Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
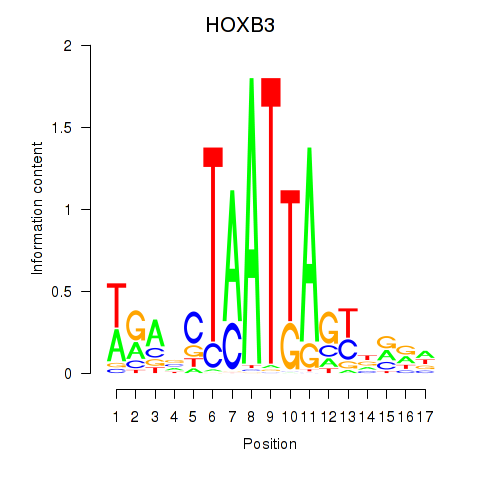
Results for HOXB3
Z-value: 1.38
Transcription factors associated with HOXB3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXB3
|
ENSG00000120093.7 | HOXB3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXB3 | hg19_v2_chr17_-_46667594_46667619 | 0.85 | 7.7e-03 | Click! |
Activity profile of HOXB3 motif
Sorted Z-values of HOXB3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_209602156 | 3.73 |
ENST00000429156.1 ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr5_+_36608422 | 2.28 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr18_+_61554932 | 2.21 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr4_+_40198527 | 2.20 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr8_-_41166953 | 2.08 |
ENST00000220772.3 |
SFRP1 |
secreted frizzled-related protein 1 |
| chr3_-_151034734 | 2.07 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr12_-_123201337 | 1.92 |
ENST00000528880.2 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
| chr1_+_156030937 | 1.90 |
ENST00000361084.5 |
RAB25 |
RAB25, member RAS oncogene family |
| chr1_+_62439037 | 1.90 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr12_-_123187890 | 1.87 |
ENST00000328880.5 |
HCAR2 |
hydroxycarboxylic acid receptor 2 |
| chr11_-_118134997 | 1.86 |
ENST00000278937.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr19_-_36001113 | 1.78 |
ENST00000434389.1 |
DMKN |
dermokine |
| chr11_-_119991589 | 1.62 |
ENST00000526881.1 |
TRIM29 |
tripartite motif containing 29 |
| chr3_-_112565703 | 1.54 |
ENST00000488794.1 |
CD200R1L |
CD200 receptor 1-like |
| chr7_+_26331541 | 1.53 |
ENST00000416246.1 ENST00000338523.4 ENST00000412416.1 |
SNX10 |
sorting nexin 10 |
| chr1_+_28261492 | 1.48 |
ENST00000373894.3 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr11_-_118135160 | 1.43 |
ENST00000438295.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr1_+_152486950 | 1.43 |
ENST00000368790.3 |
CRCT1 |
cysteine-rich C-terminal 1 |
| chr3_-_112564797 | 1.29 |
ENST00000398214.1 ENST00000448932.1 |
CD200R1L |
CD200 receptor 1-like |
| chr17_-_27418537 | 1.16 |
ENST00000408971.2 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr2_-_70780572 | 1.16 |
ENST00000450929.1 |
TGFA |
transforming growth factor, alpha |
| chr11_-_102651343 | 1.13 |
ENST00000279441.4 ENST00000539681.1 |
MMP10 |
matrix metallopeptidase 10 (stromelysin 2) |
| chr6_+_26501449 | 1.12 |
ENST00000244513.6 |
BTN1A1 |
butyrophilin, subfamily 1, member A1 |
| chr11_+_121447469 | 1.10 |
ENST00000532694.1 ENST00000534286.1 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr1_+_160370344 | 1.10 |
ENST00000368061.2 |
VANGL2 |
VANGL planar cell polarity protein 2 |
| chr11_-_124190184 | 1.06 |
ENST00000357438.2 |
OR8D2 |
olfactory receptor, family 8, subfamily D, member 2 |
| chr12_-_6740802 | 1.04 |
ENST00000431922.1 |
LPAR5 |
lysophosphatidic acid receptor 5 |
| chr18_+_55888767 | 1.01 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr9_+_130911770 | 0.98 |
ENST00000372998.1 |
LCN2 |
lipocalin 2 |
| chr4_-_25865159 | 0.94 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr12_+_20848377 | 0.92 |
ENST00000540354.1 ENST00000266509.2 ENST00000381552.1 |
SLCO1C1 |
solute carrier organic anion transporter family, member 1C1 |
| chr11_+_65554493 | 0.84 |
ENST00000335987.3 |
OVOL1 |
ovo-like zinc finger 1 |
| chrX_-_24690771 | 0.84 |
ENST00000379145.1 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr9_+_130911723 | 0.82 |
ENST00000277480.2 ENST00000373013.2 ENST00000540948.1 |
LCN2 |
lipocalin 2 |
| chrX_+_138612889 | 0.79 |
ENST00000218099.2 ENST00000394090.2 |
F9 |
coagulation factor IX |
| chr1_+_161691353 | 0.72 |
ENST00000367948.2 |
FCRLB |
Fc receptor-like B |
| chr14_+_61654271 | 0.71 |
ENST00000555185.1 ENST00000557294.1 ENST00000556778.1 |
PRKCH |
protein kinase C, eta |
| chr6_+_47666275 | 0.71 |
ENST00000327753.3 ENST00000283303.2 |
GPR115 |
G protein-coupled receptor 115 |
| chr3_+_136537911 | 0.69 |
ENST00000393079.3 |
SLC35G2 |
solute carrier family 35, member G2 |
| chr3_+_136537816 | 0.69 |
ENST00000446465.2 |
SLC35G2 |
solute carrier family 35, member G2 |
| chr12_-_6484376 | 0.69 |
ENST00000360168.3 ENST00000358945.3 |
SCNN1A |
sodium channel, non-voltage-gated 1 alpha subunit |
| chr3_-_160823040 | 0.66 |
ENST00000484127.1 ENST00000492353.1 ENST00000473142.1 ENST00000468268.1 ENST00000460353.1 ENST00000320474.4 ENST00000392781.2 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr3_-_160823158 | 0.66 |
ENST00000392779.2 ENST00000392780.1 ENST00000494173.1 |
B3GALNT1 |
beta-1,3-N-acetylgalactosaminyltransferase 1 (globoside blood group) |
| chr1_+_207226574 | 0.63 |
ENST00000367080.3 ENST00000367079.2 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr2_-_101925055 | 0.62 |
ENST00000295317.3 |
RNF149 |
ring finger protein 149 |
| chr13_+_73632897 | 0.60 |
ENST00000377687.4 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr11_-_107729887 | 0.59 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chr7_-_134896234 | 0.58 |
ENST00000354475.4 ENST00000344400.5 |
WDR91 |
WD repeat domain 91 |
| chr4_+_144354644 | 0.58 |
ENST00000512843.1 |
GAB1 |
GRB2-associated binding protein 1 |
| chr7_-_33102338 | 0.57 |
ENST00000610140.1 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
| chr8_-_17579726 | 0.57 |
ENST00000381861.3 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr11_-_59950486 | 0.56 |
ENST00000426738.2 ENST00000533023.1 ENST00000420732.2 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chr19_+_43919058 | 0.56 |
ENST00000598265.1 |
TEX101 |
testis expressed 101 |
| chr9_+_124329336 | 0.55 |
ENST00000394340.3 ENST00000436835.1 ENST00000259371.2 |
DAB2IP |
DAB2 interacting protein |
| chr7_-_33102399 | 0.54 |
ENST00000242210.7 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
| chr19_+_9296279 | 0.54 |
ENST00000344248.2 |
OR7D2 |
olfactory receptor, family 7, subfamily D, member 2 |
| chr19_-_3557570 | 0.53 |
ENST00000355415.2 |
MFSD12 |
major facilitator superfamily domain containing 12 |
| chr16_-_28621312 | 0.53 |
ENST00000314752.7 |
SULT1A1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr1_+_81771806 | 0.53 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr12_+_20848486 | 0.51 |
ENST00000545102.1 |
SLCO1C1 |
solute carrier organic anion transporter family, member 1C1 |
| chr21_-_27423339 | 0.51 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr17_-_62208169 | 0.51 |
ENST00000606895.1 |
ERN1 |
endoplasmic reticulum to nucleus signaling 1 |
| chr3_+_121774202 | 0.49 |
ENST00000469710.1 ENST00000493101.1 ENST00000330540.2 ENST00000264468.5 |
CD86 |
CD86 molecule |
| chr6_+_130339710 | 0.48 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr4_+_87515454 | 0.46 |
ENST00000427191.2 ENST00000436978.1 ENST00000502971.1 |
PTPN13 |
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
| chr12_-_91451758 | 0.46 |
ENST00000266719.3 |
KERA |
keratocan |
| chr19_+_1077393 | 0.46 |
ENST00000590577.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr12_-_8088871 | 0.45 |
ENST00000075120.7 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr4_+_123300121 | 0.44 |
ENST00000446706.1 ENST00000296513.2 |
ADAD1 |
adenosine deaminase domain containing 1 (testis-specific) |
| chr1_+_160160346 | 0.43 |
ENST00000368078.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chr2_+_228736321 | 0.43 |
ENST00000309931.2 |
DAW1 |
dynein assembly factor with WDR repeat domains 1 |
| chr9_+_470288 | 0.43 |
ENST00000382303.1 |
KANK1 |
KN motif and ankyrin repeat domains 1 |
| chr12_-_51422017 | 0.43 |
ENST00000394904.3 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr10_+_6779326 | 0.43 |
ENST00000417112.1 |
RP11-554I8.2 |
RP11-554I8.2 |
| chr3_+_97887544 | 0.41 |
ENST00000356526.2 |
OR5H15 |
olfactory receptor, family 5, subfamily H, member 15 |
| chr1_-_201140673 | 0.41 |
ENST00000367333.2 |
TMEM9 |
transmembrane protein 9 |
| chr12_+_122688090 | 0.41 |
ENST00000324189.4 ENST00000546192.1 |
B3GNT4 |
UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 4 |
| chr9_-_5339873 | 0.41 |
ENST00000223862.1 ENST00000223858.4 |
RLN1 |
relaxin 1 |
| chr7_-_24797546 | 0.40 |
ENST00000414428.1 ENST00000419307.1 ENST00000342947.3 |
DFNA5 |
deafness, autosomal dominant 5 |
| chr1_+_160160283 | 0.39 |
ENST00000368079.3 |
CASQ1 |
calsequestrin 1 (fast-twitch, skeletal muscle) |
| chrX_-_130423200 | 0.39 |
ENST00000361420.3 |
IGSF1 |
immunoglobulin superfamily, member 1 |
| chr4_-_74486109 | 0.39 |
ENST00000395777.2 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr7_-_100860851 | 0.39 |
ENST00000223127.3 |
PLOD3 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 |
| chr2_+_228736335 | 0.39 |
ENST00000440997.1 ENST00000545118.1 |
DAW1 |
dynein assembly factor with WDR repeat domains 1 |
| chr1_-_52870104 | 0.38 |
ENST00000371568.3 |
ORC1 |
origin recognition complex, subunit 1 |
| chr1_+_16090914 | 0.38 |
ENST00000441801.2 |
FBLIM1 |
filamin binding LIM protein 1 |
| chrX_+_102024075 | 0.37 |
ENST00000431616.1 ENST00000440496.1 ENST00000420471.1 ENST00000435966.1 |
LINC00630 |
long intergenic non-protein coding RNA 630 |
| chr5_-_37835929 | 0.37 |
ENST00000381826.4 ENST00000427982.1 |
GDNF |
glial cell derived neurotrophic factor |
| chr1_-_109935819 | 0.36 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr9_+_134165063 | 0.36 |
ENST00000372264.3 |
PPAPDC3 |
phosphatidic acid phosphatase type 2 domain containing 3 |
| chr22_+_21996549 | 0.36 |
ENST00000248958.4 |
SDF2L1 |
stromal cell-derived factor 2-like 1 |
| chr22_-_29107919 | 0.35 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr11_+_112047087 | 0.35 |
ENST00000526088.1 ENST00000532593.1 ENST00000531169.1 |
BCO2 |
beta-carotene oxygenase 2 |
| chr10_-_135379132 | 0.35 |
ENST00000343131.5 |
SYCE1 |
synaptonemal complex central element protein 1 |
| chr2_+_29336855 | 0.35 |
ENST00000404424.1 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
| chr3_+_147795932 | 0.34 |
ENST00000490465.1 |
RP11-639B1.1 |
RP11-639B1.1 |
| chr7_-_111032971 | 0.33 |
ENST00000450877.1 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr16_+_12059050 | 0.32 |
ENST00000396495.3 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr2_+_90077680 | 0.32 |
ENST00000390270.2 |
IGKV3D-20 |
immunoglobulin kappa variable 3D-20 |
| chr5_+_110427983 | 0.31 |
ENST00000513710.2 ENST00000505303.1 |
WDR36 |
WD repeat domain 36 |
| chr2_+_234545148 | 0.31 |
ENST00000373445.1 |
UGT1A10 |
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr2_+_168675182 | 0.31 |
ENST00000305861.1 |
B3GALT1 |
UDP-Gal:betaGlcNAc beta 1,3-galactosyltransferase, polypeptide 1 |
| chr19_+_11071546 | 0.30 |
ENST00000358026.2 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr14_+_56584414 | 0.30 |
ENST00000559044.1 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
| chr6_-_133035185 | 0.30 |
ENST00000367928.4 |
VNN1 |
vanin 1 |
| chr3_-_143567262 | 0.30 |
ENST00000474151.1 ENST00000316549.6 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr15_-_101835110 | 0.29 |
ENST00000560496.1 |
SNRPA1 |
small nuclear ribonucleoprotein polypeptide A' |
| chrX_+_41548220 | 0.29 |
ENST00000378142.4 |
GPR34 |
G protein-coupled receptor 34 |
| chr11_-_59950519 | 0.28 |
ENST00000528851.1 |
MS4A6A |
membrane-spanning 4-domains, subfamily A, member 6A |
| chrX_+_41548259 | 0.28 |
ENST00000378138.5 |
GPR34 |
G protein-coupled receptor 34 |
| chr8_-_49834299 | 0.27 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr11_+_55578854 | 0.27 |
ENST00000333973.2 |
OR5L1 |
olfactory receptor, family 5, subfamily L, member 1 |
| chr3_-_185538849 | 0.27 |
ENST00000421047.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr2_+_86669118 | 0.26 |
ENST00000427678.1 ENST00000542128.1 |
KDM3A |
lysine (K)-specific demethylase 3A |
| chr11_+_3819049 | 0.26 |
ENST00000396986.2 ENST00000300730.6 ENST00000532535.1 ENST00000396993.4 ENST00000396991.2 ENST00000532523.1 ENST00000459679.1 ENST00000464261.1 ENST00000464906.2 ENST00000464441.1 |
PGAP2 |
post-GPI attachment to proteins 2 |
| chr1_+_220267429 | 0.25 |
ENST00000366922.1 ENST00000302637.5 |
IARS2 |
isoleucyl-tRNA synthetase 2, mitochondrial |
| chr12_-_122985067 | 0.25 |
ENST00000540586.1 ENST00000543897.1 |
ZCCHC8 |
zinc finger, CCHC domain containing 8 |
| chr5_+_61874562 | 0.25 |
ENST00000334994.5 ENST00000409534.1 |
LRRC70 IPO11 |
leucine rich repeat containing 70 importin 11 |
| chr11_-_128894053 | 0.25 |
ENST00000392657.3 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr8_-_86290333 | 0.25 |
ENST00000521846.1 ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1 |
carbonic anhydrase I |
| chr5_+_150639360 | 0.24 |
ENST00000523004.1 |
GM2A |
GM2 ganglioside activator |
| chr7_-_121944491 | 0.24 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr16_-_66864806 | 0.24 |
ENST00000566336.1 ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1 |
NEDD8 activating enzyme E1 subunit 1 |
| chr14_-_54908043 | 0.23 |
ENST00000556113.1 ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1 |
cornichon family AMPA receptor auxiliary protein 1 |
| chr1_-_207226313 | 0.23 |
ENST00000367084.1 |
YOD1 |
YOD1 deubiquitinase |
| chr11_-_104817919 | 0.23 |
ENST00000533252.1 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr4_+_86525299 | 0.23 |
ENST00000512201.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr6_+_150690028 | 0.23 |
ENST00000229447.5 ENST00000344419.3 |
IYD |
iodotyrosine deiodinase |
| chr5_+_55149150 | 0.23 |
ENST00000297015.3 |
IL31RA |
interleukin 31 receptor A |
| chr19_-_55677999 | 0.22 |
ENST00000532817.1 ENST00000527223.2 ENST00000391720.4 |
DNAAF3 |
dynein, axonemal, assembly factor 3 |
| chr4_+_5527117 | 0.22 |
ENST00000505296.1 |
C4orf6 |
chromosome 4 open reading frame 6 |
| chr7_+_99202003 | 0.22 |
ENST00000609449.1 |
GS1-259H13.2 |
GS1-259H13.2 |
| chr2_+_113816685 | 0.21 |
ENST00000393200.2 |
IL36RN |
interleukin 36 receptor antagonist |
| chr8_-_49833978 | 0.21 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr4_+_5526883 | 0.21 |
ENST00000195455.2 |
C4orf6 |
chromosome 4 open reading frame 6 |
| chrX_+_144908928 | 0.21 |
ENST00000408967.2 |
TMEM257 |
transmembrane protein 257 |
| chr11_-_117748138 | 0.21 |
ENST00000527717.1 |
FXYD6 |
FXYD domain containing ion transport regulator 6 |
| chr1_-_197115818 | 0.21 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr2_+_183982238 | 0.21 |
ENST00000442895.2 ENST00000446612.1 ENST00000409798.1 |
NUP35 |
nucleoporin 35kDa |
| chr11_-_104827425 | 0.20 |
ENST00000393150.3 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr12_-_23737534 | 0.20 |
ENST00000396007.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr19_-_55677920 | 0.20 |
ENST00000524407.2 ENST00000526003.1 ENST00000534170.1 |
DNAAF3 |
dynein, axonemal, assembly factor 3 |
| chr17_-_47785504 | 0.20 |
ENST00000514907.1 ENST00000503334.1 ENST00000508520.1 |
SLC35B1 |
solute carrier family 35, member B1 |
| chr19_+_54641444 | 0.20 |
ENST00000221232.5 ENST00000358389.3 |
CNOT3 |
CCR4-NOT transcription complex, subunit 3 |
| chr2_+_113816215 | 0.19 |
ENST00000346807.3 |
IL36RN |
interleukin 36 receptor antagonist |
| chr4_-_120243545 | 0.19 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr12_-_22063787 | 0.19 |
ENST00000544039.1 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr19_+_10362882 | 0.19 |
ENST00000393733.2 ENST00000588502.1 |
MRPL4 |
mitochondrial ribosomal protein L4 |
| chr1_+_197237352 | 0.18 |
ENST00000538660.1 ENST00000367400.3 ENST00000367399.2 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chr15_-_101835414 | 0.18 |
ENST00000254193.6 |
SNRPA1 |
small nuclear ribonucleoprotein polypeptide A' |
| chr2_+_105050794 | 0.18 |
ENST00000429464.1 ENST00000414442.1 ENST00000447380.1 |
AC013402.2 |
long intergenic non-protein coding RNA 1102 |
| chr12_+_128399965 | 0.18 |
ENST00000540882.1 ENST00000542089.1 |
LINC00507 |
long intergenic non-protein coding RNA 507 |
| chr20_+_9494987 | 0.17 |
ENST00000427562.2 ENST00000246070.2 |
LAMP5 |
lysosomal-associated membrane protein family, member 5 |
| chr16_+_31885079 | 0.17 |
ENST00000300870.10 ENST00000394846.3 |
ZNF267 |
zinc finger protein 267 |
| chr22_-_30234218 | 0.17 |
ENST00000307790.3 ENST00000542393.1 ENST00000397771.2 |
ASCC2 |
activating signal cointegrator 1 complex subunit 2 |
| chr12_+_66582919 | 0.17 |
ENST00000545837.1 ENST00000457197.2 |
IRAK3 |
interleukin-1 receptor-associated kinase 3 |
| chr4_-_111563076 | 0.17 |
ENST00000354925.2 ENST00000511990.1 |
PITX2 |
paired-like homeodomain 2 |
| chr12_+_56521840 | 0.16 |
ENST00000394048.5 |
ESYT1 |
extended synaptotagmin-like protein 1 |
| chr6_-_119256279 | 0.16 |
ENST00000316068.3 |
MCM9 |
minichromosome maintenance complex component 9 |
| chrX_+_83116142 | 0.16 |
ENST00000329312.4 |
CYLC1 |
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr11_-_95522907 | 0.16 |
ENST00000358780.5 ENST00000542135.1 |
FAM76B |
family with sequence similarity 76, member B |
| chr6_+_45296391 | 0.16 |
ENST00000371436.6 ENST00000576263.1 |
RUNX2 |
runt-related transcription factor 2 |
| chr2_+_196313239 | 0.16 |
ENST00000413290.1 |
AC064834.1 |
AC064834.1 |
| chr16_+_50730910 | 0.15 |
ENST00000300589.2 |
NOD2 |
nucleotide-binding oligomerization domain containing 2 |
| chr6_+_26087509 | 0.15 |
ENST00000397022.3 ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE |
hemochromatosis |
| chr14_+_32798547 | 0.15 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr2_+_187371440 | 0.15 |
ENST00000445547.1 |
ZC3H15 |
zinc finger CCCH-type containing 15 |
| chr20_-_44600810 | 0.15 |
ENST00000322927.2 ENST00000426788.1 |
ZNF335 |
zinc finger protein 335 |
| chr19_-_6737576 | 0.15 |
ENST00000601716.1 ENST00000264080.7 |
GPR108 |
G protein-coupled receptor 108 |
| chr21_+_43823983 | 0.14 |
ENST00000291535.6 ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A |
ubiquitin associated and SH3 domain containing A |
| chr2_+_124782857 | 0.14 |
ENST00000431078.1 |
CNTNAP5 |
contactin associated protein-like 5 |
| chr3_+_186288454 | 0.14 |
ENST00000265028.3 |
DNAJB11 |
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr2_+_197577841 | 0.14 |
ENST00000409270.1 |
CCDC150 |
coiled-coil domain containing 150 |
| chr21_-_15918618 | 0.14 |
ENST00000400564.1 ENST00000400566.1 |
SAMSN1 |
SAM domain, SH3 domain and nuclear localization signals 1 |
| chr12_+_112563335 | 0.14 |
ENST00000549358.1 ENST00000257604.5 ENST00000548092.1 ENST00000552896.1 |
TRAFD1 |
TRAF-type zinc finger domain containing 1 |
| chrX_-_48824793 | 0.14 |
ENST00000376477.1 |
KCND1 |
potassium voltage-gated channel, Shal-related subfamily, member 1 |
| chr19_+_10362577 | 0.14 |
ENST00000592514.1 ENST00000307422.5 ENST00000253099.6 ENST00000590150.1 ENST00000590669.1 |
MRPL4 |
mitochondrial ribosomal protein L4 |
| chr11_-_95522639 | 0.13 |
ENST00000536839.1 |
FAM76B |
family with sequence similarity 76, member B |
| chr16_-_28621298 | 0.13 |
ENST00000566189.1 |
SULT1A1 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 1 |
| chr2_-_73869508 | 0.13 |
ENST00000272425.3 |
NAT8 |
N-acetyltransferase 8 (GCN5-related, putative) |
| chrX_-_70326455 | 0.12 |
ENST00000374251.5 |
CXorf65 |
chromosome X open reading frame 65 |
| chr19_-_17366257 | 0.12 |
ENST00000594059.1 |
AC010646.3 |
Uncharacterized protein |
| chr4_+_66536248 | 0.12 |
ENST00000514260.1 ENST00000507117.1 |
RP11-807H7.1 |
RP11-807H7.1 |
| chr12_+_128399917 | 0.12 |
ENST00000544645.1 |
LINC00507 |
long intergenic non-protein coding RNA 507 |
| chrX_+_37208540 | 0.12 |
ENST00000466533.1 ENST00000542554.1 ENST00000543642.1 ENST00000484460.1 ENST00000449135.2 ENST00000463135.1 ENST00000465127.1 |
PRRG1 TM4SF2 |
proline rich Gla (G-carboxyglutamic acid) 1 Uncharacterized protein; cDNA FLJ59144, highly similar to Tetraspanin-7 |
| chr17_-_8113542 | 0.12 |
ENST00000578549.1 ENST00000535053.1 ENST00000582368.1 |
AURKB |
aurora kinase B |
| chr12_+_28410128 | 0.12 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr12_+_22778116 | 0.11 |
ENST00000538218.1 |
ETNK1 |
ethanolamine kinase 1 |
| chr4_-_164534657 | 0.11 |
ENST00000339875.5 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chr12_+_112563303 | 0.11 |
ENST00000412615.2 |
TRAFD1 |
TRAF-type zinc finger domain containing 1 |
| chr5_-_108063949 | 0.11 |
ENST00000606054.1 |
LINC01023 |
long intergenic non-protein coding RNA 1023 |
| chr17_+_73257945 | 0.11 |
ENST00000579002.1 |
MRPS7 |
mitochondrial ribosomal protein S7 |
| chr3_-_126327398 | 0.11 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr6_-_32095968 | 0.11 |
ENST00000375203.3 ENST00000375201.4 |
ATF6B |
activating transcription factor 6 beta |
| chr7_+_129015484 | 0.11 |
ENST00000490911.1 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
| chr8_-_623547 | 0.11 |
ENST00000522893.1 |
ERICH1 |
glutamate-rich 1 |
| chr7_-_134855517 | 0.11 |
ENST00000430372.1 |
C7orf49 |
chromosome 7 open reading frame 49 |
| chr11_+_118398178 | 0.10 |
ENST00000302783.4 ENST00000539546.1 |
TTC36 |
tetratricopeptide repeat domain 36 |
| chr11_+_94706804 | 0.10 |
ENST00000335080.5 |
KDM4D |
lysine (K)-specific demethylase 4D |
| chr13_-_47471155 | 0.10 |
ENST00000543956.1 ENST00000542664.1 |
HTR2A |
5-hydroxytryptamine (serotonin) receptor 2A, G protein-coupled |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.1 | GO:1904956 | regulation of midbrain dopaminergic neuron differentiation(GO:1904956) regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.4 | 1.1 | GO:1902955 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.4 | 1.1 | GO:0060489 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.4 | 1.8 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.3 | 2.3 | GO:0006537 | glutamate biosynthetic process(GO:0006537) gamma-aminobutyric acid biosynthetic process(GO:0009449) |
| 0.3 | 1.9 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.3 | 1.8 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.3 | 0.8 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.2 | 1.0 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.2 | 0.5 | GO:0002644 | negative regulation of tolerance induction(GO:0002644) positive regulation of interleukin-4 biosynthetic process(GO:0045404) |
| 0.2 | 0.5 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.4 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.1 | 0.4 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.1 | 0.4 | GO:0046947 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.1 | 1.6 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.4 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.4 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.1 | 2.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.1 | 0.5 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.1 | 0.3 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.6 | GO:0033133 | fructose 2,6-bisphosphate metabolic process(GO:0006003) positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.1 | 0.4 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.1 | 1.4 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.1 | 0.5 | GO:0071874 | cellular response to norepinephrine stimulus(GO:0071874) |
| 0.1 | 0.8 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.1 | 1.9 | GO:0031268 | pseudopodium organization(GO:0031268) |
| 0.1 | 0.4 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.1 | 0.3 | GO:0006428 | isoleucyl-tRNA aminoacylation(GO:0006428) |
| 0.1 | 0.7 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 0.8 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.1 | 1.1 | GO:0046085 | adenosine metabolic process(GO:0046085) |
| 0.1 | 0.8 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 1.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 1.6 | GO:0009312 | oligosaccharide biosynthetic process(GO:0009312) |
| 0.1 | 0.2 | GO:0072334 | UDP-galactose transport(GO:0015785) UDP-galactose transmembrane transport(GO:0072334) |
| 0.1 | 0.6 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.5 | GO:0036289 | peptidyl-serine autophosphorylation(GO:0036289) |
| 0.1 | 0.4 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.1 | 0.3 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
| 0.1 | 0.4 | GO:1902714 | negative regulation of interferon-gamma secretion(GO:1902714) |
| 0.1 | 2.2 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.1 | 0.2 | GO:1990167 | protein K27-linked deubiquitination(GO:1990167) protein K33-linked deubiquitination(GO:1990168) |
| 0.1 | 0.2 | GO:0060127 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.1 | 1.5 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.1 | 0.3 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.1 | 0.2 | GO:0060584 | detection of peptidoglycan(GO:0032499) activation of MAPK activity involved in innate immune response(GO:0035419) regulation of prostaglandin-endoperoxide synthase activity(GO:0060584) positive regulation of prostaglandin-endoperoxide synthase activity(GO:0060585) positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.1 | 0.5 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.1 | 0.3 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.2 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.0 | 0.5 | GO:0051552 | flavone metabolic process(GO:0051552) |
| 0.0 | 0.2 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.2 | GO:1902261 | positive regulation of ryanodine-sensitive calcium-release channel activity(GO:0060316) positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.5 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.0 | 2.2 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.4 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.5 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.7 | GO:0009812 | flavonoid metabolic process(GO:0009812) |
| 0.0 | 0.2 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.2 | GO:0010936 | negative regulation of macrophage cytokine production(GO:0010936) |
| 0.0 | 0.4 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.3 | GO:0033089 | pantothenate metabolic process(GO:0015939) positive regulation of T cell differentiation in thymus(GO:0033089) positive regulation of thymocyte aggregation(GO:2000400) |
| 0.0 | 0.2 | GO:0006689 | ganglioside catabolic process(GO:0006689) |
| 0.0 | 0.1 | GO:0000454 | snoRNA guided rRNA pseudouridine synthesis(GO:0000454) |
| 0.0 | 0.4 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.2 | GO:0055059 | asymmetric neuroblast division(GO:0055059) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 2.1 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.1 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.2 | GO:0033314 | mitotic DNA replication checkpoint(GO:0033314) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.2 | GO:0006570 | tyrosine metabolic process(GO:0006570) |
| 0.0 | 0.3 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0000973 | posttranscriptional tethering of RNA polymerase II gene DNA at nuclear periphery(GO:0000973) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.2 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.2 | GO:0043031 | negative regulation of macrophage activation(GO:0043031) |
| 0.0 | 0.4 | GO:0007130 | synaptonemal complex assembly(GO:0007130) |
| 0.0 | 2.5 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.2 | GO:0019433 | triglyceride catabolic process(GO:0019433) |
| 0.0 | 0.4 | GO:0044458 | motile cilium assembly(GO:0044458) |
| 0.0 | 0.2 | GO:0006307 | DNA dealkylation involved in DNA repair(GO:0006307) |
| 0.0 | 1.5 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.1 | GO:0035995 | skeletal muscle myosin thick filament assembly(GO:0030241) detection of muscle stretch(GO:0035995) |
| 0.0 | 1.0 | GO:0030574 | collagen catabolic process(GO:0030574) |
| 0.0 | 0.1 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.7 | GO:0055078 | sodium ion homeostasis(GO:0055078) |
| 0.0 | 0.1 | GO:0007089 | traversing start control point of mitotic cell cycle(GO:0007089) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.2 | 1.5 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.2 | 0.5 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.2 | 0.5 | GO:1990597 | AIP1-IRE1 complex(GO:1990597) |
| 0.1 | 0.3 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.1 | 1.6 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.1 | 0.8 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.1 | 0.4 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.1 | 1.9 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.1 | 0.2 | GO:0005889 | hydrogen:potassium-exchanging ATPase complex(GO:0005889) |
| 0.1 | 0.5 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.1 | 0.2 | GO:0043159 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.1 | 0.3 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.2 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.2 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
| 0.0 | 2.2 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.4 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.2 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.5 | GO:0034663 | endoplasmic reticulum chaperone complex(GO:0034663) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.6 | GO:0034706 | sodium channel complex(GO:0034706) |
| 0.0 | 0.2 | GO:0014701 | junctional sarcoplasmic reticulum membrane(GO:0014701) |
| 0.0 | 0.2 | GO:0030015 | CCR4-NOT core complex(GO:0030015) |
| 0.0 | 0.5 | GO:0005686 | U2 snRNP(GO:0005686) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0044615 | nuclear pore nuclear basket(GO:0044615) |
| 0.0 | 1.0 | GO:0005771 | multivesicular body(GO:0005771) |
| 0.0 | 1.4 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.6 | GO:0002080 | acrosomal membrane(GO:0002080) |
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 1.4 | GO:0035580 | specific granule lumen(GO:0035580) |
| 0.0 | 0.1 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.0 | 0.4 | GO:0030867 | rough endoplasmic reticulum membrane(GO:0030867) |
| 0.0 | 0.1 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.0 | GO:1990742 | microvesicle(GO:1990742) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.4 | 1.3 | GO:0047273 | galactosylgalactosylglucosylceramide beta-D-acetylgalactosaminyltransferase activity(GO:0047273) |
| 0.3 | 2.3 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.2 | 1.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.2 | 0.8 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.2 | 1.4 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.1 | 0.4 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.1 | 0.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.4 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.1 | 1.6 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 1.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.1 | 2.6 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.7 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.6 | GO:0004331 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) |
| 0.1 | 0.4 | GO:0008532 | N-acetyllactosaminide beta-1,3-N-acetylglucosaminyltransferase activity(GO:0008532) |
| 0.1 | 0.3 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.1 | 2.2 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.1 | 0.4 | GO:0015094 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.1 | 0.3 | GO:0004822 | isoleucine-tRNA ligase activity(GO:0004822) |
| 0.1 | 2.1 | GO:0042813 | Wnt-activated receptor activity(GO:0042813) |
| 0.1 | 0.4 | GO:0005152 | interleukin-1 receptor antagonist activity(GO:0005152) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 1.0 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.1 | 0.2 | GO:1904455 | ubiquitin-specific protease activity involved in negative regulation of ERAD pathway(GO:1904455) |
| 0.1 | 0.4 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.2 | GO:0005459 | UDP-galactose transmembrane transporter activity(GO:0005459) |
| 0.1 | 0.2 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.1 | 0.3 | GO:0017159 | pantetheine hydrolase activity(GO:0017159) |
| 0.1 | 0.4 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.1 | 1.5 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.1 | 1.6 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.5 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 1.5 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.4 | GO:0034711 | inhibin binding(GO:0034711) |
| 0.0 | 0.5 | GO:0030620 | U2 snRNA binding(GO:0030620) |
| 0.0 | 1.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.7 | GO:0015280 | ligand-gated sodium channel activity(GO:0015280) |
| 0.0 | 0.4 | GO:0016702 | oxidoreductase activity, acting on single donors with incorporation of molecular oxygen(GO:0016701) oxidoreductase activity, acting on single donors with incorporation of molecular oxygen, incorporation of two atoms of oxygen(GO:0016702) |
| 0.0 | 0.2 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.5 | GO:0036312 | phosphatidylinositol 3-kinase regulatory subunit binding(GO:0036312) |
| 0.0 | 0.7 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.2 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.2 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.0 | 0.4 | GO:0004000 | adenosine deaminase activity(GO:0004000) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.5 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 1.2 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.5 | GO:0005161 | platelet-derived growth factor receptor binding(GO:0005161) |
| 0.0 | 0.2 | GO:0008641 | small protein activating enzyme activity(GO:0008641) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.3 | GO:0030957 | Tat protein binding(GO:0030957) |
| 0.0 | 2.3 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.6 | GO:0061630 | ubiquitin protein ligase activity(GO:0061630) |
| 0.0 | 0.1 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.3 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.0 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.2 | GO:1990459 | transferrin receptor binding(GO:1990459) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.2 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.5 | GO:0015026 | coreceptor activity(GO:0015026) |
| 0.0 | 0.0 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 2.1 | ST WNT BETA CATENIN PATHWAY | Wnt/beta-catenin Pathway |
| 0.0 | 1.2 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.5 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
| 0.0 | 1.8 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 3.9 | NABA ECM REGULATORS | Genes encoding enzymes and their regulators involved in the remodeling of the extracellular matrix |
| 0.0 | 0.6 | PID IL12 STAT4 PATHWAY | IL12 signaling mediated by STAT4 |
| 0.0 | 0.5 | PID RAS PATHWAY | Regulation of Ras family activation |
| 0.0 | 0.4 | PID HIF2PATHWAY | HIF-2-alpha transcription factor network |
| 0.0 | 0.5 | PID EPHRINB REV PATHWAY | Ephrin B reverse signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.1 | 0.8 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 1.9 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.6 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.4 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.0 | 0.8 | REACTOME SYNTHESIS OF PC | Genes involved in Synthesis of PC |
| 0.0 | 1.9 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.7 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.4 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.5 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 1.7 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.6 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.4 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.2 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE BIOSYNTHESIS | Genes involved in Keratan sulfate biosynthesis |
| 0.0 | 0.6 | REACTOME NOD1 2 SIGNALING PATHWAY | Genes involved in NOD1/2 Signaling Pathway |
| 0.0 | 3.6 | REACTOME G ALPHA I SIGNALLING EVENTS | Genes involved in G alpha (i) signalling events |
| 0.0 | 0.4 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.1 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.6 | REACTOME GLYCOLYSIS | Genes involved in Glycolysis |
| 0.0 | 0.1 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |