Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for HOXA10_HOXB9
Z-value: 0.52
Transcription factors associated with HOXA10_HOXB9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
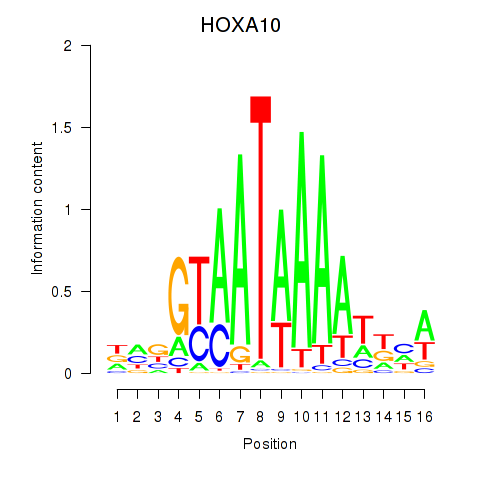
HOXA10
|
ENSG00000253293.3 | HOXA10 |
|
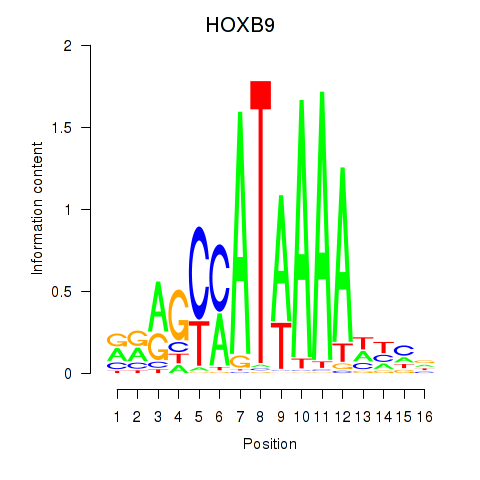
HOXB9
|
ENSG00000170689.8 | HOXB9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXA10 | hg19_v2_chr7_-_27219849_27219880 | 0.67 | 7.2e-02 | Click! |
| HOXB9 | hg19_v2_chr17_-_46703826_46703845 | 0.31 | 4.5e-01 | Click! |
Activity profile of HOXA10_HOXB9 motif
Sorted Z-values of HOXA10_HOXB9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXA10_HOXB9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_41742697 | 0.90 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chrX_+_105937068 | 0.86 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_153003671 | 0.71 |
ENST00000307098.4 |
SPRR1B |
small proline-rich protein 1B |
| chr1_+_160370344 | 0.58 |
ENST00000368061.2 |
VANGL2 |
VANGL planar cell polarity protein 2 |
| chr2_-_216878305 | 0.49 |
ENST00000263268.6 |
MREG |
melanoregulin |
| chr17_-_39646116 | 0.48 |
ENST00000328119.6 |
KRT36 |
keratin 36 |
| chr7_+_128784712 | 0.44 |
ENST00000289407.4 |
TSPAN33 |
tetraspanin 33 |
| chr10_-_129691195 | 0.37 |
ENST00000368671.3 |
CLRN3 |
clarin 3 |
| chr5_+_140261703 | 0.36 |
ENST00000409494.1 ENST00000289272.2 |
PCDHA13 |
protocadherin alpha 13 |
| chr14_-_72458326 | 0.34 |
ENST00000542853.1 |
AC005477.1 |
AC005477.1 |
| chrX_-_117119243 | 0.32 |
ENST00000539496.1 ENST00000469946.1 |
KLHL13 |
kelch-like family member 13 |
| chr3_+_189507432 | 0.31 |
ENST00000354600.5 |
TP63 |
tumor protein p63 |
| chr7_-_143105941 | 0.29 |
ENST00000275815.3 |
EPHA1 |
EPH receptor A1 |
| chr4_-_72649763 | 0.28 |
ENST00000513476.1 |
GC |
group-specific component (vitamin D binding protein) |
| chr18_-_19994830 | 0.27 |
ENST00000525417.1 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
| chr8_-_81083890 | 0.26 |
ENST00000518937.1 |
TPD52 |
tumor protein D52 |
| chr3_-_172241250 | 0.25 |
ENST00000420541.2 ENST00000241261.2 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
| chr11_+_63606373 | 0.25 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr17_-_39274606 | 0.24 |
ENST00000391413.2 |
KRTAP4-11 |
keratin associated protein 4-11 |
| chr1_+_46152886 | 0.24 |
ENST00000372025.4 |
TMEM69 |
transmembrane protein 69 |
| chr5_+_140165876 | 0.24 |
ENST00000504120.2 ENST00000394633.3 ENST00000378133.3 |
PCDHA1 |
protocadherin alpha 1 |
| chr7_+_35756092 | 0.24 |
ENST00000458087.3 |
AC018647.3 |
AC018647.3 |
| chr7_+_35756186 | 0.23 |
ENST00000430518.1 |
AC018647.3 |
AC018647.3 |
| chr11_+_10471836 | 0.22 |
ENST00000444303.2 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr10_+_96698406 | 0.21 |
ENST00000260682.6 |
CYP2C9 |
cytochrome P450, family 2, subfamily C, polypeptide 9 |
| chr3_-_150690471 | 0.20 |
ENST00000468836.1 ENST00000328863.4 |
CLRN1 |
clarin 1 |
| chr8_-_81083731 | 0.19 |
ENST00000379096.5 |
TPD52 |
tumor protein D52 |
| chr20_+_10015678 | 0.19 |
ENST00000378392.1 ENST00000378380.3 |
ANKEF1 |
ankyrin repeat and EF-hand domain containing 1 |
| chr18_-_52989217 | 0.19 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr2_-_69180012 | 0.18 |
ENST00000481498.1 |
GKN2 |
gastrokine 2 |
| chrX_-_133792480 | 0.18 |
ENST00000359237.4 |
PLAC1 |
placenta-specific 1 |
| chr4_+_71263599 | 0.17 |
ENST00000399575.2 |
PROL1 |
proline rich, lacrimal 1 |
| chr6_+_43738444 | 0.17 |
ENST00000324450.6 ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA |
vascular endothelial growth factor A |
| chr18_-_52989525 | 0.17 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr18_-_74839891 | 0.16 |
ENST00000581878.1 |
MBP |
myelin basic protein |
| chr6_+_43737939 | 0.16 |
ENST00000372067.3 |
VEGFA |
vascular endothelial growth factor A |
| chr1_+_23345943 | 0.16 |
ENST00000400181.4 ENST00000542151.1 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr5_+_98109322 | 0.16 |
ENST00000513185.1 |
RGMB |
repulsive guidance molecule family member b |
| chr18_-_44561988 | 0.16 |
ENST00000332567.4 |
TCEB3B |
transcription elongation factor B polypeptide 3B (elongin A2) |
| chr1_+_23345930 | 0.16 |
ENST00000356634.3 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr1_-_186430222 | 0.16 |
ENST00000391997.2 |
PDC |
phosducin |
| chr12_+_20963632 | 0.15 |
ENST00000540853.1 ENST00000261196.2 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr12_+_20963647 | 0.15 |
ENST00000381545.3 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr3_+_189507523 | 0.15 |
ENST00000437221.1 ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63 |
tumor protein p63 |
| chr2_-_69180083 | 0.15 |
ENST00000328895.4 |
GKN2 |
gastrokine 2 |
| chr5_+_34915444 | 0.14 |
ENST00000336767.5 |
BRIX1 |
BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
| chr10_+_85933494 | 0.14 |
ENST00000372126.3 |
C10orf99 |
chromosome 10 open reading frame 99 |
| chr2_-_51259229 | 0.14 |
ENST00000405472.3 |
NRXN1 |
neurexin 1 |
| chr12_+_1800179 | 0.14 |
ENST00000357103.4 |
ADIPOR2 |
adiponectin receptor 2 |
| chr3_+_125687987 | 0.14 |
ENST00000514116.1 ENST00000251776.4 ENST00000504401.1 ENST00000513830.1 ENST00000508088.1 |
ROPN1B |
rhophilin associated tail protein 1B |
| chr2_-_51259292 | 0.14 |
ENST00000401669.2 |
NRXN1 |
neurexin 1 |
| chr13_-_103719196 | 0.14 |
ENST00000245312.3 |
SLC10A2 |
solute carrier family 10 (sodium/bile acid cotransporter), member 2 |
| chr10_+_24755416 | 0.14 |
ENST00000396446.1 ENST00000396445.1 ENST00000376451.2 |
KIAA1217 |
KIAA1217 |
| chr9_+_124413873 | 0.13 |
ENST00000408936.3 |
DAB2IP |
DAB2 interacting protein |
| chr1_+_67773044 | 0.13 |
ENST00000262345.1 ENST00000371000.1 |
IL12RB2 |
interleukin 12 receptor, beta 2 |
| chr3_+_57875711 | 0.13 |
ENST00000442599.2 |
SLMAP |
sarcolemma associated protein |
| chr3_+_140981456 | 0.13 |
ENST00000504264.1 |
ACPL2 |
acid phosphatase-like 2 |
| chr8_-_21669826 | 0.13 |
ENST00000517328.1 |
GFRA2 |
GDNF family receptor alpha 2 |
| chr1_+_32379174 | 0.13 |
ENST00000391369.1 |
AL136115.1 |
HCG2032337; PRO1848; Uncharacterized protein |
| chr3_+_57875738 | 0.13 |
ENST00000417128.1 ENST00000438794.1 |
SLMAP |
sarcolemma associated protein |
| chr4_-_76957214 | 0.13 |
ENST00000306621.3 |
CXCL11 |
chemokine (C-X-C motif) ligand 11 |
| chr12_+_70219052 | 0.13 |
ENST00000552032.2 ENST00000547771.2 |
MYRFL |
myelin regulatory factor-like |
| chr10_+_133747955 | 0.13 |
ENST00000455566.1 |
PPP2R2D |
protein phosphatase 2, regulatory subunit B, delta |
| chr10_-_73848531 | 0.12 |
ENST00000373109.2 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr1_+_244515930 | 0.12 |
ENST00000366537.1 ENST00000308105.4 |
C1orf100 |
chromosome 1 open reading frame 100 |
| chr5_-_87516448 | 0.12 |
ENST00000511218.1 |
TMEM161B |
transmembrane protein 161B |
| chr16_-_75498553 | 0.11 |
ENST00000569276.1 ENST00000357613.4 ENST00000561878.1 ENST00000566980.1 ENST00000567194.1 |
TMEM170A RP11-77K12.1 |
transmembrane protein 170A Uncharacterized protein |
| chr21_-_15583165 | 0.11 |
ENST00000536861.1 |
LIPI |
lipase, member I |
| chr2_+_234668894 | 0.11 |
ENST00000305208.5 ENST00000608383.1 ENST00000360418.3 |
UGT1A8 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr2_+_113885138 | 0.11 |
ENST00000409930.3 |
IL1RN |
interleukin 1 receptor antagonist |
| chr2_-_71062938 | 0.11 |
ENST00000410009.3 |
CD207 |
CD207 molecule, langerin |
| chr6_+_131958436 | 0.11 |
ENST00000357639.3 ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3 |
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr13_+_28813645 | 0.10 |
ENST00000282391.5 |
PAN3 |
PAN3 poly(A) specific ribonuclease subunit homolog (S. cerevisiae) |
| chr6_+_111408698 | 0.10 |
ENST00000368851.5 |
SLC16A10 |
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr1_-_226926864 | 0.10 |
ENST00000429204.1 ENST00000366784.1 |
ITPKB |
inositol-trisphosphate 3-kinase B |
| chr19_+_4402659 | 0.10 |
ENST00000301280.5 ENST00000585854.1 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
| chr15_-_22448819 | 0.10 |
ENST00000604066.1 |
IGHV1OR15-1 |
immunoglobulin heavy variable 1/OR15-1 (non-functional) |
| chr6_+_25279651 | 0.10 |
ENST00000329474.6 |
LRRC16A |
leucine rich repeat containing 16A |
| chr6_+_121756809 | 0.10 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr15_+_40886199 | 0.10 |
ENST00000346991.5 ENST00000528975.1 ENST00000527044.1 |
CASC5 |
cancer susceptibility candidate 5 |
| chr15_+_52155001 | 0.09 |
ENST00000544199.1 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chr9_-_85678043 | 0.09 |
ENST00000376447.3 ENST00000340717.4 |
RASEF |
RAS and EF-hand domain containing |
| chr2_+_27440229 | 0.09 |
ENST00000264705.4 ENST00000403525.1 |
CAD |
carbamoyl-phosphate synthetase 2, aspartate transcarbamylase, and dihydroorotase |
| chr3_-_196911002 | 0.09 |
ENST00000452595.1 |
DLG1 |
discs, large homolog 1 (Drosophila) |
| chr10_-_24770632 | 0.09 |
ENST00000596413.1 |
AL353583.1 |
AL353583.1 |
| chr1_+_248201474 | 0.09 |
ENST00000366479.2 |
OR2L2 |
olfactory receptor, family 2, subfamily L, member 2 |
| chr12_-_71148357 | 0.09 |
ENST00000378778.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr17_-_10452929 | 0.09 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr12_-_71148413 | 0.09 |
ENST00000440835.2 ENST00000549308.1 ENST00000550661.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chrX_+_41548259 | 0.09 |
ENST00000378138.5 |
GPR34 |
G protein-coupled receptor 34 |
| chr18_-_53069419 | 0.09 |
ENST00000570177.2 |
TCF4 |
transcription factor 4 |
| chr4_+_155484155 | 0.08 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr4_+_88896819 | 0.08 |
ENST00000237623.7 ENST00000395080.3 ENST00000508233.1 ENST00000360804.4 |
SPP1 |
secreted phosphoprotein 1 |
| chr4_-_90759440 | 0.08 |
ENST00000336904.3 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr5_+_147582387 | 0.08 |
ENST00000325630.2 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr3_+_15045419 | 0.08 |
ENST00000406272.2 |
NR2C2 |
nuclear receptor subfamily 2, group C, member 2 |
| chr6_+_42584847 | 0.08 |
ENST00000372883.3 |
UBR2 |
ubiquitin protein ligase E3 component n-recognin 2 |
| chr17_-_38821373 | 0.08 |
ENST00000394052.3 |
KRT222 |
keratin 222 |
| chr10_-_73848086 | 0.08 |
ENST00000536168.1 |
SPOCK2 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 2 |
| chr8_+_1993173 | 0.08 |
ENST00000523438.1 |
MYOM2 |
myomesin 2 |
| chr17_-_29624343 | 0.07 |
ENST00000247271.4 |
OMG |
oligodendrocyte myelin glycoprotein |
| chrX_+_65384182 | 0.07 |
ENST00000441993.2 ENST00000419594.1 |
HEPH |
hephaestin |
| chr5_-_16738451 | 0.07 |
ENST00000274203.9 ENST00000515803.1 |
MYO10 |
myosin X |
| chr14_+_56584414 | 0.07 |
ENST00000559044.1 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
| chr12_-_18243119 | 0.07 |
ENST00000538724.1 ENST00000229002.2 |
RERGL |
RERG/RAS-like |
| chr7_-_25268104 | 0.07 |
ENST00000222674.2 |
NPVF |
neuropeptide VF precursor |
| chr1_-_161207953 | 0.07 |
ENST00000367982.4 |
NR1I3 |
nuclear receptor subfamily 1, group I, member 3 |
| chr4_+_70861647 | 0.07 |
ENST00000246895.4 ENST00000381060.2 |
STATH |
statherin |
| chr19_-_19626838 | 0.07 |
ENST00000360913.3 |
TSSK6 |
testis-specific serine kinase 6 |
| chr8_+_1993152 | 0.07 |
ENST00000262113.4 |
MYOM2 |
myomesin 2 |
| chr3_-_129612394 | 0.07 |
ENST00000505616.1 ENST00000426664.2 |
TMCC1 |
transmembrane and coiled-coil domain family 1 |
| chr9_-_104145795 | 0.07 |
ENST00000259407.2 |
BAAT |
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr1_-_23342340 | 0.07 |
ENST00000566855.1 |
C1orf234 |
chromosome 1 open reading frame 234 |
| chr2_+_197577841 | 0.07 |
ENST00000409270.1 |
CCDC150 |
coiled-coil domain containing 150 |
| chr4_+_56815102 | 0.07 |
ENST00000257287.4 |
CEP135 |
centrosomal protein 135kDa |
| chr16_-_15180257 | 0.07 |
ENST00000540462.1 |
RRN3 |
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chrX_+_65384052 | 0.07 |
ENST00000336279.5 ENST00000458621.1 |
HEPH |
hephaestin |
| chr4_+_95972822 | 0.07 |
ENST00000509540.1 ENST00000440890.2 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr9_+_134065506 | 0.07 |
ENST00000483497.2 |
NUP214 |
nucleoporin 214kDa |
| chr20_+_42984330 | 0.07 |
ENST00000316673.4 ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A |
hepatocyte nuclear factor 4, alpha |
| chr2_-_209010874 | 0.07 |
ENST00000260988.4 |
CRYGB |
crystallin, gamma B |
| chr19_-_3500635 | 0.07 |
ENST00000250937.3 |
DOHH |
deoxyhypusine hydroxylase/monooxygenase |
| chr9_+_130026756 | 0.07 |
ENST00000314904.5 ENST00000373387.4 |
GARNL3 |
GTPase activating Rap/RanGAP domain-like 3 |
| chr1_-_94147385 | 0.06 |
ENST00000260502.6 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr12_+_64173583 | 0.06 |
ENST00000261234.6 |
TMEM5 |
transmembrane protein 5 |
| chr12_+_75728419 | 0.06 |
ENST00000378695.4 ENST00000312442.2 |
GLIPR1L1 |
GLI pathogenesis-related 1 like 1 |
| chr2_-_40657397 | 0.06 |
ENST00000408028.2 ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr1_-_68962805 | 0.06 |
ENST00000370966.5 |
DEPDC1 |
DEP domain containing 1 |
| chr17_+_61086917 | 0.06 |
ENST00000424789.2 ENST00000389520.4 |
TANC2 |
tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| chr13_-_47012325 | 0.06 |
ENST00000409879.2 |
KIAA0226L |
KIAA0226-like |
| chr4_-_89951028 | 0.06 |
ENST00000506913.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr4_-_123377880 | 0.06 |
ENST00000226730.4 |
IL2 |
interleukin 2 |
| chr6_+_44215603 | 0.06 |
ENST00000371554.1 |
HSP90AB1 |
heat shock protein 90kDa alpha (cytosolic), class B member 1 |
| chr15_-_58571445 | 0.06 |
ENST00000558231.1 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr17_-_10560619 | 0.06 |
ENST00000583535.1 |
MYH3 |
myosin, heavy chain 3, skeletal muscle, embryonic |
| chr9_-_95640218 | 0.06 |
ENST00000395506.3 ENST00000375495.3 ENST00000332591.6 |
ZNF484 |
zinc finger protein 484 |
| chr11_-_118972575 | 0.06 |
ENST00000432443.2 |
DPAGT1 |
dolichyl-phosphate (UDP-N-acetylglucosamine) N-acetylglucosaminephosphotransferase 1 (GlcNAc-1-P transferase) |
| chr4_+_88529681 | 0.05 |
ENST00000399271.1 |
DSPP |
dentin sialophosphoprotein |
| chr17_+_7211656 | 0.05 |
ENST00000416016.2 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr6_-_10115007 | 0.05 |
ENST00000485268.1 |
OFCC1 |
orofacial cleft 1 candidate 1 |
| chr10_+_102106829 | 0.05 |
ENST00000370355.2 |
SCD |
stearoyl-CoA desaturase (delta-9-desaturase) |
| chr7_-_50633078 | 0.05 |
ENST00000444124.2 |
DDC |
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chr20_+_30697298 | 0.05 |
ENST00000398022.2 |
TM9SF4 |
transmembrane 9 superfamily protein member 4 |
| chr4_+_71337834 | 0.05 |
ENST00000304887.5 |
MUC7 |
mucin 7, secreted |
| chr3_-_123710893 | 0.05 |
ENST00000467907.1 ENST00000459660.1 ENST00000495093.1 ENST00000460743.1 ENST00000405845.3 ENST00000484329.1 ENST00000479867.1 ENST00000496145.1 |
ROPN1 |
rhophilin associated tail protein 1 |
| chr9_+_125132803 | 0.05 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr9_+_77112244 | 0.05 |
ENST00000376896.3 |
RORB |
RAR-related orphan receptor B |
| chr12_-_24102576 | 0.05 |
ENST00000537393.1 ENST00000309359.1 ENST00000381381.2 ENST00000451604.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr8_+_145215928 | 0.05 |
ENST00000528919.1 |
MROH1 |
maestro heat-like repeat family member 1 |
| chr3_-_139396853 | 0.05 |
ENST00000406164.1 ENST00000406824.1 |
NMNAT3 |
nicotinamide nucleotide adenylyltransferase 3 |
| chr2_+_32390925 | 0.05 |
ENST00000440718.1 ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6 |
solute carrier family 30 (zinc transporter), member 6 |
| chr10_-_103599591 | 0.05 |
ENST00000348850.5 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr17_-_39324424 | 0.05 |
ENST00000391356.2 |
KRTAP4-3 |
keratin associated protein 4-3 |
| chr15_+_66797455 | 0.05 |
ENST00000446801.2 |
ZWILCH |
zwilch kinetochore protein |
| chr20_-_50384864 | 0.05 |
ENST00000311637.5 ENST00000402822.1 |
ATP9A |
ATPase, class II, type 9A |
| chrX_-_15619076 | 0.05 |
ENST00000252519.3 |
ACE2 |
angiotensin I converting enzyme 2 |
| chr13_-_47471155 | 0.05 |
ENST00000543956.1 ENST00000542664.1 |
HTR2A |
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
| chr12_+_12510352 | 0.05 |
ENST00000298571.6 |
LOH12CR1 |
loss of heterozygosity, 12, chromosomal region 1 |
| chr13_-_46626847 | 0.05 |
ENST00000242848.4 ENST00000282007.3 |
ZC3H13 |
zinc finger CCCH-type containing 13 |
| chr14_+_22675388 | 0.04 |
ENST00000390461.2 |
TRAV34 |
T cell receptor alpha variable 34 |
| chr19_+_1440838 | 0.04 |
ENST00000594262.1 |
AC027307.3 |
Uncharacterized protein |
| chr4_+_186317133 | 0.04 |
ENST00000507753.1 |
ANKRD37 |
ankyrin repeat domain 37 |
| chr10_+_115614370 | 0.04 |
ENST00000369301.3 |
NHLRC2 |
NHL repeat containing 2 |
| chr2_+_28718921 | 0.04 |
ENST00000327757.5 ENST00000422425.2 ENST00000404858.1 |
PLB1 |
phospholipase B1 |
| chr14_+_31046959 | 0.04 |
ENST00000547532.1 ENST00000555429.1 |
G2E3 |
G2/M-phase specific E3 ubiquitin protein ligase |
| chr4_-_110723194 | 0.04 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr1_-_86848760 | 0.04 |
ENST00000460698.2 |
ODF2L |
outer dense fiber of sperm tails 2-like |
| chr3_-_87325728 | 0.04 |
ENST00000350375.2 |
POU1F1 |
POU class 1 homeobox 1 |
| chr3_-_139396801 | 0.04 |
ENST00000413939.2 ENST00000339837.5 ENST00000512391.1 |
NMNAT3 |
nicotinamide nucleotide adenylyltransferase 3 |
| chrY_-_24038660 | 0.04 |
ENST00000382677.3 |
RBMY1D |
RNA binding motif protein, Y-linked, family 1, member D |
| chr11_+_61717279 | 0.04 |
ENST00000378043.4 |
BEST1 |
bestrophin 1 |
| chrY_+_23698778 | 0.04 |
ENST00000303902.5 |
RBMY1A1 |
RNA binding motif protein, Y-linked, family 1, member A1 |
| chr10_-_50396425 | 0.04 |
ENST00000374148.1 |
C10orf128 |
chromosome 10 open reading frame 128 |
| chr4_+_68424434 | 0.04 |
ENST00000265404.2 ENST00000396225.1 |
STAP1 |
signal transducing adaptor family member 1 |
| chr15_+_66797627 | 0.04 |
ENST00000565627.1 ENST00000564179.1 |
ZWILCH |
zwilch kinetochore protein |
| chr3_-_58196688 | 0.04 |
ENST00000486455.1 |
DNASE1L3 |
deoxyribonuclease I-like 3 |
| chr1_-_17766198 | 0.04 |
ENST00000375436.4 |
RCC2 |
regulator of chromosome condensation 2 |
| chr2_-_231989808 | 0.04 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr8_+_107738343 | 0.04 |
ENST00000521592.1 |
OXR1 |
oxidation resistance 1 |
| chr1_-_54355430 | 0.04 |
ENST00000371399.1 ENST00000072644.1 ENST00000412288.1 |
YIPF1 |
Yip1 domain family, member 1 |
| chr12_-_95945246 | 0.04 |
ENST00000258499.3 |
USP44 |
ubiquitin specific peptidase 44 |
| chr2_+_173686303 | 0.04 |
ENST00000397087.3 |
RAPGEF4 |
Rap guanine nucleotide exchange factor (GEF) 4 |
| chr21_+_34398153 | 0.04 |
ENST00000382357.3 ENST00000430860.1 ENST00000333337.3 |
OLIG2 |
oligodendrocyte lineage transcription factor 2 |
| chr3_-_12200851 | 0.04 |
ENST00000287814.4 |
TIMP4 |
TIMP metallopeptidase inhibitor 4 |
| chr15_+_40886439 | 0.04 |
ENST00000532056.1 ENST00000399668.2 |
CASC5 |
cancer susceptibility candidate 5 |
| chr4_-_100356551 | 0.04 |
ENST00000209665.4 |
ADH7 |
alcohol dehydrogenase 7 (class IV), mu or sigma polypeptide |
| chr12_-_57146095 | 0.04 |
ENST00000550770.1 ENST00000338193.6 |
PRIM1 |
primase, DNA, polypeptide 1 (49kDa) |
| chr8_+_128426535 | 0.04 |
ENST00000465342.2 |
POU5F1B |
POU class 5 homeobox 1B |
| chr11_+_61717336 | 0.04 |
ENST00000378042.3 |
BEST1 |
bestrophin 1 |
| chr19_-_29704448 | 0.04 |
ENST00000304863.4 |
UQCRFS1 |
ubiquinol-cytochrome c reductase, Rieske iron-sulfur polypeptide 1 |
| chr3_+_149191723 | 0.03 |
ENST00000305354.4 |
TM4SF4 |
transmembrane 4 L six family member 4 |
| chr1_-_161207986 | 0.03 |
ENST00000506209.1 ENST00000367980.2 |
NR1I3 |
nuclear receptor subfamily 1, group I, member 3 |
| chr6_+_26156551 | 0.03 |
ENST00000304218.3 |
HIST1H1E |
histone cluster 1, H1e |
| chr11_-_5345582 | 0.03 |
ENST00000328813.2 |
OR51B2 |
olfactory receptor, family 51, subfamily B, member 2 |
| chr17_-_73663245 | 0.03 |
ENST00000584999.1 ENST00000317905.5 ENST00000420326.2 ENST00000340830.5 |
RECQL5 |
RecQ protein-like 5 |
| chr4_+_86525299 | 0.03 |
ENST00000512201.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr14_-_106453155 | 0.03 |
ENST00000390594.2 |
IGHV1-2 |
immunoglobulin heavy variable 1-2 |
| chr4_+_155484103 | 0.03 |
ENST00000302068.4 |
FGB |
fibrinogen beta chain |
| chr12_-_118628350 | 0.03 |
ENST00000537952.1 ENST00000537822.1 |
TAOK3 |
TAO kinase 3 |
| chr13_-_79979919 | 0.03 |
ENST00000267229.7 |
RBM26 |
RNA binding motif protein 26 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.2 | 0.6 | GO:0060488 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.5 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.3 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.9 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.1 | 0.3 | GO:0046098 | guanine metabolic process(GO:0046098) |
| 0.0 | 0.2 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.3 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.2 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.0 | 0.2 | GO:0008616 | queuosine biosynthetic process(GO:0008616) queuosine metabolic process(GO:0046116) |
| 0.0 | 0.3 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.1 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.2 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.1 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.1 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0070980 | biphenyl catabolic process(GO:0070980) |
| 0.0 | 0.1 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:1903964 | monounsaturated fatty acid metabolic process(GO:1903964) monounsaturated fatty acid biosynthetic process(GO:1903966) |
| 0.0 | 0.1 | GO:0033076 | isoquinoline alkaloid metabolic process(GO:0033076) phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0051620 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.1 | GO:0043950 | positive regulation of cAMP-mediated signaling(GO:0043950) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0019355 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.2 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.1 | GO:1903660 | negative regulation of complement-dependent cytotoxicity(GO:1903660) |
| 0.0 | 0.0 | GO:1903978 | negative regulation of macrophage chemotaxis(GO:0010760) negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) regulation of microglial cell activation(GO:1903978) |
| 0.0 | 0.1 | GO:0001550 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.0 | GO:0021529 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.1 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.0 | GO:0010430 | fatty acid omega-oxidation(GO:0010430) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.9 | GO:0018149 | peptide cross-linking(GO:0018149) |
| 0.0 | 0.1 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.1 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.0 | 0.5 | GO:0051904 | melanosome transport(GO:0032402) pigment granule transport(GO:0051904) |
| 0.0 | 0.1 | GO:0008612 | peptidyl-lysine modification to peptidyl-hypusine(GO:0008612) |
| 0.0 | 0.0 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.1 | GO:0010606 | positive regulation of cytoplasmic mRNA processing body assembly(GO:0010606) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.6 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.4 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.0 | 0.1 | GO:0031251 | PAN complex(GO:0031251) |
| 0.0 | 0.2 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.1 | GO:1990423 | RZZ complex(GO:1990423) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.3 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.3 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0033695 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.3 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0005150 | interleukin-1, Type I receptor binding(GO:0005150) |
| 0.0 | 0.5 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0008508 | bile acid:sodium symporter activity(GO:0008508) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.4 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.5 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0004087 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.2 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0048248 | CXCR3 chemokine receptor binding(GO:0048248) |
| 0.0 | 0.2 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) vascular endothelial growth factor receptor 2 binding(GO:0043184) |
| 0.0 | 0.1 | GO:0001884 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) sulfonylurea receptor binding(GO:0017098) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0016167 | glial cell-derived neurotrophic factor receptor activity(GO:0016167) |
| 0.0 | 0.3 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:0070087 | chromo shadow domain binding(GO:0070087) |
| 0.0 | 0.3 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.1 | GO:0016623 | oxidoreductase activity, acting on the aldehyde or oxo group of donors, oxygen as acceptor(GO:0016623) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.2 | GO:0050321 | tau-protein kinase activity(GO:0050321) |
| 0.0 | 0.1 | GO:0004768 | stearoyl-CoA 9-desaturase activity(GO:0004768) acyl-CoA desaturase activity(GO:0016215) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.5 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME XENOBIOTICS | Genes involved in Xenobiotics |