Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
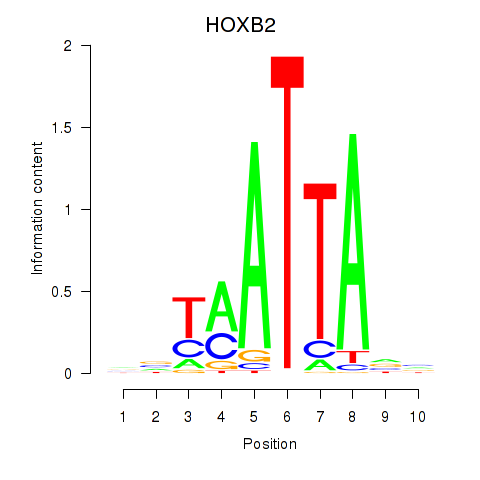
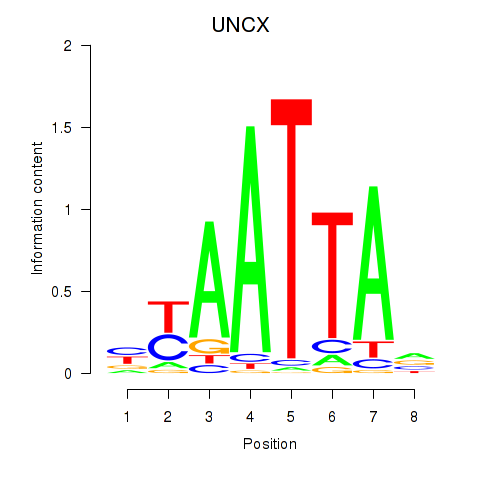
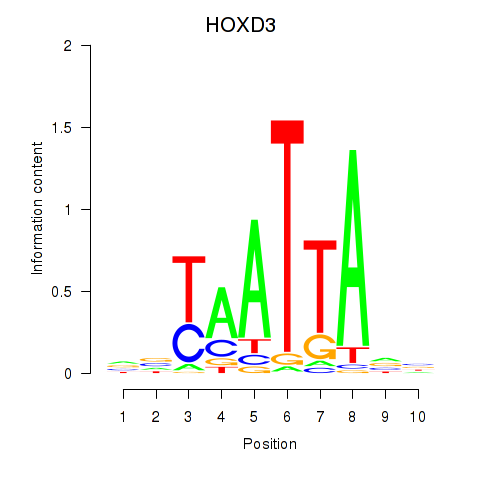
Results for HOXB2_UNCX_HOXD3
Z-value: 0.57
Transcription factors associated with HOXB2_UNCX_HOXD3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB2
|
ENSG00000173917.9 | HOXB2 |
|
UNCX
|
ENSG00000164853.8 | UNCX |
|
HOXD3
|
ENSG00000128652.7 | HOXD3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD3 | hg19_v2_chr2_+_177015122_177015122 | 0.12 | 7.8e-01 | Click! |
| HOXB2 | hg19_v2_chr17_-_46623441_46623441 | 0.08 | 8.5e-01 | Click! |
Activity profile of HOXB2_UNCX_HOXD3 motif
Sorted Z-values of HOXB2_UNCX_HOXD3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB2_UNCX_HOXD3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr9_+_12693336 | 1.07 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr3_-_151034734 | 0.86 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr18_+_29027696 | 0.77 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr19_-_51522955 | 0.73 |
ENST00000358789.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr4_+_40198527 | 0.70 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr8_-_42234745 | 0.60 |
ENST00000220812.2 |
DKK4 |
dickkopf WNT signaling pathway inhibitor 4 |
| chr1_+_62439037 | 0.56 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr12_+_107712173 | 0.50 |
ENST00000280758.5 ENST00000420571.2 |
BTBD11 |
BTB (POZ) domain containing 11 |
| chr1_-_242612779 | 0.48 |
ENST00000427495.1 |
PLD5 |
phospholipase D family, member 5 |
| chr1_+_160370344 | 0.46 |
ENST00000368061.2 |
VANGL2 |
VANGL planar cell polarity protein 2 |
| chr1_+_209602771 | 0.45 |
ENST00000440276.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr8_+_94241867 | 0.42 |
ENST00000598428.1 |
AC016885.1 |
Uncharacterized protein |
| chr5_-_82969405 | 0.42 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr18_+_61554932 | 0.40 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr19_-_36001113 | 0.39 |
ENST00000434389.1 |
DMKN |
dermokine |
| chr2_+_68961905 | 0.38 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr7_-_25268104 | 0.38 |
ENST00000222674.2 |
NPVF |
neuropeptide VF precursor |
| chr5_+_66300446 | 0.37 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr2_+_68961934 | 0.34 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr12_-_28123206 | 0.34 |
ENST00000542963.1 ENST00000535992.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr12_-_28122980 | 0.33 |
ENST00000395868.3 ENST00000534890.1 |
PTHLH |
parathyroid hormone-like hormone |
| chr9_-_5304432 | 0.33 |
ENST00000416837.1 ENST00000308420.3 |
RLN2 |
relaxin 2 |
| chr6_+_130339710 | 0.31 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr15_+_101420028 | 0.31 |
ENST00000557963.1 ENST00000346623.6 |
ALDH1A3 |
aldehyde dehydrogenase 1 family, member A3 |
| chr10_+_24497704 | 0.30 |
ENST00000376456.4 ENST00000458595.1 |
KIAA1217 |
KIAA1217 |
| chr5_+_140227048 | 0.27 |
ENST00000532602.1 |
PCDHA9 |
protocadherin alpha 9 |
| chr6_-_136847610 | 0.27 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr2_-_70780770 | 0.26 |
ENST00000444975.1 ENST00000445399.1 ENST00000418333.2 |
TGFA |
transforming growth factor, alpha |
| chr2_+_191045656 | 0.26 |
ENST00000443551.2 |
C2orf88 |
chromosome 2 open reading frame 88 |
| chr10_+_6779326 | 0.26 |
ENST00000417112.1 |
RP11-554I8.2 |
RP11-554I8.2 |
| chr17_-_54893250 | 0.26 |
ENST00000397862.2 |
C17orf67 |
chromosome 17 open reading frame 67 |
| chr21_-_42219065 | 0.25 |
ENST00000400454.1 |
DSCAM |
Down syndrome cell adhesion molecule |
| chr6_-_32157947 | 0.25 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
| chr9_+_71986182 | 0.24 |
ENST00000303068.7 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr14_+_29236269 | 0.24 |
ENST00000313071.4 |
FOXG1 |
forkhead box G1 |
| chr5_+_36608422 | 0.23 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr2_+_68962014 | 0.23 |
ENST00000467265.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr20_-_46415297 | 0.22 |
ENST00000467815.1 ENST00000359930.4 |
SULF2 |
sulfatase 2 |
| chr20_-_46415341 | 0.22 |
ENST00000484875.1 ENST00000361612.4 |
SULF2 |
sulfatase 2 |
| chr12_-_25055177 | 0.22 |
ENST00000538118.1 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr18_-_61329118 | 0.22 |
ENST00000332821.8 ENST00000283752.5 |
SERPINB3 |
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr9_+_125281420 | 0.21 |
ENST00000340750.1 |
OR1J4 |
olfactory receptor, family 1, subfamily J, member 4 |
| chr10_-_105845674 | 0.21 |
ENST00000353479.5 ENST00000369733.3 |
COL17A1 |
collagen, type XVII, alpha 1 |
| chr10_+_118187379 | 0.21 |
ENST00000369230.3 |
PNLIPRP3 |
pancreatic lipase-related protein 3 |
| chr12_+_20963647 | 0.20 |
ENST00000381545.3 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr16_+_12059050 | 0.20 |
ENST00000396495.3 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr19_-_10697895 | 0.20 |
ENST00000591240.1 ENST00000589684.1 ENST00000591676.1 ENST00000250244.6 ENST00000590923.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr11_+_55578854 | 0.20 |
ENST00000333973.2 |
OR5L1 |
olfactory receptor, family 5, subfamily L, member 1 |
| chr3_+_111717600 | 0.20 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr4_-_153601136 | 0.20 |
ENST00000504064.1 ENST00000304385.3 |
TMEM154 |
transmembrane protein 154 |
| chr3_+_189349162 | 0.19 |
ENST00000264731.3 ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63 |
tumor protein p63 |
| chr12_+_20963632 | 0.19 |
ENST00000540853.1 ENST00000261196.2 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr4_-_143227088 | 0.19 |
ENST00000511838.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr12_-_22063787 | 0.19 |
ENST00000544039.1 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr3_+_111717511 | 0.18 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr3_-_112564797 | 0.18 |
ENST00000398214.1 ENST00000448932.1 |
CD200R1L |
CD200 receptor 1-like |
| chr5_-_150948414 | 0.18 |
ENST00000261800.5 |
FAT2 |
FAT atypical cadherin 2 |
| chr8_+_101170563 | 0.18 |
ENST00000520508.1 ENST00000388798.2 |
SPAG1 |
sperm associated antigen 1 |
| chr12_-_52867569 | 0.18 |
ENST00000252250.6 |
KRT6C |
keratin 6C |
| chr3_+_111718173 | 0.18 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr3_-_191000172 | 0.18 |
ENST00000427544.2 |
UTS2B |
urotensin 2B |
| chr18_+_61254534 | 0.17 |
ENST00000269489.5 |
SERPINB13 |
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr11_-_124190184 | 0.17 |
ENST00000357438.2 |
OR8D2 |
olfactory receptor, family 8, subfamily D, member 2 |
| chr4_-_143226979 | 0.17 |
ENST00000514525.1 |
INPP4B |
inositol polyphosphate-4-phosphatase, type II, 105kDa |
| chr13_-_86373536 | 0.17 |
ENST00000400286.2 |
SLITRK6 |
SLIT and NTRK-like family, member 6 |
| chr4_-_74486217 | 0.17 |
ENST00000335049.5 ENST00000307439.5 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr6_+_150920999 | 0.17 |
ENST00000367328.1 ENST00000367326.1 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr5_+_89770664 | 0.17 |
ENST00000503973.1 ENST00000399107.1 |
POLR3G |
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr11_+_128563652 | 0.17 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr12_-_39734783 | 0.17 |
ENST00000552961.1 |
KIF21A |
kinesin family member 21A |
| chr21_+_25801041 | 0.16 |
ENST00000453784.2 ENST00000423581.1 |
AP000476.1 |
AP000476.1 |
| chr6_+_125540951 | 0.16 |
ENST00000524679.1 |
TPD52L1 |
tumor protein D52-like 1 |
| chr1_+_70876926 | 0.16 |
ENST00000370938.3 ENST00000346806.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr14_-_104181771 | 0.16 |
ENST00000554913.1 ENST00000554974.1 ENST00000553361.1 ENST00000555055.1 ENST00000555964.1 ENST00000556682.1 ENST00000445556.1 ENST00000553332.1 ENST00000352127.7 |
XRCC3 |
X-ray repair complementing defective repair in Chinese hamster cells 3 |
| chr5_+_89770696 | 0.16 |
ENST00000504930.1 ENST00000514483.1 |
POLR3G |
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr13_-_46716969 | 0.16 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr12_+_4385230 | 0.16 |
ENST00000536537.1 |
CCND2 |
cyclin D2 |
| chr4_+_69313145 | 0.16 |
ENST00000305363.4 |
TMPRSS11E |
transmembrane protease, serine 11E |
| chr7_-_22234381 | 0.16 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr12_+_81110684 | 0.16 |
ENST00000228644.3 |
MYF5 |
myogenic factor 5 |
| chr11_-_101000445 | 0.16 |
ENST00000534013.1 |
PGR |
progesterone receptor |
| chr2_-_113594279 | 0.16 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr17_-_38956205 | 0.16 |
ENST00000306658.7 |
KRT28 |
keratin 28 |
| chr19_-_51523412 | 0.16 |
ENST00000391805.1 ENST00000599077.1 |
KLK10 |
kallikrein-related peptidase 10 |
| chr4_-_87028478 | 0.15 |
ENST00000515400.1 ENST00000395157.3 |
MAPK10 |
mitogen-activated protein kinase 10 |
| chr4_+_77356248 | 0.15 |
ENST00000296043.6 |
SHROOM3 |
shroom family member 3 |
| chr1_+_152881014 | 0.15 |
ENST00000368764.3 ENST00000392667.2 |
IVL |
involucrin |
| chr18_-_40857493 | 0.15 |
ENST00000255224.3 |
SYT4 |
synaptotagmin IV |
| chr1_+_28261621 | 0.15 |
ENST00000549094.1 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr6_-_136788001 | 0.15 |
ENST00000544465.1 |
MAP7 |
microtubule-associated protein 7 |
| chrX_-_24690771 | 0.15 |
ENST00000379145.1 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr14_-_23652849 | 0.15 |
ENST00000316902.7 ENST00000469263.1 ENST00000525062.1 ENST00000524758.1 |
SLC7A8 |
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr19_-_51523275 | 0.15 |
ENST00000309958.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr8_-_57026541 | 0.15 |
ENST00000311923.1 |
MOS |
v-mos Moloney murine sarcoma viral oncogene homolog |
| chr8_-_41166953 | 0.15 |
ENST00000220772.3 |
SFRP1 |
secreted frizzled-related protein 1 |
| chr14_+_61654271 | 0.15 |
ENST00000555185.1 ENST00000557294.1 ENST00000556778.1 |
PRKCH |
protein kinase C, eta |
| chr4_+_110749143 | 0.14 |
ENST00000317735.4 |
RRH |
retinal pigment epithelium-derived rhodopsin homolog |
| chr6_+_131571535 | 0.14 |
ENST00000474850.2 |
AKAP7 |
A kinase (PRKA) anchor protein 7 |
| chr6_+_111408698 | 0.14 |
ENST00000368851.5 |
SLC16A10 |
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr6_-_32908792 | 0.14 |
ENST00000418107.2 |
HLA-DMB |
major histocompatibility complex, class II, DM beta |
| chr4_+_71458012 | 0.14 |
ENST00000449493.2 |
AMBN |
ameloblastin (enamel matrix protein) |
| chr3_-_190167571 | 0.14 |
ENST00000354905.2 |
TMEM207 |
transmembrane protein 207 |
| chr4_-_89951028 | 0.14 |
ENST00000506913.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr20_-_29978383 | 0.14 |
ENST00000339144.3 ENST00000376321.3 |
DEFB119 |
defensin, beta 119 |
| chr16_+_76587314 | 0.14 |
ENST00000563764.1 |
RP11-58C22.1 |
Uncharacterized protein |
| chr12_+_41831485 | 0.14 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr3_-_143567262 | 0.14 |
ENST00000474151.1 ENST00000316549.6 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr3_+_98699880 | 0.14 |
ENST00000473756.1 |
LINC00973 |
long intergenic non-protein coding RNA 973 |
| chr6_+_134758827 | 0.14 |
ENST00000431422.1 |
LINC01010 |
long intergenic non-protein coding RNA 1010 |
| chr18_-_19994830 | 0.14 |
ENST00000525417.1 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
| chr18_+_61254570 | 0.13 |
ENST00000344731.5 |
SERPINB13 |
serpin peptidase inhibitor, clade B (ovalbumin), member 13 |
| chr6_-_136847099 | 0.13 |
ENST00000438100.2 |
MAP7 |
microtubule-associated protein 7 |
| chr1_-_24469602 | 0.13 |
ENST00000270800.1 |
IL22RA1 |
interleukin 22 receptor, alpha 1 |
| chr12_-_12715266 | 0.13 |
ENST00000228862.2 |
DUSP16 |
dual specificity phosphatase 16 |
| chr6_+_151042224 | 0.13 |
ENST00000358517.2 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr15_+_58430567 | 0.13 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr6_-_11779174 | 0.13 |
ENST00000379413.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr17_-_39306054 | 0.13 |
ENST00000343246.4 |
KRTAP4-5 |
keratin associated protein 4-5 |
| chr5_+_147582387 | 0.13 |
ENST00000325630.2 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr5_-_135290705 | 0.13 |
ENST00000274507.1 |
LECT2 |
leukocyte cell-derived chemotaxin 2 |
| chr12_+_44229846 | 0.13 |
ENST00000551577.1 ENST00000266534.3 |
TMEM117 |
transmembrane protein 117 |
| chr10_+_24738355 | 0.13 |
ENST00000307544.6 |
KIAA1217 |
KIAA1217 |
| chr3_-_190040223 | 0.13 |
ENST00000295522.3 |
CLDN1 |
claudin 1 |
| chr1_+_156254070 | 0.13 |
ENST00000405535.2 ENST00000456810.1 |
TMEM79 |
transmembrane protein 79 |
| chr3_-_197300194 | 0.13 |
ENST00000358186.2 ENST00000431056.1 |
BDH1 |
3-hydroxybutyrate dehydrogenase, type 1 |
| chr18_-_44181442 | 0.13 |
ENST00000398722.4 |
LOXHD1 |
lipoxygenase homology domains 1 |
| chr2_-_89327228 | 0.13 |
ENST00000483158.1 |
IGKV3-11 |
immunoglobulin kappa variable 3-11 |
| chr11_-_118023490 | 0.13 |
ENST00000324727.4 |
SCN4B |
sodium channel, voltage-gated, type IV, beta subunit |
| chr6_-_25874440 | 0.13 |
ENST00000361703.6 ENST00000397060.4 |
SLC17A3 |
solute carrier family 17 (organic anion transporter), member 3 |
| chr9_+_71944241 | 0.12 |
ENST00000257515.8 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr3_-_33686743 | 0.12 |
ENST00000333778.6 ENST00000539981.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr8_+_105235572 | 0.12 |
ENST00000523362.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr10_+_695888 | 0.12 |
ENST00000441152.2 |
PRR26 |
proline rich 26 |
| chr17_-_5321549 | 0.12 |
ENST00000572809.1 |
NUP88 |
nucleoporin 88kDa |
| chr2_+_207804278 | 0.12 |
ENST00000272852.3 |
CPO |
carboxypeptidase O |
| chr10_+_47746929 | 0.12 |
ENST00000340243.6 ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2 AL603965.1 |
annexin A8-like 2 Protein LOC100996760 |
| chr14_-_72458326 | 0.12 |
ENST00000542853.1 |
AC005477.1 |
AC005477.1 |
| chr6_-_132967142 | 0.12 |
ENST00000275216.1 |
TAAR1 |
trace amine associated receptor 1 |
| chr3_-_33686925 | 0.12 |
ENST00000485378.2 ENST00000313350.6 ENST00000487200.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr17_+_55173933 | 0.12 |
ENST00000539273.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr1_-_94586651 | 0.12 |
ENST00000535735.1 ENST00000370225.3 |
ABCA4 |
ATP-binding cassette, sub-family A (ABC1), member 4 |
| chr5_-_1882858 | 0.12 |
ENST00000511126.1 ENST00000231357.2 |
IRX4 |
iroquois homeobox 4 |
| chr9_+_72002837 | 0.12 |
ENST00000377216.3 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr14_+_56584414 | 0.12 |
ENST00000559044.1 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
| chr6_-_32806506 | 0.12 |
ENST00000374897.2 ENST00000452392.2 |
TAP2 TAP2 |
transporter 2, ATP-binding cassette, sub-family B (MDR/TAP) Uncharacterized protein |
| chr1_+_186798073 | 0.12 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr11_-_102668879 | 0.12 |
ENST00000315274.6 |
MMP1 |
matrix metallopeptidase 1 (interstitial collagenase) |
| chr2_-_136678123 | 0.12 |
ENST00000422708.1 |
DARS |
aspartyl-tRNA synthetase |
| chr17_-_9694614 | 0.11 |
ENST00000330255.5 ENST00000571134.1 |
DHRS7C |
dehydrogenase/reductase (SDR family) member 7C |
| chr7_+_107224364 | 0.11 |
ENST00000491150.1 |
BCAP29 |
B-cell receptor-associated protein 29 |
| chr19_-_51456198 | 0.11 |
ENST00000594846.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr7_+_18535786 | 0.11 |
ENST00000406072.1 |
HDAC9 |
histone deacetylase 9 |
| chr18_-_61311485 | 0.11 |
ENST00000436264.1 ENST00000356424.6 ENST00000341074.5 |
SERPINB4 |
serpin peptidase inhibitor, clade B (ovalbumin), member 4 |
| chr3_+_124223586 | 0.11 |
ENST00000393496.1 |
KALRN |
kalirin, RhoGEF kinase |
| chrX_+_138612889 | 0.11 |
ENST00000218099.2 ENST00000394090.2 |
F9 |
coagulation factor IX |
| chrX_+_105445717 | 0.11 |
ENST00000372552.1 |
MUM1L1 |
melanoma associated antigen (mutated) 1-like 1 |
| chr17_-_4458616 | 0.11 |
ENST00000381556.2 |
MYBBP1A |
MYB binding protein (P160) 1a |
| chr12_-_95510743 | 0.11 |
ENST00000551521.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr19_+_7030589 | 0.11 |
ENST00000329753.5 |
MBD3L5 |
methyl-CpG binding domain protein 3-like 5 |
| chr12_-_10978957 | 0.11 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr11_-_72070206 | 0.11 |
ENST00000544382.1 |
CLPB |
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr9_+_125376948 | 0.11 |
ENST00000297913.2 |
OR1Q1 |
olfactory receptor, family 1, subfamily Q, member 1 |
| chr8_-_86253888 | 0.11 |
ENST00000522389.1 ENST00000432364.2 ENST00000517618.1 |
CA1 |
carbonic anhydrase I |
| chr8_-_125577940 | 0.11 |
ENST00000519168.1 ENST00000395508.2 |
MTSS1 |
metastasis suppressor 1 |
| chrX_-_84634737 | 0.11 |
ENST00000262753.4 |
POF1B |
premature ovarian failure, 1B |
| chr17_-_39274606 | 0.11 |
ENST00000391413.2 |
KRTAP4-11 |
keratin associated protein 4-11 |
| chr7_-_139763521 | 0.11 |
ENST00000263549.3 |
PARP12 |
poly (ADP-ribose) polymerase family, member 12 |
| chr7_-_22233442 | 0.11 |
ENST00000401957.2 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr2_-_216003127 | 0.11 |
ENST00000412081.1 ENST00000272895.7 |
ABCA12 |
ATP-binding cassette, sub-family A (ABC1), member 12 |
| chr18_+_28956740 | 0.11 |
ENST00000308128.4 ENST00000359747.4 |
DSG4 |
desmoglein 4 |
| chr1_-_183560011 | 0.11 |
ENST00000367536.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr1_-_190446759 | 0.11 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr4_-_74486347 | 0.11 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr1_+_70876891 | 0.10 |
ENST00000411986.2 |
CTH |
cystathionase (cystathionine gamma-lyase) |
| chr10_+_32873190 | 0.10 |
ENST00000375025.4 |
C10orf68 |
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr19_-_7040190 | 0.10 |
ENST00000381394.4 |
MBD3L4 |
methyl-CpG binding domain protein 3-like 4 |
| chr11_+_121447469 | 0.10 |
ENST00000532694.1 ENST00000534286.1 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr10_-_50396425 | 0.10 |
ENST00000374148.1 |
C10orf128 |
chromosome 10 open reading frame 128 |
| chr2_-_70780572 | 0.10 |
ENST00000450929.1 |
TGFA |
transforming growth factor, alpha |
| chr12_-_15103621 | 0.10 |
ENST00000536592.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr19_-_33360647 | 0.10 |
ENST00000590341.1 ENST00000587772.1 ENST00000023064.4 |
SLC7A9 |
solute carrier family 7 (amino acid transporter light chain, bo,+ system), member 9 |
| chr3_+_189507523 | 0.10 |
ENST00000437221.1 ENST00000392463.2 ENST00000392461.3 ENST00000449992.1 ENST00000456148.1 |
TP63 |
tumor protein p63 |
| chr3_+_111718036 | 0.10 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr4_-_76944621 | 0.10 |
ENST00000306602.1 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
| chr11_+_117947724 | 0.10 |
ENST00000534111.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr5_-_24645078 | 0.10 |
ENST00000264463.4 |
CDH10 |
cadherin 10, type 2 (T2-cadherin) |
| chr5_-_87516448 | 0.10 |
ENST00000511218.1 |
TMEM161B |
transmembrane protein 161B |
| chr17_-_39324424 | 0.10 |
ENST00000391356.2 |
KRTAP4-3 |
keratin associated protein 4-3 |
| chr7_+_142378566 | 0.10 |
ENST00000390398.3 |
TRBV25-1 |
T cell receptor beta variable 25-1 |
| chr7_+_73245193 | 0.10 |
ENST00000340958.2 |
CLDN4 |
claudin 4 |
| chr15_+_32907691 | 0.10 |
ENST00000361627.3 ENST00000567348.1 ENST00000563864.1 ENST00000543522.1 |
ARHGAP11A |
Rho GTPase activating protein 11A |
| chr12_-_22094336 | 0.10 |
ENST00000326684.4 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr11_+_112047087 | 0.10 |
ENST00000526088.1 ENST00000532593.1 ENST00000531169.1 |
BCO2 |
beta-carotene oxygenase 2 |
| chr16_-_66864806 | 0.10 |
ENST00000566336.1 ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1 |
NEDD8 activating enzyme E1 subunit 1 |
| chr18_+_71815743 | 0.10 |
ENST00000169551.6 ENST00000580087.1 |
TIMM21 |
translocase of inner mitochondrial membrane 21 homolog (yeast) |
| chr8_+_32579341 | 0.10 |
ENST00000519240.1 ENST00000539990.1 |
NRG1 |
neuregulin 1 |
| chr12_+_113354341 | 0.10 |
ENST00000553152.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr3_-_126327398 | 0.10 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chrX_+_149887090 | 0.10 |
ENST00000538506.1 |
MTMR1 |
myotubularin related protein 1 |
| chr3_-_69129501 | 0.10 |
ENST00000540295.1 ENST00000415609.2 ENST00000361055.4 ENST00000349511.4 |
UBA3 |
ubiquitin-like modifier activating enzyme 3 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.1 | 0.3 | GO:0060166 | olfactory pit development(GO:0060166) |
| 0.1 | 0.4 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.7 | GO:1900378 | positive regulation of melanin biosynthetic process(GO:0048023) positive regulation of secondary metabolite biosynthetic process(GO:1900378) |
| 0.1 | 0.6 | GO:0051799 | negative regulation of hair follicle development(GO:0051799) |
| 0.1 | 0.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.3 | GO:0018352 | protein-pyridoxal-5-phosphate linkage(GO:0018352) |
| 0.1 | 0.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.2 | GO:2001190 | positive regulation of T cell activation via T cell receptor contact with antigen bound to MHC molecule on antigen presenting cell(GO:2001190) |
| 0.1 | 0.1 | GO:0042270 | protection from natural killer cell mediated cytotoxicity(GO:0042270) |
| 0.1 | 0.2 | GO:0071139 | resolution of recombination intermediates(GO:0071139) resolution of mitotic recombination intermediates(GO:0071140) |
| 0.1 | 0.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.1 | 0.2 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.1 | 0.2 | GO:2000670 | positive regulation of dendritic cell apoptotic process(GO:2000670) |
| 0.1 | 0.2 | GO:0018153 | isopeptide cross-linking via N6-(L-isoglutamyl)-L-lysine(GO:0018153) isopeptide cross-linking(GO:0018262) |
| 0.0 | 0.1 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.0 | GO:0021828 | cerebral cortex tangential migration using cell-axon interactions(GO:0021824) gonadotrophin-releasing hormone neuronal migration to the hypothalamus(GO:0021828) hypothalamic tangential migration using cell-axon interactions(GO:0021856) facioacoustic ganglion development(GO:1903375) |
| 0.0 | 0.5 | GO:0032277 | negative regulation of gonadotropin secretion(GO:0032277) |
| 0.0 | 0.2 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:1903348 | positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.0 | 0.1 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) mesodermal to mesenchymal transition involved in gastrulation(GO:0060809) |
| 0.0 | 0.2 | GO:0060748 | tertiary branching involved in mammary gland duct morphogenesis(GO:0060748) |
| 0.0 | 0.1 | GO:0002476 | antigen processing and presentation of endogenous peptide antigen via MHC class Ib(GO:0002476) |
| 0.0 | 0.2 | GO:0009449 | gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0030186 | melatonin metabolic process(GO:0030186) melatonin biosynthetic process(GO:0030187) |
| 0.0 | 0.9 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.2 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:1902955 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.0 | 0.2 | GO:0071460 | cellular response to cell-matrix adhesion(GO:0071460) |
| 0.0 | 0.1 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) |
| 0.0 | 0.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.1 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.0 | 0.0 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.0 | 0.1 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.0 | 0.2 | GO:2000490 | negative regulation of hepatic stellate cell activation(GO:2000490) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.1 | GO:0046952 | ketone body catabolic process(GO:0046952) |
| 0.0 | 0.1 | GO:1903625 | negative regulation of DNA catabolic process(GO:1903625) |
| 0.0 | 0.1 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0055099 | response to high density lipoprotein particle(GO:0055099) |
| 0.0 | 0.4 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 0.1 | GO:0060722 | spongiotrophoblast cell proliferation(GO:0060720) regulation of spongiotrophoblast cell proliferation(GO:0060721) cell proliferation involved in embryonic placenta development(GO:0060722) regulation of cell proliferation involved in embryonic placenta development(GO:0060723) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0035627 | ceramide transport(GO:0035627) |
| 0.0 | 0.3 | GO:1902172 | keratinocyte apoptotic process(GO:0097283) regulation of keratinocyte apoptotic process(GO:1902172) |
| 0.0 | 0.1 | GO:0052331 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.1 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0014806 | negative regulation of muscle hyperplasia(GO:0014740) smooth muscle hyperplasia(GO:0014806) |
| 0.0 | 0.1 | GO:0050883 | musculoskeletal movement, spinal reflex action(GO:0050883) |
| 0.0 | 0.1 | GO:0031247 | actin rod assembly(GO:0031247) |
| 0.0 | 0.4 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0021773 | striatal medium spiny neuron differentiation(GO:0021773) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0042335 | cuticle development(GO:0042335) |
| 0.0 | 0.1 | GO:1901529 | positive regulation of anion channel activity(GO:1901529) |
| 0.0 | 0.1 | GO:0032304 | negative regulation of icosanoid secretion(GO:0032304) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.0 | GO:0042663 | regulation of endodermal cell fate specification(GO:0042663) |
| 0.0 | 0.1 | GO:0006667 | sphinganine metabolic process(GO:0006667) |
| 0.0 | 0.3 | GO:0048842 | positive regulation of axon extension involved in axon guidance(GO:0048842) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.2 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.2 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.1 | GO:0050955 | thermoception(GO:0050955) |
| 0.0 | 0.1 | GO:0042542 | response to hydrogen peroxide(GO:0042542) |
| 0.0 | 0.0 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.0 | 0.1 | GO:0015811 | L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.6 | GO:0002076 | osteoblast development(GO:0002076) |
| 0.0 | 0.1 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.0 | 0.1 | GO:0032459 | regulation of protein oligomerization(GO:0032459) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.2 | GO:0090132 | epithelial cell migration(GO:0010631) epithelium migration(GO:0090132) |
| 0.0 | 0.2 | GO:0048743 | positive regulation of skeletal muscle fiber development(GO:0048743) |
| 0.0 | 0.0 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.0 | GO:0042223 | regulation of interleukin-3 production(GO:0032672) positive regulation of interleukin-3 production(GO:0032752) interleukin-3 biosynthetic process(GO:0042223) regulation of interleukin-3 biosynthetic process(GO:0045399) positive regulation of interleukin-3 biosynthetic process(GO:0045401) |
| 0.0 | 0.0 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0006668 | sphinganine-1-phosphate metabolic process(GO:0006668) |
| 0.0 | 0.1 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.1 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.0 | 0.0 | GO:0089700 | protein kinase D signaling(GO:0089700) |
| 0.0 | 0.1 | GO:0036229 | glutamine secretion(GO:0010585) L-glutamine import(GO:0036229) L-glutamine import into cell(GO:1903803) |
| 0.0 | 0.1 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.0 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.0 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.0 | 0.0 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.1 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.0 | 0.1 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.0 | 0.0 | GO:0031938 | pre-replicative complex assembly involved in nuclear cell cycle DNA replication(GO:0006267) regulation of chromatin silencing at telomere(GO:0031938) pre-replicative complex assembly(GO:0036388) pre-replicative complex assembly involved in cell cycle DNA replication(GO:1902299) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.1 | GO:2001206 | positive regulation of osteoclast development(GO:2001206) |
| 0.0 | 0.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.0 | GO:0044210 | 'de novo' CTP biosynthetic process(GO:0044210) |
| 0.0 | 0.1 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0000055 | ribosomal large subunit export from nucleus(GO:0000055) ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:0048496 | maintenance of organ identity(GO:0048496) |
| 0.0 | 0.1 | GO:0050680 | negative regulation of epithelial cell proliferation(GO:0050680) |
| 0.0 | 0.0 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.1 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.0 | GO:0006710 | androgen catabolic process(GO:0006710) |
| 0.0 | 0.0 | GO:0072321 | chaperone-mediated protein transport(GO:0072321) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0015801 | aromatic amino acid transport(GO:0015801) |
| 0.0 | 0.1 | GO:1900425 | negative regulation of defense response to bacterium(GO:1900425) |
| 0.0 | 0.5 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.0 | GO:0002428 | antigen processing and presentation of peptide antigen via MHC class Ib(GO:0002428) |
| 0.0 | 0.1 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0006438 | valyl-tRNA aminoacylation(GO:0006438) |
| 0.0 | 0.0 | GO:1904440 | positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) |
| 0.0 | 0.1 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.0 | GO:0000957 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0043305 | negative regulation of mast cell degranulation(GO:0043305) |
| 0.0 | 0.2 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.0 | GO:0098923 | retrograde trans-synaptic signaling by soluble gas(GO:0098923) trans-synaptic signaling by soluble gas(GO:0099543) trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0051086 | chaperone mediated protein folding independent of cofactor(GO:0051086) |
| 0.0 | 0.2 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.5 | GO:0015721 | bile acid and bile salt transport(GO:0015721) |
| 0.0 | 0.0 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.0 | GO:0035553 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.0 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.0 | 0.1 | GO:0010819 | regulation of T cell chemotaxis(GO:0010819) |
| 0.0 | 0.7 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.0 | GO:0010216 | maintenance of DNA methylation(GO:0010216) |
| 0.0 | 0.0 | GO:0010730 | negative regulation of hydrogen peroxide biosynthetic process(GO:0010730) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:0010571 | positive regulation of nuclear cell cycle DNA replication(GO:0010571) |
| 0.0 | 1.0 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0048742 | regulation of skeletal muscle fiber development(GO:0048742) |
| 0.0 | 0.0 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.1 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.0 | GO:1901535 | regulation of DNA demethylation(GO:1901535) |
| 0.0 | 0.0 | GO:1903490 | regulation of cytokinetic process(GO:0032954) regulation of mitotic cytokinetic process(GO:1903436) positive regulation of mitotic cytokinetic process(GO:1903438) positive regulation of mitotic cytokinesis(GO:1903490) |
| 0.0 | 0.0 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.2 | GO:0007597 | blood coagulation, intrinsic pathway(GO:0007597) |
| 0.0 | 0.1 | GO:0070842 | aggresome assembly(GO:0070842) |
| 0.0 | 0.0 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:0033184 | positive regulation of histone ubiquitination(GO:0033184) |
| 0.0 | 0.3 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.0 | GO:0071955 | recycling endosome to Golgi transport(GO:0071955) |
| 0.0 | 0.0 | GO:0060164 | negative regulation of auditory receptor cell differentiation(GO:0045608) regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0060187 | cell pole(GO:0060187) |
| 0.1 | 0.3 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.1 | 0.2 | GO:0032127 | dense core granule membrane(GO:0032127) |
| 0.0 | 0.9 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.2 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.2 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 1.0 | GO:0030057 | desmosome(GO:0030057) |
| 0.0 | 0.2 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.2 | GO:0005828 | kinetochore microtubule(GO:0005828) |
| 0.0 | 0.1 | GO:0042825 | TAP complex(GO:0042825) |
| 0.0 | 0.1 | GO:0000120 | RNA polymerase I transcription factor complex(GO:0000120) |
| 0.0 | 0.0 | GO:0033150 | cytoskeletal calyx(GO:0033150) |
| 0.0 | 0.1 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.4 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.0 | 0.0 | GO:0036387 | nuclear pre-replicative complex(GO:0005656) pre-replicative complex(GO:0036387) |
| 0.0 | 0.1 | GO:0031261 | DNA replication preinitiation complex(GO:0031261) |
| 0.0 | 0.0 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.4 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.0 | GO:0030849 | autosome(GO:0030849) |
| 0.0 | 0.7 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.0 | GO:0042824 | MHC class I peptide loading complex(GO:0042824) |
| 0.0 | 0.0 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.4 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.4 | GO:0017161 | phosphatidylinositol-3,4-bisphosphate 4-phosphatase activity(GO:0016316) inositol-1,3,4-trisphosphate 4-phosphatase activity(GO:0017161) inositol-3,4-bisphosphate 4-phosphatase activity(GO:0052828) |
| 0.1 | 0.2 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0034040 | lipid-transporting ATPase activity(GO:0034040) |
| 0.0 | 0.1 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.2 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.0 | 0.9 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.2 | GO:0004084 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.1 | GO:0047888 | fatty acid peroxidase activity(GO:0047888) |
| 0.0 | 0.1 | GO:0070283 | lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.0 | 0.3 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0036313 | phosphatidylinositol 3-kinase catalytic subunit binding(GO:0036313) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.0 | 0.1 | GO:0010348 | lithium:proton antiporter activity(GO:0010348) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.7 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.3 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.0 | 0.2 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.0 | 0.2 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0015616 | DNA translocase activity(GO:0015616) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0015433 | peptide antigen-transporting ATPase activity(GO:0015433) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.5 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.6 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 0.1 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0016846 | carbon-sulfur lyase activity(GO:0016846) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.6 | GO:0051428 | peptide hormone receptor binding(GO:0051428) |
| 0.0 | 0.1 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.1 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.1 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.0 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.0 | 0.1 | GO:0005148 | prolactin receptor binding(GO:0005148) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.2 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.0 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.1 | GO:0047757 | chondroitin-glucuronate 5-epimerase activity(GO:0047757) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) |
| 0.0 | 0.0 | GO:0004088 | carbamoyl-phosphate synthase (ammonia) activity(GO:0004087) carbamoyl-phosphate synthase (glutamine-hydrolyzing) activity(GO:0004088) |
| 0.0 | 0.0 | GO:0003883 | CTP synthase activity(GO:0003883) |
| 0.0 | 0.1 | GO:0050509 | N-acetylglucosaminyl-proteoglycan 4-beta-glucuronosyltransferase activity(GO:0050509) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.1 | GO:0050473 | arachidonate 15-lipoxygenase activity(GO:0050473) |
| 0.0 | 0.0 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.0 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.0 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.0 | GO:0003870 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0004832 | valine-tRNA ligase activity(GO:0004832) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0000293 | ferric-chelate reductase activity(GO:0000293) |
| 0.0 | 0.1 | GO:0015186 | L-glutamine transmembrane transporter activity(GO:0015186) |
| 0.0 | 0.0 | GO:0016715 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced ascorbate as one donor, and incorporation of one atom of oxygen(GO:0016715) |
| 0.0 | 0.0 | GO:0002134 | pyrimidine nucleoside binding(GO:0001884) UTP binding(GO:0002134) pyrimidine ribonucleoside binding(GO:0032551) |
| 0.0 | 0.1 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.0 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.0 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.1 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.0 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.0 | 0.0 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.1 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 1.0 | GO:0005507 | copper ion binding(GO:0005507) |
| 0.0 | 0.2 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.1 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | PID FRA PATHWAY | Validated transcriptional targets of AP1 family members Fra1 and Fra2 |
| 0.0 | 0.1 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 0.6 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.6 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.7 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 0.0 | PID TRAIL PATHWAY | TRAIL signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.6 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PIPS AT THE EARLY ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the early endosome membrane |
| 0.0 | 0.2 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.3 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME PLATELET ADHESION TO EXPOSED COLLAGEN | Genes involved in Platelet Adhesion to exposed collagen |
| 0.0 | 0.2 | REACTOME ACTIVATION OF THE AP1 FAMILY OF TRANSCRIPTION FACTORS | Genes involved in Activation of the AP-1 family of transcription factors |
| 0.0 | 1.0 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |