiquitin-dependent endocytosis(GO:2000395) positive regulation of ubiquitin-dependent endocytosis(GO:2000397) |
| 0.0 | 0.4 | GO:0006528 | asparagine metabolic process(GO:0006528) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:2000434 | regulation of protein neddylation(GO:2000434) negative regulation of protein neddylation(GO:2000435) |
| 0.0 | 0.5 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.3 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.2 | GO:0019303 | D-ribose catabolic process(GO:0019303) |
| 0.0 | 0.3 | GO:2000270 | negative regulation of fibroblast apoptotic process(GO:2000270) |
| 0.0 | 0.2 | GO:1902730 | regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010908) positive regulation of heparan sulfate proteoglycan biosynthetic process(GO:0010909) canonical Wnt signaling pathway involved in positive regulation of epithelial to mesenchymal transition(GO:0044334) positive regulation of proteoglycan biosynthetic process(GO:1902730) |
| 0.0 | 0.1 | GO:0031508 | pericentric heterochromatin assembly(GO:0031508) |
| 0.0 | 0.2 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.1 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.1 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.4 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.1 | GO:0032242 | regulation of nucleoside transport(GO:0032242) positive regulation of necroptotic process(GO:0060545) |
| 0.0 | 0.2 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 1.2 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.3 | GO:2000111 | senescence-associated heterochromatin focus assembly(GO:0035986) positive regulation of macrophage apoptotic process(GO:2000111) |
| 0.0 | 0.3 | GO:0046146 | tetrahydrobiopterin biosynthetic process(GO:0006729) tetrahydrobiopterin metabolic process(GO:0046146) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.2 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:0009183 | purine deoxyribonucleoside diphosphate biosynthetic process(GO:0009183) |
| 0.0 | 0.7 | GO:0060972 | left/right pattern formation(GO:0060972) |
| 0.0 | 0.2 | GO:0006741 | NADP biosynthetic process(GO:0006741) |
| 0.0 | 0.0 | GO:0050728 | negative regulation of inflammatory response(GO:0050728) |
| 0.0 | 0.2 | GO:0006659 | phosphatidylserine biosynthetic process(GO:0006659) regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000371) positive regulation of DNA topoisomerase (ATP-hydrolyzing) activity(GO:2000373) |
| 0.0 | 0.1 | GO:0001970 | positive regulation of activation of membrane attack complex(GO:0001970) |
| 0.0 | 0.1 | GO:0002424 | plasmacytoid dendritic cell activation(GO:0002270) T cell mediated immune response to tumor cell(GO:0002424) regulation of T cell mediated immune response to tumor cell(GO:0002840) regulation of restriction endodeoxyribonuclease activity(GO:0032072) |
| 0.0 | 0.7 | GO:0006293 | nucleotide-excision repair, preincision complex stabilization(GO:0006293) |
| 0.0 | 0.1 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:0006617 | SRP-dependent cotranslational protein targeting to membrane, signal sequence recognition(GO:0006617) |
| 0.0 | 0.4 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.3 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.1 | GO:0090170 | regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.1 | GO:0052551 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.5 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.3 | GO:0030854 | positive regulation of granulocyte differentiation(GO:0030854) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0032071 | regulation of endodeoxyribonuclease activity(GO:0032071) |
| 0.0 | 0.6 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.1 | GO:0060087 | relaxation of vascular smooth muscle(GO:0060087) |
| 0.0 | 0.1 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.0 | 0.1 | GO:1902513 | regulation of organelle transport along microtubule(GO:1902513) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.3 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.2 | GO:0046784 | viral mRNA export from host cell nucleus(GO:0046784) |
| 0.0 | 0.2 | GO:2000271 | positive regulation of fibroblast apoptotic process(GO:2000271) |
| 0.0 | 0.7 | GO:0021522 | spinal cord motor neuron differentiation(GO:0021522) |
| 0.0 | 0.3 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0045144 | meiotic sister chromatid segregation(GO:0045144) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.1 | GO:2000382 | positive regulation of mesoderm development(GO:2000382) |
| 0.0 | 0.1 | GO:1900063 | regulation of peroxisome organization(GO:1900063) |
| 0.0 | 0.0 | GO:0048213 | Golgi vesicle prefusion complex stabilization(GO:0048213) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.0 | GO:0035377 | transepithelial water transport(GO:0035377) positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.2 | GO:2000643 | positive regulation of early endosome to late endosome transport(GO:2000643) |
| 0.0 | 0.1 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.3 | GO:0007213 | G-protein coupled acetylcholine receptor signaling pathway(GO:0007213) |
| 0.0 | 0.1 | GO:0010637 | negative regulation of mitochondrial fusion(GO:0010637) |
| 0.0 | 0.1 | GO:0031536 | positive regulation of exit from mitosis(GO:0031536) |
| 0.0 | 0.1 | GO:0070585 | protein localization to mitochondrion(GO:0070585) |
| 0.0 | 0.0 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.1 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.0 | 0.2 | GO:0048934 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.0 | 0.1 | GO:0034465 | response to carbon monoxide(GO:0034465) smooth muscle contraction involved in micturition(GO:0060083) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.0 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.0 | 0.1 | GO:0033211 | positive regulation of activin receptor signaling pathway(GO:0032927) adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:0043562 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.1 | GO:1990822 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.1 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.2 | GO:0086069 | bundle of His cell to Purkinje myocyte communication(GO:0086069) |
| 0.0 | 0.0 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.0 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.7 | GO:0044292 | dendrite terminus(GO:0044292) |
| 0.0 | 0.1 | GO:0000939 | condensed chromosome inner kinetochore(GO:0000939) |
| 0.0 | 0.2 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.2 | GO:0070369 | beta-catenin-TCF7L2 complex(GO:0070369) |
| 0.0 | 0.3 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.3 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.2 | GO:0000347 | THO complex(GO:0000347) THO complex part of transcription export complex(GO:0000445) |
| 0.0 | 0.2 | GO:0032593 | insulin-responsive compartment(GO:0032593) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.2 | GO:0005947 | mitochondrial alpha-ketoglutarate dehydrogenase complex(GO:0005947) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.2 | GO:0005742 | mitochondrial outer membrane translocase complex(GO:0005742) |
| 0.0 | 0.2 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.1 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.1 | GO:0030891 | VCB complex(GO:0030891) |
| 0.0 | 0.2 | GO:0000243 | commitment complex(GO:0000243) |
| 0.0 | 0.3 | GO:0005662 | DNA replication factor A complex(GO:0005662) |
| 0.0 | 0.0 | GO:0071598 | neuronal ribonucleoprotein granule(GO:0071598) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.1 | GO:1990130 | Iml1 complex(GO:1990130) |
| 0.0 | 0.3 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.1 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.3 | GO:0097386 | glial cell projection(GO:0097386) |
| 0.0 | 0.1 | GO:0000275 | mitochondrial proton-transporting ATP synthase complex, catalytic core F(1)(GO:0000275) |
| 0.0 | 1.7 | GO:0031093 | platelet alpha granule lumen(GO:0031093) |
| 0.0 | 0.1 | GO:0036128 | CatSper complex(GO:0036128) |
| 0.0 | 0.0 | GO:0019008 | molybdopterin synthase complex(GO:0019008) |
| 0.0 | 0.1 | GO:0005956 | protein kinase CK2 complex(GO:0005956) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:0010370 | perinucleolar chromocenter(GO:0010370) |
| 0.0 | 0.3 | GO:0030992 | intraciliary transport particle B(GO:0030992) |
| 0.0 | 0.1 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
| 0.0 | 1.0 | GO:0031463 | Cul3-RING ubiquitin ligase complex(GO:0031463) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.1 | 0.4 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 0.3 | GO:0004356 | glutamate-ammonia ligase activity(GO:0004356) ammonia ligase activity(GO:0016211) acid-ammonia (or amide) ligase activity(GO:0016880) |
| 0.1 | 0.3 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.1 | 0.3 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.1 | 0.2 | GO:0004967 | glucagon receptor activity(GO:0004967) |
| 0.1 | 0.2 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.1 | 0.3 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.3 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.1 | 0.3 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 0.2 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 0.5 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.1 | 0.2 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.1 | 0.2 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.2 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 1.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.0 | 0.8 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.2 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.2 | GO:0044715 | 8-oxo-dGDP phosphatase activity(GO:0044715) |
| 0.0 | 0.4 | GO:0051525 | NFAT protein binding(GO:0051525) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.4 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 0.1 | GO:0004692 | cGMP-dependent protein kinase activity(GO:0004692) |
| 0.0 | 0.1 | GO:0004739 | pyruvate dehydrogenase (acetyl-transferring) activity(GO:0004739) |
| 0.0 | 0.1 | GO:0032795 | heterotrimeric G-protein binding(GO:0032795) |
| 0.0 | 0.1 | GO:0008969 | phosphohistidine phosphatase activity(GO:0008969) |
| 0.0 | 0.5 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.2 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0004749 | ribose phosphate diphosphokinase activity(GO:0004749) |
| 0.0 | 1.0 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.5 | GO:0034236 | protein kinase A catalytic subunit binding(GO:0034236) |
| 0.0 | 0.1 | GO:0008670 | 2,4-dienoyl-CoA reductase (NADPH) activity(GO:0008670) |
| 0.0 | 0.1 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.2 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0061628 | H3K27me3 modified histone binding(GO:0061628) |
| 0.0 | 0.2 | GO:0042834 | peptidoglycan binding(GO:0042834) |
| 0.0 | 0.1 | GO:0060072 | large conductance calcium-activated potassium channel activity(GO:0060072) |
| 0.0 | 0.2 | GO:0016290 | palmitoyl-CoA hydrolase activity(GO:0016290) |
| 0.0 | 0.0 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.0 | 0.3 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0055104 | ligase inhibitor activity(GO:0055104) ubiquitin ligase inhibitor activity(GO:1990948) |
| 0.0 | 0.2 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.2 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.6 | GO:0061631 | ubiquitin conjugating enzyme activity(GO:0061631) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.0 | 0.2 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.0 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.1 | GO:0050291 | sphingosine N-acyltransferase activity(GO:0050291) |
| 0.0 | 0.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.0 | GO:0004082 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.7 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 0.5 | SA G1 AND S PHASES | Cdk2, 4, and 6 bind cyclin D in G1, while cdk2/cyclin E promotes the G1/S transition. |
| 0.0 | 0.3 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME ADENYLATE CYCLASE INHIBITORY PATHWAY | Genes involved in Adenylate cyclase inhibitory pathway |
| 0.0 | 0.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.5 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME REMOVAL OF THE FLAP INTERMEDIATE FROM THE C STRAND | Genes involved in Removal of the Flap Intermediate from the C-strand |
| 0.0 | 0.4 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.6 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME GLUCAGON TYPE LIGAND RECEPTORS | Genes involved in Glucagon-type ligand receptors |
| 0.0 | 1.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 0.3 | REACTOME GLUCAGON SIGNALING IN METABOLIC REGULATION | Genes involved in Glucagon signaling in metabolic regulation |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.2 | REACTOME MITOCHONDRIAL FATTY ACID BETA OXIDATION | Genes involved in Mitochondrial Fatty Acid Beta-Oxidation |
| 0.0 | 0.3 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.3 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.2 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.1 | REACTOME ELONGATION ARREST AND RECOVERY | Genes involved in Elongation arrest and recovery |
| 0.0 | 0.1 | REACTOME MEMBRANE BINDING AND TARGETTING OF GAG PROTEINS | Genes involved in Membrane binding and targetting of GAG proteins |