Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for HOXB6_PRRX2
Z-value: 1.83
Transcription factors associated with HOXB6_PRRX2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
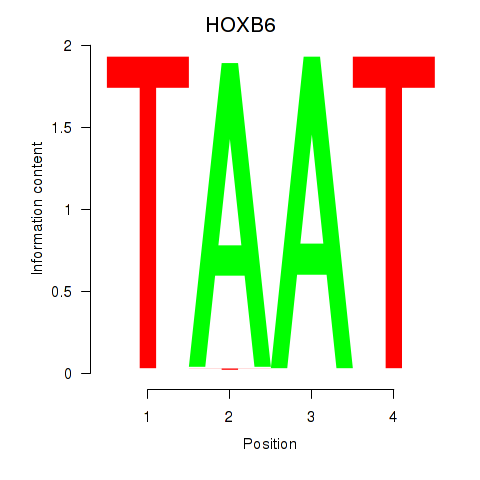
HOXB6
|
ENSG00000108511.8 | HOXB6 |
|
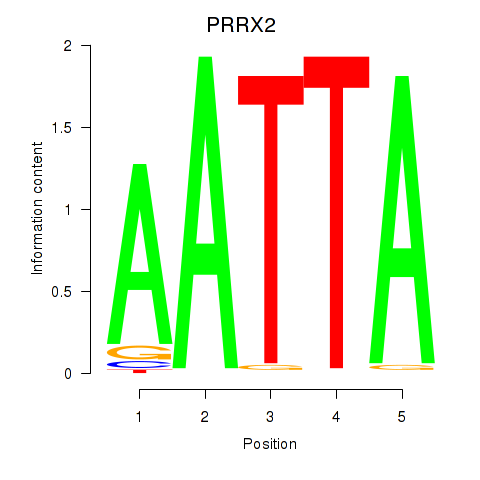
PRRX2
|
ENSG00000167157.9 | PRRX2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PRRX2 | hg19_v2_chr9_+_132427883_132427951 | -0.79 | 1.9e-02 | Click! |
| HOXB6 | hg19_v2_chr17_-_46682321_46682362 | -0.19 | 6.6e-01 | Click! |
Activity profile of HOXB6_PRRX2 motif
Sorted Z-values of HOXB6_PRRX2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXB6_PRRX2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_92413353 | 9.23 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr12_-_91576750 | 8.41 |
ENST00000228329.5 ENST00000303320.3 ENST00000052754.5 |
DCN |
decorin |
| chr12_-_91576561 | 8.25 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr14_-_92413727 | 7.78 |
ENST00000267620.10 |
FBLN5 |
fibulin 5 |
| chr12_-_91576429 | 7.76 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr2_-_145278475 | 7.38 |
ENST00000558170.2 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr3_+_141105705 | 4.05 |
ENST00000513258.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr2_-_145277569 | 3.92 |
ENST00000303660.4 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr16_+_53133070 | 3.54 |
ENST00000565832.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr1_-_57045228 | 3.48 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr13_+_102104980 | 3.39 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr4_+_160188889 | 3.24 |
ENST00000264431.4 |
RAPGEF2 |
Rap guanine nucleotide exchange factor (GEF) 2 |
| chr13_-_67802549 | 3.17 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr13_+_102104952 | 3.06 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr12_-_91574142 | 2.97 |
ENST00000547937.1 |
DCN |
decorin |
| chr14_-_92414055 | 2.89 |
ENST00000342058.4 |
FBLN5 |
fibulin 5 |
| chr12_-_91546926 | 2.78 |
ENST00000550758.1 |
DCN |
decorin |
| chr12_-_91539918 | 2.77 |
ENST00000548218.1 |
DCN |
decorin |
| chr5_-_111093759 | 2.70 |
ENST00000509979.1 ENST00000513100.1 ENST00000508161.1 ENST00000455559.2 |
NREP |
neuronal regeneration related protein |
| chr15_+_93443419 | 2.54 |
ENST00000557381.1 ENST00000420239.2 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
| chr9_-_95186739 | 2.52 |
ENST00000375550.4 |
OMD |
osteomodulin |
| chr5_-_88119580 | 2.47 |
ENST00000539796.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr2_+_210444748 | 2.46 |
ENST00000392194.1 |
MAP2 |
microtubule-associated protein 2 |
| chr7_-_14026063 | 2.44 |
ENST00000443608.1 ENST00000438956.1 |
ETV1 |
ets variant 1 |
| chr3_+_69928256 | 2.44 |
ENST00000394355.2 |
MITF |
microphthalmia-associated transcription factor |
| chr5_-_20575959 | 2.35 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr13_-_38172863 | 2.34 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr3_+_69985734 | 2.34 |
ENST00000314557.6 ENST00000394351.3 |
MITF |
microphthalmia-associated transcription factor |
| chr5_+_156712372 | 2.28 |
ENST00000541131.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr2_-_145277640 | 2.27 |
ENST00000539609.3 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr13_+_102142296 | 2.26 |
ENST00000376162.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr2_+_210444142 | 2.25 |
ENST00000360351.4 ENST00000361559.4 |
MAP2 |
microtubule-associated protein 2 |
| chr4_-_138453606 | 2.24 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr3_+_141106458 | 2.22 |
ENST00000509883.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr5_-_111092930 | 2.21 |
ENST00000257435.7 |
NREP |
neuronal regeneration related protein |
| chr8_-_120651020 | 2.20 |
ENST00000522826.1 ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr3_+_69812877 | 2.17 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr13_+_76334498 | 2.16 |
ENST00000534657.1 |
LMO7 |
LIM domain 7 |
| chr11_-_27723158 | 2.10 |
ENST00000395980.2 |
BDNF |
brain-derived neurotrophic factor |
| chr3_+_69812701 | 2.09 |
ENST00000472437.1 |
MITF |
microphthalmia-associated transcription factor |
| chr6_-_87804815 | 2.03 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr8_+_77593474 | 2.01 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr5_-_111093167 | 1.99 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr9_-_20622478 | 1.96 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr2_-_175711133 | 1.93 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr6_-_46293378 | 1.92 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr7_-_14026123 | 1.89 |
ENST00000420159.2 ENST00000399357.3 ENST00000403527.1 |
ETV1 |
ets variant 1 |
| chr16_-_51185172 | 1.86 |
ENST00000251020.4 |
SALL1 |
spalt-like transcription factor 1 |
| chr2_+_33359646 | 1.85 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr3_-_114477787 | 1.85 |
ENST00000464560.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr8_+_77593448 | 1.84 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr4_+_55095264 | 1.82 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr2_+_33359687 | 1.82 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr8_-_122653630 | 1.75 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr20_-_34025999 | 1.70 |
ENST00000374369.3 |
GDF5 |
growth differentiation factor 5 |
| chr6_+_136172820 | 1.68 |
ENST00000308191.6 |
PDE7B |
phosphodiesterase 7B |
| chr8_-_108510224 | 1.68 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr3_-_114477962 | 1.63 |
ENST00000471418.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr10_-_49812997 | 1.62 |
ENST00000417912.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr9_+_2159850 | 1.62 |
ENST00000416751.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr13_+_76334795 | 1.61 |
ENST00000526202.1 ENST00000465261.2 |
LMO7 |
LIM domain 7 |
| chr7_-_27142290 | 1.60 |
ENST00000222718.5 |
HOXA2 |
homeobox A2 |
| chr2_-_224467093 | 1.58 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chr6_-_85474219 | 1.57 |
ENST00000369663.5 |
TBX18 |
T-box 18 |
| chr5_+_95066823 | 1.57 |
ENST00000506817.1 ENST00000379982.3 |
RHOBTB3 |
Rho-related BTB domain containing 3 |
| chr5_+_67586465 | 1.55 |
ENST00000336483.5 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr13_+_76378357 | 1.49 |
ENST00000489941.2 ENST00000525373.1 |
LMO7 |
LIM domain 7 |
| chr8_-_93107443 | 1.48 |
ENST00000360348.2 ENST00000520428.1 ENST00000518992.1 ENST00000520556.1 ENST00000518317.1 ENST00000521319.1 ENST00000521375.1 ENST00000518449.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr6_+_155537771 | 1.45 |
ENST00000275246.7 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
| chr5_-_111092873 | 1.43 |
ENST00000509025.1 ENST00000515855.1 |
NREP |
neuronal regeneration related protein |
| chr7_-_27183263 | 1.42 |
ENST00000222726.3 |
HOXA5 |
homeobox A5 |
| chr1_-_232651312 | 1.41 |
ENST00000262861.4 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr5_+_82767583 | 1.40 |
ENST00000512590.2 ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN |
versican |
| chr5_+_156696362 | 1.39 |
ENST00000377576.3 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr10_-_49813090 | 1.37 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr14_-_61116168 | 1.36 |
ENST00000247182.6 |
SIX1 |
SIX homeobox 1 |
| chr3_-_112360116 | 1.35 |
ENST00000206423.3 ENST00000439685.2 |
CCDC80 |
coiled-coil domain containing 80 |
| chr3_+_157154578 | 1.35 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr5_+_173316341 | 1.35 |
ENST00000520867.1 ENST00000334035.5 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
| chr13_+_76334567 | 1.34 |
ENST00000321797.8 |
LMO7 |
LIM domain 7 |
| chr1_+_162602244 | 1.33 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chr16_+_53242350 | 1.32 |
ENST00000565442.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr5_+_82767487 | 1.32 |
ENST00000343200.5 ENST00000342785.4 |
VCAN |
versican |
| chr5_+_145718587 | 1.32 |
ENST00000230732.4 |
POU4F3 |
POU class 4 homeobox 3 |
| chrX_+_9431324 | 1.31 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr18_-_25616519 | 1.28 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chr8_+_97597148 | 1.26 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr2_+_54785485 | 1.26 |
ENST00000333896.5 |
SPTBN1 |
spectrin, beta, non-erythrocytic 1 |
| chr5_+_119799927 | 1.25 |
ENST00000407149.2 ENST00000379551.2 |
PRR16 |
proline rich 16 |
| chr1_+_168250194 | 1.24 |
ENST00000367821.3 |
TBX19 |
T-box 19 |
| chr9_-_95244781 | 1.21 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr10_-_106240032 | 1.21 |
ENST00000447860.1 |
RP11-127O4.3 |
RP11-127O4.3 |
| chr9_-_123812542 | 1.20 |
ENST00000223642.1 |
C5 |
complement component 5 |
| chr7_-_14029283 | 1.20 |
ENST00000433547.1 ENST00000405192.2 |
ETV1 |
ets variant 1 |
| chr3_+_12392971 | 1.19 |
ENST00000287820.6 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr4_+_129730947 | 1.18 |
ENST00000452328.2 ENST00000504089.1 |
PHF17 |
jade family PHD finger 1 |
| chr13_+_76378305 | 1.18 |
ENST00000526371.1 ENST00000526528.1 |
LMO7 |
LIM domain 7 |
| chr5_-_124081008 | 1.18 |
ENST00000306315.5 |
ZNF608 |
zinc finger protein 608 |
| chr5_-_61031495 | 1.18 |
ENST00000506550.1 ENST00000512882.2 |
CTD-2170G1.2 |
CTD-2170G1.2 |
| chr1_+_155829286 | 1.15 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr16_+_86612112 | 1.14 |
ENST00000320241.3 |
FOXL1 |
forkhead box L1 |
| chr3_+_141105235 | 1.13 |
ENST00000503809.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr1_-_232598163 | 1.12 |
ENST00000308942.4 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr17_-_56082455 | 1.11 |
ENST00000578794.1 |
RP11-159D12.5 |
Uncharacterized protein |
| chr2_-_200322723 | 1.08 |
ENST00000417098.1 |
SATB2 |
SATB homeobox 2 |
| chr4_+_129730779 | 1.07 |
ENST00000226319.6 |
PHF17 |
jade family PHD finger 1 |
| chr3_+_141106643 | 1.07 |
ENST00000514251.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr3_+_148447887 | 1.06 |
ENST00000475347.1 ENST00000474935.1 ENST00000461609.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr1_+_65730385 | 1.04 |
ENST00000263441.7 ENST00000395325.3 |
DNAJC6 |
DnaJ (Hsp40) homolog, subfamily C, member 6 |
| chr5_-_19988339 | 1.03 |
ENST00000382275.1 |
CDH18 |
cadherin 18, type 2 |
| chr5_-_111093081 | 1.02 |
ENST00000453526.2 ENST00000509427.1 |
NREP |
neuronal regeneration related protein |
| chr10_-_33623310 | 1.00 |
ENST00000395995.1 ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1 |
neuropilin 1 |
| chr2_-_45236540 | 0.99 |
ENST00000303077.6 |
SIX2 |
SIX homeobox 2 |
| chr4_+_129730839 | 0.99 |
ENST00000511647.1 |
PHF17 |
jade family PHD finger 1 |
| chr2_+_152214098 | 0.98 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr2_+_33172012 | 0.98 |
ENST00000404816.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr3_+_69985792 | 0.98 |
ENST00000531774.1 |
MITF |
microphthalmia-associated transcription factor |
| chr1_+_223101757 | 0.97 |
ENST00000284476.6 |
DISP1 |
dispatched homolog 1 (Drosophila) |
| chr3_-_71353892 | 0.97 |
ENST00000484350.1 |
FOXP1 |
forkhead box P1 |
| chr5_-_41510725 | 0.96 |
ENST00000328457.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr5_-_41510656 | 0.96 |
ENST00000377801.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr2_+_33172221 | 0.95 |
ENST00000354476.3 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr10_-_92681033 | 0.95 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
| chr1_+_174844645 | 0.94 |
ENST00000486220.1 |
RABGAP1L |
RAB GTPase activating protein 1-like |
| chr5_-_111091948 | 0.94 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr6_-_152639479 | 0.94 |
ENST00000356820.4 |
SYNE1 |
spectrin repeat containing, nuclear envelope 1 |
| chr12_+_54422142 | 0.93 |
ENST00000243108.4 |
HOXC6 |
homeobox C6 |
| chr3_+_12330560 | 0.93 |
ENST00000397026.2 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr8_-_120685608 | 0.92 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr11_+_20044096 | 0.92 |
ENST00000533917.1 |
NAV2 |
neuron navigator 2 |
| chr13_+_36050881 | 0.91 |
ENST00000537702.1 |
NBEA |
neurobeachin |
| chr3_+_69915385 | 0.91 |
ENST00000314589.5 |
MITF |
microphthalmia-associated transcription factor |
| chr3_-_114790179 | 0.91 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr5_-_124080203 | 0.91 |
ENST00000504926.1 |
ZNF608 |
zinc finger protein 608 |
| chr10_+_69865866 | 0.91 |
ENST00000354393.2 |
MYPN |
myopalladin |
| chr5_+_59783540 | 0.90 |
ENST00000515734.2 |
PART1 |
prostate androgen-regulated transcript 1 (non-protein coding) |
| chr6_+_72926145 | 0.90 |
ENST00000425662.2 ENST00000453976.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr10_-_21786179 | 0.87 |
ENST00000377113.5 |
CASC10 |
cancer susceptibility candidate 10 |
| chr5_-_111093340 | 0.87 |
ENST00000508870.1 |
NREP |
neuronal regeneration related protein |
| chr8_-_93107696 | 0.86 |
ENST00000436581.2 ENST00000520583.1 ENST00000519061.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr17_-_39623681 | 0.85 |
ENST00000225899.3 |
KRT32 |
keratin 32 |
| chr8_+_70404996 | 0.84 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr11_+_12766583 | 0.84 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr16_-_3350614 | 0.84 |
ENST00000268674.2 |
TIGD7 |
tigger transposable element derived 7 |
| chr1_-_227505289 | 0.83 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr4_-_41884620 | 0.82 |
ENST00000504870.1 |
LINC00682 |
long intergenic non-protein coding RNA 682 |
| chr2_-_175462934 | 0.82 |
ENST00000392546.2 ENST00000436221.1 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr18_-_70532906 | 0.80 |
ENST00000299430.2 ENST00000397929.1 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr9_+_2158443 | 0.80 |
ENST00000302401.3 ENST00000324954.5 ENST00000423555.1 ENST00000382186.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr9_+_2015335 | 0.80 |
ENST00000349721.2 ENST00000357248.2 ENST00000450198.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr18_-_30353025 | 0.78 |
ENST00000359358.4 |
KLHL14 |
kelch-like family member 14 |
| chr5_-_111093406 | 0.78 |
ENST00000379671.3 |
NREP |
neuronal regeneration related protein |
| chr9_-_13165457 | 0.78 |
ENST00000542239.1 ENST00000538841.1 ENST00000433359.2 |
MPDZ |
multiple PDZ domain protein |
| chr1_+_104293028 | 0.77 |
ENST00000370079.3 |
AMY1C |
amylase, alpha 1C (salivary) |
| chr8_-_72268889 | 0.76 |
ENST00000388742.4 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr4_+_170581213 | 0.75 |
ENST00000507875.1 |
CLCN3 |
chloride channel, voltage-sensitive 3 |
| chr9_+_2158485 | 0.75 |
ENST00000417599.1 ENST00000382185.1 ENST00000382183.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr5_+_32711419 | 0.74 |
ENST00000265074.8 |
NPR3 |
natriuretic peptide receptor C/guanylate cyclase C (atrionatriuretic peptide receptor C) |
| chr5_+_174151536 | 0.73 |
ENST00000239243.6 ENST00000507785.1 |
MSX2 |
msh homeobox 2 |
| chr4_-_138453559 | 0.73 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr12_+_79258444 | 0.73 |
ENST00000261205.4 |
SYT1 |
synaptotagmin I |
| chr2_-_99279928 | 0.72 |
ENST00000414521.2 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr2_-_145277882 | 0.72 |
ENST00000465070.1 ENST00000444559.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr2_-_190044480 | 0.72 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr12_+_109785708 | 0.72 |
ENST00000310903.5 |
MYO1H |
myosin IH |
| chr11_-_96076334 | 0.72 |
ENST00000524717.1 |
MAML2 |
mastermind-like 2 (Drosophila) |
| chr16_+_53241854 | 0.71 |
ENST00000565803.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr2_-_158300556 | 0.71 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chr8_-_25281747 | 0.71 |
ENST00000421054.2 |
GNRH1 |
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr1_-_68698222 | 0.70 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr8_-_72268968 | 0.69 |
ENST00000388740.3 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr1_-_241799232 | 0.69 |
ENST00000366553.1 |
CHML |
choroideremia-like (Rab escort protein 2) |
| chr14_-_53331239 | 0.69 |
ENST00000553663.1 |
FERMT2 |
fermitin family member 2 |
| chr14_-_37051798 | 0.68 |
ENST00000258829.5 |
NKX2-8 |
NK2 homeobox 8 |
| chr1_+_61869748 | 0.67 |
ENST00000357977.5 |
NFIA |
nuclear factor I/A |
| chr19_+_2476116 | 0.67 |
ENST00000215631.4 ENST00000587345.1 |
GADD45B |
growth arrest and DNA-damage-inducible, beta |
| chr15_+_96869165 | 0.67 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr4_+_62067860 | 0.67 |
ENST00000514591.1 |
LPHN3 |
latrophilin 3 |
| chr5_+_140792614 | 0.66 |
ENST00000398610.2 |
PCDHGA10 |
protocadherin gamma subfamily A, 10 |
| chr12_+_79258547 | 0.66 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chr3_+_141103634 | 0.66 |
ENST00000507722.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr6_+_12717892 | 0.66 |
ENST00000379350.1 |
PHACTR1 |
phosphatase and actin regulator 1 |
| chr7_-_80141328 | 0.64 |
ENST00000398291.3 |
GNAT3 |
guanine nucleotide binding protein, alpha transducing 3 |
| chr4_+_88754113 | 0.64 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chrX_+_100333709 | 0.63 |
ENST00000372930.4 |
TMEM35 |
transmembrane protein 35 |
| chr3_+_152017924 | 0.63 |
ENST00000465907.2 ENST00000492948.1 ENST00000485509.1 ENST00000464596.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr11_+_120973375 | 0.63 |
ENST00000264037.2 |
TECTA |
tectorin alpha |
| chr3_-_71294304 | 0.62 |
ENST00000498215.1 |
FOXP1 |
forkhead box P1 |
| chr2_+_179318295 | 0.62 |
ENST00000442710.1 |
DFNB59 |
deafness, autosomal recessive 59 |
| chr7_+_90339169 | 0.62 |
ENST00000436577.2 |
CDK14 |
cyclin-dependent kinase 14 |
| chr1_-_68698197 | 0.61 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr1_-_104238912 | 0.61 |
ENST00000330330.5 |
AMY1B |
amylase, alpha 1B (salivary) |
| chr4_+_71457970 | 0.61 |
ENST00000322937.6 |
AMBN |
ameloblastin (enamel matrix protein) |
| chr5_+_140810132 | 0.61 |
ENST00000252085.3 |
PCDHGA12 |
protocadherin gamma subfamily A, 12 |
| chr12_+_60058458 | 0.60 |
ENST00000548610.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chrX_+_22050546 | 0.60 |
ENST00000379374.4 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr14_+_51955831 | 0.60 |
ENST00000356218.4 |
FRMD6 |
FERM domain containing 6 |
| chr5_-_121413974 | 0.59 |
ENST00000231004.4 |
LOX |
lysyl oxidase |
| chr17_-_39191107 | 0.59 |
ENST00000344363.5 |
KRTAP1-3 |
keratin associated protein 1-3 |
| chr3_+_63953415 | 0.59 |
ENST00000484332.1 |
ATXN7 |
ataxin 7 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 14.3 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 2.2 | 32.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 1.5 | 20.0 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 1.2 | 3.6 | GO:0048022 | negative regulation of melanin biosynthetic process(GO:0048022) negative regulation of secondary metabolite biosynthetic process(GO:1900377) |
| 0.7 | 2.1 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.7 | 2.0 | GO:0035283 | rhombomere 3 development(GO:0021569) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.6 | 1.9 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.6 | 1.8 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.6 | 1.8 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.6 | 5.6 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.5 | 3.8 | GO:1902897 | regulation of postsynaptic density protein 95 clustering(GO:1902897) |
| 0.5 | 2.1 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.5 | 1.6 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.5 | 1.4 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.5 | 2.3 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.5 | 2.3 | GO:0032625 | interleukin-21 production(GO:0032625) interleukin-21 secretion(GO:0072619) |
| 0.5 | 0.9 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.5 | 1.8 | GO:0072092 | ureteric bud invasion(GO:0072092) |
| 0.4 | 1.7 | GO:0003185 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.4 | 1.7 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.4 | 3.7 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.4 | 1.2 | GO:1990927 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.4 | 1.4 | GO:0009956 | radial pattern formation(GO:0009956) |
| 0.3 | 1.4 | GO:1900042 | positive regulation of interleukin-2 secretion(GO:1900042) |
| 0.3 | 1.3 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.3 | 2.9 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.3 | 1.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.2 | 1.0 | GO:0072137 | condensed mesenchymal cell proliferation(GO:0072137) |
| 0.2 | 0.7 | GO:0051795 | positive regulation of catagen(GO:0051795) activation of meiosis(GO:0090427) |
| 0.2 | 1.2 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.2 | 1.7 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.2 | 0.7 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 1.1 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) |
| 0.2 | 1.3 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.2 | 10.2 | GO:0030318 | melanocyte differentiation(GO:0030318) |
| 0.2 | 1.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 0.8 | GO:1904059 | regulation of locomotor rhythm(GO:1904059) |
| 0.2 | 5.8 | GO:0048935 | peripheral nervous system neuron differentiation(GO:0048934) peripheral nervous system neuron development(GO:0048935) |
| 0.2 | 0.6 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.2 | 1.0 | GO:1904835 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.2 | 3.2 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 1.8 | GO:0002740 | negative regulation of cytokine secretion involved in immune response(GO:0002740) |
| 0.2 | 0.8 | GO:0018032 | protein amidation(GO:0018032) |
| 0.2 | 0.6 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.2 | 0.7 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.2 | 1.6 | GO:0003025 | regulation of systemic arterial blood pressure by baroreceptor feedback(GO:0003025) |
| 0.2 | 3.2 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.2 | 1.0 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.2 | 0.8 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.2 | 0.2 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.2 | 2.7 | GO:0030207 | chondroitin sulfate catabolic process(GO:0030207) |
| 0.2 | 0.2 | GO:1901979 | regulation of inward rectifier potassium channel activity(GO:1901979) |
| 0.1 | 0.9 | GO:0048388 | endosomal lumen acidification(GO:0048388) |
| 0.1 | 0.4 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.6 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.1 | 0.9 | GO:2000795 | negative regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000795) |
| 0.1 | 0.7 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.6 | GO:0060023 | soft palate development(GO:0060023) |
| 0.1 | 1.0 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.1 | 0.6 | GO:0060353 | regulation of cell adhesion molecule production(GO:0060353) positive regulation of cell adhesion molecule production(GO:0060355) |
| 0.1 | 0.5 | GO:1904450 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.1 | 0.3 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.4 | GO:0060729 | intestinal epithelial structure maintenance(GO:0060729) |
| 0.1 | 0.3 | GO:0071879 | positive regulation of adrenergic receptor signaling pathway(GO:0071879) |
| 0.1 | 0.9 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.1 | 0.4 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.1 | 0.4 | GO:0061743 | motor learning(GO:0061743) |
| 0.1 | 0.4 | GO:0042427 | serotonin biosynthetic process(GO:0042427) |
| 0.1 | 0.5 | GO:1904383 | response to sodium phosphate(GO:1904383) |
| 0.1 | 1.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.1 | 0.4 | GO:0035262 | gonad morphogenesis(GO:0035262) |
| 0.1 | 0.9 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.1 | 2.0 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.1 | 1.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.9 | GO:0090292 | nuclear matrix anchoring at nuclear membrane(GO:0090292) |
| 0.1 | 0.2 | GO:2001202 | negative regulation of transforming growth factor-beta secretion(GO:2001202) |
| 0.1 | 5.3 | GO:0032728 | positive regulation of interferon-beta production(GO:0032728) |
| 0.1 | 0.6 | GO:0060385 | axonogenesis involved in innervation(GO:0060385) |
| 0.1 | 0.3 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.1 | 1.4 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.1 | 0.3 | GO:0097187 | dentinogenesis(GO:0097187) regulation of sodium-dependent phosphate transport(GO:2000118) |
| 0.1 | 0.4 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.1 | 0.6 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.1 | 0.3 | GO:0021650 | vestibulocochlear nerve morphogenesis(GO:0021648) vestibulocochlear nerve formation(GO:0021650) |
| 0.1 | 0.8 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.1 | 0.6 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 0.1 | GO:1900111 | positive regulation of histone H3-K9 dimethylation(GO:1900111) |
| 0.1 | 0.4 | GO:2001271 | negative regulation of cysteine-type endopeptidase activity involved in execution phase of apoptosis(GO:2001271) |
| 0.1 | 0.2 | GO:1902036 | regulation of hematopoietic stem cell differentiation(GO:1902036) |
| 0.1 | 0.2 | GO:0046543 | development of secondary female sexual characteristics(GO:0046543) |
| 0.1 | 0.3 | GO:0071699 | olfactory placode formation(GO:0030910) olfactory placode development(GO:0071698) olfactory placode morphogenesis(GO:0071699) |
| 0.1 | 0.3 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.1 | 0.7 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.1 | 0.7 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.3 | GO:0021812 | neuronal-glial interaction involved in cerebral cortex radial glia guided migration(GO:0021812) |
| 0.1 | 0.6 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.1 | 0.6 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.1 | 1.4 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.1 | 1.2 | GO:0060539 | diaphragm development(GO:0060539) |
| 0.1 | 0.9 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.1 | 0.8 | GO:0048739 | cardiac muscle fiber development(GO:0048739) |
| 0.1 | 1.0 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.1 | 0.4 | GO:1903527 | positive regulation of membrane tubulation(GO:1903527) |
| 0.1 | 0.4 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.1 | 0.1 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.1 | 0.1 | GO:0061373 | mammillary body development(GO:0021767) mammillary axonal complex development(GO:0061373) |
| 0.1 | 0.5 | GO:1901843 | positive regulation of high voltage-gated calcium channel activity(GO:1901843) |
| 0.1 | 0.2 | GO:2000301 | negative regulation of synaptic vesicle exocytosis(GO:2000301) |
| 0.1 | 0.7 | GO:0002158 | osteoclast proliferation(GO:0002158) |
| 0.1 | 0.2 | GO:0035701 | hematopoietic stem cell migration(GO:0035701) |
| 0.1 | 0.2 | GO:0051180 | vitamin transport(GO:0051180) |
| 0.1 | 0.9 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.1 | 0.8 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.1 | 0.4 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.1 | 0.5 | GO:0001886 | endothelial cell morphogenesis(GO:0001886) |
| 0.1 | 1.2 | GO:0070168 | negative regulation of bone mineralization(GO:0030502) negative regulation of biomineral tissue development(GO:0070168) |
| 0.1 | 0.1 | GO:2000645 | negative regulation of receptor catabolic process(GO:2000645) |
| 0.1 | 0.6 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 | 0.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.1 | 0.1 | GO:2000538 | T-helper 1 cell activation(GO:0035711) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.2 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.1 | 0.5 | GO:0010724 | regulation of definitive erythrocyte differentiation(GO:0010724) |
| 0.1 | 3.1 | GO:0045599 | negative regulation of fat cell differentiation(GO:0045599) |
| 0.1 | 0.4 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.1 | 1.1 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.1 | 0.4 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.6 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.1 | 0.7 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 0.2 | GO:0003358 | noradrenergic neuron development(GO:0003358) |
| 0.1 | 0.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.0 | 0.3 | GO:0043418 | homocysteine catabolic process(GO:0043418) |
| 0.0 | 0.2 | GO:0086021 | SA node cell to atrial cardiac muscle cell communication by electrical coupling(GO:0086021) |
| 0.0 | 2.2 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.6 | GO:0060272 | embryonic skeletal joint morphogenesis(GO:0060272) |
| 0.0 | 0.2 | GO:0045872 | regulation of rhodopsin gene expression(GO:0007468) positive regulation of rhodopsin gene expression(GO:0045872) |
| 0.0 | 0.1 | GO:0090119 | vesicle-mediated cholesterol transport(GO:0090119) |
| 0.0 | 0.4 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 1.7 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.4 | GO:0043402 | glucocorticoid mediated signaling pathway(GO:0043402) |
| 0.0 | 4.8 | GO:0001578 | microtubule bundle formation(GO:0001578) |
| 0.0 | 0.2 | GO:0098728 | germ-line stem cell division(GO:0042078) male germ-line stem cell asymmetric division(GO:0048133) germline stem cell asymmetric division(GO:0098728) |
| 0.0 | 0.0 | GO:0070432 | regulation of nucleotide-binding oligomerization domain containing 2 signaling pathway(GO:0070432) |
| 0.0 | 0.2 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.8 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.5 | GO:0030210 | heparin metabolic process(GO:0030202) heparin biosynthetic process(GO:0030210) |
| 0.0 | 0.2 | GO:0061113 | pancreas morphogenesis(GO:0061113) |
| 0.0 | 0.5 | GO:0090324 | negative regulation of oxidative phosphorylation(GO:0090324) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0043324 | eye pigment biosynthetic process(GO:0006726) eye pigment metabolic process(GO:0042441) pigment metabolic process involved in developmental pigmentation(GO:0043324) pigment metabolic process involved in pigmentation(GO:0043474) |
| 0.0 | 0.2 | GO:1903593 | regulation of histamine secretion by mast cell(GO:1903593) |
| 0.0 | 0.3 | GO:0045656 | negative regulation of monocyte differentiation(GO:0045656) |
| 0.0 | 0.1 | GO:0060164 | regulation of timing of neuron differentiation(GO:0060164) |
| 0.0 | 2.5 | GO:0014047 | glutamate secretion(GO:0014047) |
| 0.0 | 1.5 | GO:0001937 | negative regulation of endothelial cell proliferation(GO:0001937) |
| 0.0 | 0.5 | GO:0060628 | regulation of ER to Golgi vesicle-mediated transport(GO:0060628) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 1.4 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.4 | GO:0007016 | cytoskeletal anchoring at plasma membrane(GO:0007016) |
| 0.0 | 1.0 | GO:1902895 | positive regulation of pri-miRNA transcription from RNA polymerase II promoter(GO:1902895) |
| 0.0 | 0.3 | GO:0034214 | protein hexamerization(GO:0034214) |
| 0.0 | 4.6 | GO:0007286 | spermatid development(GO:0007286) |
| 0.0 | 0.7 | GO:1900745 | positive regulation of p38MAPK cascade(GO:1900745) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
| 0.0 | 0.1 | GO:1900245 | positive regulation of MDA-5 signaling pathway(GO:1900245) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.3 | GO:0034197 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.7 | GO:0030325 | adrenal gland development(GO:0030325) |
| 0.0 | 0.1 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 1.1 | GO:0070884 | regulation of calcineurin-NFAT signaling cascade(GO:0070884) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.6 | GO:0072189 | ureter development(GO:0072189) |
| 0.0 | 0.1 | GO:1903367 | positive regulation of fear response(GO:1903367) positive regulation of behavioral fear response(GO:2000987) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 0.1 | GO:1902882 | regulation of response to oxidative stress(GO:1902882) |
| 0.0 | 0.0 | GO:0021730 | trigeminal sensory nucleus development(GO:0021730) principal sensory nucleus of trigeminal nerve development(GO:0021740) |
| 0.0 | 0.1 | GO:0009946 | proximal/distal axis specification(GO:0009946) neuroblast differentiation(GO:0014016) |
| 0.0 | 0.5 | GO:0001696 | gastric acid secretion(GO:0001696) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 2.6 | GO:0032508 | DNA duplex unwinding(GO:0032508) |
| 0.0 | 0.1 | GO:0006295 | nucleotide-excision repair, DNA incision, 3'-to lesion(GO:0006295) |
| 0.0 | 0.2 | GO:0032525 | somite rostral/caudal axis specification(GO:0032525) |
| 0.0 | 0.0 | GO:0023016 | signal transduction by trans-phosphorylation(GO:0023016) |
| 0.0 | 0.1 | GO:0021846 | cell proliferation in forebrain(GO:0021846) |
| 0.0 | 0.3 | GO:0071670 | smooth muscle cell chemotaxis(GO:0071670) |
| 0.0 | 5.6 | GO:0006275 | regulation of DNA replication(GO:0006275) |
| 0.0 | 0.4 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0034238 | macrophage fusion(GO:0034238) regulation of macrophage fusion(GO:0034239) positive regulation of macrophage fusion(GO:0034241) |
| 0.0 | 0.3 | GO:0045838 | positive regulation of membrane potential(GO:0045838) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.7 | GO:0090162 | establishment of epithelial cell polarity(GO:0090162) |
| 0.0 | 0.2 | GO:0048341 | paraxial mesoderm formation(GO:0048341) |
| 0.0 | 0.2 | GO:0048278 | vesicle docking involved in exocytosis(GO:0006904) membrane docking(GO:0022406) vesicle docking(GO:0048278) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.4 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.0 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:0097210 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.4 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.4 | GO:0097067 | cellular response to thyroid hormone stimulus(GO:0097067) |
| 0.0 | 0.6 | GO:0008045 | motor neuron axon guidance(GO:0008045) |
| 0.0 | 0.2 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.1 | GO:0006627 | protein processing involved in protein targeting to mitochondrion(GO:0006627) |
| 0.0 | 1.8 | GO:0042147 | retrograde transport, endosome to Golgi(GO:0042147) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0035150 | regulation of tube size(GO:0035150) regulation of blood vessel size(GO:0050880) |
| 0.0 | 0.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.1 | GO:0060736 | prostate gland growth(GO:0060736) |
| 0.0 | 0.3 | GO:0031269 | pseudopodium assembly(GO:0031269) |
| 0.0 | 0.6 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.1 | GO:0002863 | positive regulation of inflammatory response to antigenic stimulus(GO:0002863) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.9 | GO:0007528 | neuromuscular junction development(GO:0007528) |
| 0.0 | 0.8 | GO:0019228 | neuronal action potential(GO:0019228) |
| 0.0 | 0.3 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0007494 | midgut development(GO:0007494) |
| 0.0 | 0.1 | GO:0060484 | lung-associated mesenchyme development(GO:0060484) |
| 0.0 | 0.9 | GO:0008542 | visual learning(GO:0008542) |
| 0.0 | 0.5 | GO:0002323 | natural killer cell activation involved in immune response(GO:0002323) |
| 0.0 | 0.2 | GO:1904776 | regulation of protein localization to cell cortex(GO:1904776) positive regulation of protein localization to cell cortex(GO:1904778) |
| 0.0 | 0.1 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 7.1 | GO:0000209 | protein polyubiquitination(GO:0000209) |
| 0.0 | 0.3 | GO:0033622 | integrin activation(GO:0033622) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.2 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.1 | GO:0019227 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.4 | GO:0043968 | histone H2A acetylation(GO:0043968) |
| 0.0 | 1.2 | GO:0050830 | defense response to Gram-positive bacterium(GO:0050830) |
| 0.0 | 0.1 | GO:0021554 | optic nerve development(GO:0021554) |
| 0.0 | 6.0 | GO:0019216 | regulation of lipid metabolic process(GO:0019216) |
| 0.0 | 2.6 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.0 | GO:2000628 | regulation of miRNA metabolic process(GO:2000628) |
| 0.0 | 0.2 | GO:0014065 | phosphatidylinositol 3-kinase signaling(GO:0014065) |
| 0.0 | 0.1 | GO:0046548 | retinal rod cell development(GO:0046548) |
| 0.0 | 0.4 | GO:0006123 | mitochondrial electron transport, cytochrome c to oxygen(GO:0006123) |
| 0.0 | 0.3 | GO:0010569 | regulation of double-strand break repair via homologous recombination(GO:0010569) |
| 0.0 | 0.1 | GO:0043154 | negative regulation of cysteine-type endopeptidase activity involved in apoptotic process(GO:0043154) |
| 0.0 | 0.3 | GO:0032204 | regulation of telomere maintenance(GO:0032204) |
| 0.0 | 0.1 | GO:0019566 | arabinose metabolic process(GO:0019566) L-arabinose metabolic process(GO:0046373) |
| 0.0 | 0.4 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.2 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.1 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.2 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.0 | GO:0042320 | regulation of circadian sleep/wake cycle, REM sleep(GO:0042320) positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0035873 | lactate transport(GO:0015727) lactate transmembrane transport(GO:0035873) |
| 0.0 | 0.2 | GO:0032793 | positive regulation of CREB transcription factor activity(GO:0032793) |
| 0.0 | 0.0 | GO:0061009 | common bile duct development(GO:0061009) |
| 0.0 | 0.1 | GO:0097094 | craniofacial suture morphogenesis(GO:0097094) |
| 0.0 | 0.2 | GO:0075522 | IRES-dependent viral translational initiation(GO:0075522) |
| 0.0 | 0.1 | GO:0051603 | proteolysis involved in cellular protein catabolic process(GO:0051603) |
| 0.0 | 0.0 | GO:0038188 | cholecystokinin signaling pathway(GO:0038188) |
| 0.0 | 0.2 | GO:0016577 | histone demethylation(GO:0016577) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 2.4 | 32.9 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 2.1 | 21.2 | GO:0071953 | elastic fiber(GO:0071953) |
| 1.2 | 4.7 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.5 | 2.0 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.4 | 1.9 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.3 | 1.2 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.2 | 0.7 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.2 | 1.2 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.2 | 5.1 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 1.4 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.2 | 0.6 | GO:0042585 | germinal vesicle(GO:0042585) |
| 0.2 | 0.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.2 | 4.0 | GO:0071564 | npBAF complex(GO:0071564) |
| 0.1 | 0.7 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.1 | 1.1 | GO:0005579 | membrane attack complex(GO:0005579) |
| 0.1 | 0.5 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.1 | 0.4 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.1 | 2.6 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.1 | 1.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.3 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.1 | 1.0 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.5 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.1 | 0.9 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.1 | 0.6 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.1 | 0.7 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.1 | 0.9 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.1 | 0.5 | GO:0031519 | PcG protein complex(GO:0031519) |
| 0.1 | 1.1 | GO:0030478 | actin cap(GO:0030478) |
| 0.1 | 1.9 | GO:0010369 | chromocenter(GO:0010369) |
| 0.1 | 9.3 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.1 | 1.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.1 | 0.5 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 1.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.1 | 1.8 | GO:0032839 | dendrite cytoplasm(GO:0032839) |
| 0.1 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.1 | 0.9 | GO:0034992 | microtubule organizing center attachment site(GO:0034992) LINC complex(GO:0034993) |
| 0.1 | 0.9 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.1 | 0.2 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 1.1 | GO:0043220 | Schmidt-Lanterman incisure(GO:0043220) |
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.6 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 0.4 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.8 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.7 | GO:0042589 | zymogen granule membrane(GO:0042589) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.6 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 2.3 | GO:0043198 | dendritic shaft(GO:0043198) |
| 0.0 | 0.2 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 7.6 | GO:0072562 | blood microparticle(GO:0072562) |
| 0.0 | 0.6 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.3 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.3 | GO:0008091 | spectrin(GO:0008091) |
| 0.0 | 0.1 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 1.0 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.4 | GO:0044300 | cerebellar mossy fiber(GO:0044300) |
| 0.0 | 4.9 | GO:0016363 | nuclear matrix(GO:0016363) |
| 0.0 | 2.8 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 2.3 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 1.0 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.5 | GO:0031258 | lamellipodium membrane(GO:0031258) |
| 0.0 | 0.3 | GO:0036157 | outer dynein arm(GO:0036157) |
| 0.0 | 0.2 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.3 | GO:0070776 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 1.3 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.0 | GO:0060171 | stereocilium membrane(GO:0060171) |
| 0.0 | 1.5 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 0.4 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 1.2 | GO:0005876 | spindle microtubule(GO:0005876) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.4 | GO:0002102 | podosome(GO:0002102) |
| 0.0 | 1.0 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 1.6 | GO:0005884 | actin filament(GO:0005884) |
| 0.0 | 0.1 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.7 | GO:0031430 | M band(GO:0031430) |
| 0.0 | 0.4 | GO:0045277 | respiratory chain complex IV(GO:0045277) |
| 0.0 | 0.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.2 | GO:0001917 | photoreceptor inner segment(GO:0001917) |
| 0.0 | 0.1 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.4 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.1 | GO:0043679 | axon terminus(GO:0043679) |
| 0.0 | 0.1 | GO:0005677 | chromatin silencing complex(GO:0005677) |
| 0.0 | 0.5 | GO:0034399 | nuclear periphery(GO:0034399) |
| 0.0 | 4.0 | GO:0005770 | late endosome(GO:0005770) |
| 0.0 | 0.7 | GO:0016235 | aggresome(GO:0016235) |
| 0.0 | 3.1 | GO:0005802 | trans-Golgi network(GO:0005802) |
| 0.0 | 0.1 | GO:0044305 | calyx of Held(GO:0044305) |
| 0.0 | 5.8 | GO:0016607 | nuclear speck(GO:0016607) |
| 0.0 | 0.0 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 1.2 | GO:0001669 | acrosomal vesicle(GO:0001669) |
| 0.0 | 0.1 | GO:0005583 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 5.6 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.8 | 3.1 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.7 | 2.1 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.6 | 3.2 | GO:0031697 | beta-1 adrenergic receptor binding(GO:0031697) |
| 0.4 | 1.8 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.4 | 1.8 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.4 | 1.7 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.4 | 32.1 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.4 | 1.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.3 | 2.0 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.3 | 1.0 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.3 | 15.1 | GO:0001205 | transcriptional activator activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001205) |
| 0.3 | 1.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.3 | 1.6 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.3 | 2.3 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.2 | 9.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.2 | 2.0 | GO:0043559 | insulin binding(GO:0043559) |
| 0.2 | 1.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.2 | 0.4 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.2 | 0.8 | GO:0004504 | peptidylglycine monooxygenase activity(GO:0004504) peptidylamidoglycolate lyase activity(GO:0004598) |
| 0.2 | 1.5 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.2 | 0.5 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.2 | 0.6 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.9 | GO:0072320 | volume-sensitive chloride channel activity(GO:0072320) |
| 0.1 | 1.0 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.4 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.1 | 0.9 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.7 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.1 | 0.5 | GO:0016160 | amylase activity(GO:0016160) |
| 0.1 | 0.7 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.6 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.1 | 3.6 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 0.5 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.1 | 0.6 | GO:0008113 | peptide-methionine (S)-S-oxide reductase activity(GO:0008113) |
| 0.1 | 0.8 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.1 | 1.3 | GO:0031852 | mu-type opioid receptor binding(GO:0031852) |
| 0.1 | 0.6 | GO:0005534 | galactose binding(GO:0005534) |
| 0.1 | 0.2 | GO:0086020 | gap junction channel activity involved in SA node cell-atrial cardiac muscle cell electrical coupling(GO:0086020) |
| 0.1 | 1.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 0.4 | GO:0031690 | adrenergic receptor binding(GO:0031690) |
| 0.1 | 1.2 | GO:0036122 | BMP binding(GO:0036122) |
| 0.1 | 1.0 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.1 | 0.4 | GO:0050693 | LBD domain binding(GO:0050693) |
| 0.1 | 0.5 | GO:0015016 | [heparan sulfate]-glucosamine N-sulfotransferase activity(GO:0015016) |
| 0.1 | 1.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.1 | 4.0 | GO:0001106 | RNA polymerase II transcription corepressor activity(GO:0001106) |
| 0.1 | 13.2 | GO:0005178 | integrin binding(GO:0005178) |
| 0.1 | 0.5 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.1 | 1.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.1 | 0.4 | GO:0038051 | glucocorticoid receptor activity(GO:0004883) glucocorticoid-activated RNA polymerase II transcription factor binding transcription factor activity(GO:0038051) |
| 0.1 | 1.0 | GO:0015197 | peptide transporter activity(GO:0015197) |
| 0.1 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.1 | 0.3 | GO:0004528 | phosphodiesterase I activity(GO:0004528) |
| 0.1 | 3.6 | GO:0001105 | RNA polymerase II transcription coactivator activity(GO:0001105) |
| 0.1 | 1.4 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.1 | 0.4 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.1 | 0.4 | GO:0048101 | calcium- and calmodulin-regulated 3',5'-cyclic-GMP phosphodiesterase activity(GO:0048101) |
| 0.1 | 19.3 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.1 | 0.7 | GO:0005078 | MAP-kinase scaffold activity(GO:0005078) |
| 0.1 | 1.7 | GO:0001223 | transcription coactivator binding(GO:0001223) |
| 0.1 | 1.4 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.1 | 0.3 | GO:0055077 | gap junction hemi-channel activity(GO:0055077) |
| 0.1 | 0.5 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.1 | 0.2 | GO:0070698 | type I activin receptor binding(GO:0070698) |
| 0.1 | 2.6 | GO:0004003 | ATP-dependent DNA helicase activity(GO:0004003) |
| 0.1 | 0.2 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.1 | 0.3 | GO:1904315 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 1.5 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.4 | GO:0016714 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced pteridine as one donor, and incorporation of one atom of oxygen(GO:0016714) |
| 0.0 | 1.5 | GO:0000900 | translation repressor activity, nucleic acid binding(GO:0000900) |
| 0.0 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.0 | 0.2 | GO:1990829 | C-rich single-stranded DNA binding(GO:1990829) |
| 0.0 | 0.4 | GO:0005004 | GPI-linked ephrin receptor activity(GO:0005004) |
| 0.0 | 0.3 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.0 | 0.3 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 1.4 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 1.1 | GO:0038187 | signaling pattern recognition receptor activity(GO:0008329) pattern recognition receptor activity(GO:0038187) |
| 0.0 | 0.7 | GO:0030247 | pattern binding(GO:0001871) polysaccharide binding(GO:0030247) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.1 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.5 | GO:0015038 | glutathione disulfide oxidoreductase activity(GO:0015038) |
| 0.0 | 1.1 | GO:0051371 | muscle alpha-actinin binding(GO:0051371) |
| 0.0 | 0.4 | GO:0042043 | neurexin family protein binding(GO:0042043) |
| 0.0 | 1.7 | GO:0046875 | ephrin receptor binding(GO:0046875) |
| 0.0 | 5.8 | GO:0004386 | helicase activity(GO:0004386) |
| 0.0 | 0.9 | GO:0005521 | lamin binding(GO:0005521) |
| 0.0 | 0.1 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.2 | GO:0015277 | kainate selective glutamate receptor activity(GO:0015277) |
| 0.0 | 2.0 | GO:0050681 | androgen receptor binding(GO:0050681) |
| 0.0 | 0.4 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.9 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.5 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.4 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 0.3 | GO:0015321 | sodium-dependent phosphate transmembrane transporter activity(GO:0015321) |
| 0.0 | 0.9 | GO:0005523 | tropomyosin binding(GO:0005523) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 2.2 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.0 | 0.6 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.6 | GO:0031683 | G-protein beta/gamma-subunit complex binding(GO:0031683) |
| 0.0 | 1.3 | GO:0004693 | cyclin-dependent protein serine/threonine kinase activity(GO:0004693) |
| 0.0 | 2.4 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.2 | GO:0034452 | dynactin binding(GO:0034452) |
| 0.0 | 0.6 | GO:0005248 | voltage-gated sodium channel activity(GO:0005248) voltage-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1905030) |
| 0.0 | 0.3 | GO:0008271 | secondary active sulfate transmembrane transporter activity(GO:0008271) |
| 0.0 | 0.2 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 0.2 | GO:0042731 | PH domain binding(GO:0042731) |
| 0.0 | 1.2 | GO:0005518 | collagen binding(GO:0005518) |
| 0.0 | 0.3 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.6 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 1.4 | GO:0070063 | RNA polymerase binding(GO:0070063) |
| 0.0 | 1.0 | GO:0048487 | beta-tubulin binding(GO:0048487) |
| 0.0 | 0.3 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 9.6 | GO:0030695 | GTPase regulator activity(GO:0030695) |
| 0.0 | 0.1 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 4.2 | GO:0003714 | transcription corepressor activity(GO:0003714) |
| 0.0 | 0.9 | GO:0008009 | chemokine activity(GO:0008009) |
| 0.0 | 0.4 | GO:0005547 | phosphatidylinositol-3,4,5-trisphosphate binding(GO:0005547) |
| 0.0 | 0.7 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.6 | GO:0001671 | ATPase activator activity(GO:0001671) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) methylumbelliferyl-acetate deacetylase activity(GO:0047374) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 1.7 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.2 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 2.0 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0017151 | DEAD/H-box RNA helicase binding(GO:0017151) transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.2 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.6 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.0 | 0.2 | GO:0051378 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.2 | GO:0047035 | alcohol dehydrogenase (NAD) activity(GO:0004022) testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.0 | GO:0003918 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.4 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.9 | GO:0004402 | histone acetyltransferase activity(GO:0004402) |
| 0.0 | 0.2 | GO:0070402 | NADPH binding(GO:0070402) |
| 0.0 | 0.2 | GO:0019966 | interleukin-1 binding(GO:0019966) |
| 0.0 | 0.5 | GO:0051019 | mitogen-activated protein kinase binding(GO:0051019) |
| 0.0 | 0.1 | GO:0003691 | double-stranded telomeric DNA binding(GO:0003691) |
| 0.0 | 0.1 | GO:0015129 | lactate transmembrane transporter activity(GO:0015129) |
| 0.0 | 0.1 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.0 | 0.1 | GO:0046556 | alpha-L-arabinofuranosidase activity(GO:0046556) |
| 0.0 | 0.3 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0004974 | leukotriene receptor activity(GO:0004974) |
| 0.0 | 0.3 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.3 | GO:0043395 | heparan sulfate proteoglycan binding(GO:0043395) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:1902282 | voltage-gated potassium channel activity involved in ventricular cardiac muscle cell action potential repolarization(GO:1902282) |
| 0.0 | 0.3 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.2 | GO:0008381 | mechanically-gated ion channel activity(GO:0008381) mechanically gated channel activity(GO:0022833) |
| 0.0 | 0.9 | GO:0008201 | heparin binding(GO:0008201) |
| 0.0 | 0.4 | GO:0015002 | cytochrome-c oxidase activity(GO:0004129) heme-copper terminal oxidase activity(GO:0015002) oxidoreductase activity, acting on a heme group of donors, oxygen as acceptor(GO:0016676) |
| 0.0 | 0.0 | GO:0004951 | cholecystokinin receptor activity(GO:0004951) |
| 0.0 | 0.3 | GO:0030374 | ligand-dependent nuclear receptor transcription coactivator activity(GO:0030374) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 40.1 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.2 | 6.1 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.2 | 12.5 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
| 0.1 | 3.6 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.1 | 2.0 | PID TCR CALCIUM PATHWAY | Calcium signaling in the CD4+ TCR pathway |
| 0.1 | 27.4 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.1 | 4.3 | PID LKB1 PATHWAY | LKB1 signaling events |
| 0.1 | 4.2 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 1.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 3.6 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.3 | ST GRANULE CELL SURVIVAL PATHWAY | Granule Cell Survival Pathway is a specific case of more general PAC1 Receptor Pathway. |
| 0.0 | 3.0 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 1.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 2.9 | PID NFAT TFPATHWAY | Calcineurin-regulated NFAT-dependent transcription in lymphocytes |
| 0.0 | 1.4 | ST ADRENERGIC | Adrenergic Pathway |
| 0.0 | 0.7 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
| 0.0 | 1.8 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 1.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 2.6 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 0.1 | ST DIFFERENTIATION PATHWAY IN PC12 CELLS | Differentiation Pathway in PC12 Cells; this is a specific case of PAC1 Receptor Pathway. |
| 0.0 | 0.5 | PID REELIN PATHWAY | Reelin signaling pathway |
| 0.0 | 0.5 | PID RB 1PATHWAY | Regulation of retinoblastoma protein |
| 0.0 | 0.1 | PID S1P S1P2 PATHWAY | S1P2 pathway |
| 0.0 | 0.7 | PID AURORA A PATHWAY | Aurora A signaling |
| 0.0 | 0.8 | PID INTEGRIN1 PATHWAY | Beta1 integrin cell surface interactions |
| 0.0 | 1.1 | PID TGFBR PATHWAY | TGF-beta receptor signaling |
| 0.0 | 0.5 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
| 0.0 | 0.4 | PID ARF 3PATHWAY | Arf1 pathway |
| 0.0 | 0.6 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 1.0 | PID REG GR PATHWAY | Glucocorticoid receptor regulatory network |
| 0.0 | 0.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.0 | 0.3 | ST JNK MAPK PATHWAY | JNK MAPK Pathway |
| 0.0 | 0.8 | PID FCER1 PATHWAY | Fc-epsilon receptor I signaling in mast cells |
| 0.0 | 0.3 | PID CIRCADIAN PATHWAY | Circadian rhythm pathway |
| 0.0 | 0.3 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.4 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 0.3 | PID VEGFR1 2 PATHWAY | Signaling events mediated by VEGFR1 and VEGFR2 |
| 0.0 | 0.1 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.3 | PID EPHA FWDPATHWAY | EPHA forward signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.3 | 35.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.2 | 4.4 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.2 | 3.2 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.2 | 3.1 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |
| 0.2 | 2.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 3.8 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.1 | 3.4 | REACTOME NEPHRIN INTERACTIONS | Genes involved in Nephrin interactions |
| 0.1 | 3.5 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.1 | 1.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.1 | 0.9 | REACTOME NOTCH HLH TRANSCRIPTION PATHWAY | Genes involved in Notch-HLH transcription pathway |
| 0.1 | 1.3 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.1 | 2.0 | REACTOME YAP1 AND WWTR1 TAZ STIMULATED GENE EXPRESSION | Genes involved in YAP1- and WWTR1 (TAZ)-stimulated gene expression |
| 0.0 | 0.3 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 3.7 | REACTOME TRANSCRIPTIONAL REGULATION OF WHITE ADIPOCYTE DIFFERENTIATION | Genes involved in Transcriptional Regulation of White Adipocyte Differentiation |
| 0.0 | 1.0 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.7 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 2.3 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 1.6 | REACTOME CIRCADIAN CLOCK | Genes involved in Circadian Clock |
| 0.0 | 0.4 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |
| 0.0 | 0.5 | REACTOME GABA SYNTHESIS RELEASE REUPTAKE AND DEGRADATION | Genes involved in GABA synthesis, release, reuptake and degradation |
| 0.0 | 0.9 | REACTOME RNA POL III TRANSCRIPTION TERMINATION | Genes involved in RNA Polymerase III Transcription Termination |
| 0.0 | 1.0 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.5 | REACTOME CREB PHOSPHORYLATION THROUGH THE ACTIVATION OF CAMKII | Genes involved in CREB phosphorylation through the activation of CaMKII |
| 0.0 | 0.7 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.4 | REACTOME ACYL CHAIN REMODELLING OF PG | Genes involved in Acyl chain remodelling of PG |
| 0.0 | 0.4 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.7 | REACTOME CASPASE MEDIATED CLEAVAGE OF CYTOSKELETAL PROTEINS | Genes involved in Caspase-mediated cleavage of cytoskeletal proteins |
| 0.0 | 0.3 | REACTOME AMINE LIGAND BINDING RECEPTORS | Genes involved in Amine ligand-binding receptors |
| 0.0 | 3.9 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.5 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.5 | REACTOME SEMA4D IN SEMAPHORIN SIGNALING | Genes involved in Sema4D in semaphorin signaling |
| 0.0 | 0.8 | REACTOME SMOOTH MUSCLE CONTRACTION | Genes involved in Smooth Muscle Contraction |
| 0.0 | 0.8 | REACTOME COMPLEMENT CASCADE | Genes involved in Complement cascade |
| 0.0 | 0.5 | REACTOME SYNTHESIS AND INTERCONVERSION OF NUCLEOTIDE DI AND TRIPHOSPHATES | Genes involved in Synthesis and interconversion of nucleotide di- and triphosphates |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN LIKE GROWTH FACTOR IGF ACTIVITY BY INSULIN LIKE GROWTH FACTOR BINDING PROTEINS IGFBPS | Genes involved in Regulation of Insulin-like Growth Factor (IGF) Activity by Insulin-like Growth Factor Binding Proteins (IGFBPs) |
| 0.0 | 0.3 | REACTOME HS GAG BIOSYNTHESIS | Genes involved in HS-GAG biosynthesis |
| 0.0 | 0.2 | REACTOME SEROTONIN RECEPTORS | Genes involved in Serotonin receptors |
| 0.0 | 0.1 | REACTOME CD28 DEPENDENT VAV1 PATHWAY | Genes involved in CD28 dependent Vav1 pathway |
| 0.0 | 0.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 0.2 | REACTOME G ALPHA1213 SIGNALLING EVENTS | Genes involved in G alpha (12/13) signalling events |
| 0.0 | 0.2 | REACTOME SPRY REGULATION OF FGF SIGNALING | Genes involved in Spry regulation of FGF signaling |
| 0.0 | 0.2 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.4 | REACTOME EGFR DOWNREGULATION | Genes involved in EGFR downregulation |