Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
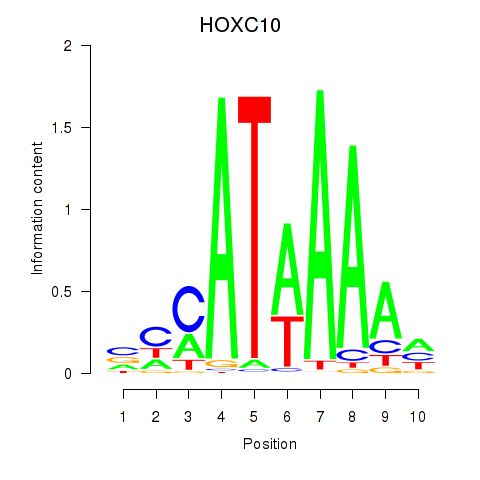
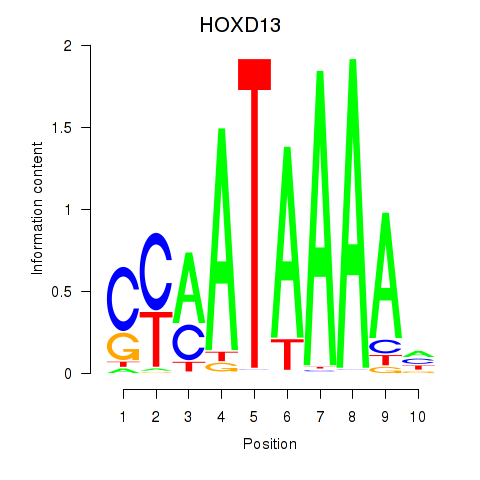
Results for HOXC10_HOXD13
Z-value: 0.77
Transcription factors associated with HOXC10_HOXD13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC10
|
ENSG00000180818.4 | HOXC10 |
|
HOXD13
|
ENSG00000128714.5 | HOXD13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC10 | hg19_v2_chr12_+_54378923_54378970 | 0.30 | 4.6e-01 | Click! |
| HOXD13 | hg19_v2_chr2_+_176957619_176957619 | 0.28 | 5.1e-01 | Click! |
Activity profile of HOXC10_HOXD13 motif
Sorted Z-values of HOXC10_HOXD13 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC10_HOXD13
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chrX_+_65382433 | 0.41 |
ENST00000374727.3 |
HEPH |
hephaestin |
| chr9_-_133814527 | 0.33 |
ENST00000451466.1 |
FIBCD1 |
fibrinogen C domain containing 1 |
| chr17_-_46806540 | 0.29 |
ENST00000290295.7 |
HOXB13 |
homeobox B13 |
| chr6_+_127898312 | 0.29 |
ENST00000329722.7 |
C6orf58 |
chromosome 6 open reading frame 58 |
| chr5_+_147582348 | 0.28 |
ENST00000514389.1 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr6_+_32812568 | 0.22 |
ENST00000414474.1 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr19_+_46367518 | 0.21 |
ENST00000302177.2 |
FOXA3 |
forkhead box A3 |
| chrX_+_65384052 | 0.21 |
ENST00000336279.5 ENST00000458621.1 |
HEPH |
hephaestin |
| chrX_+_65384182 | 0.20 |
ENST00000441993.2 ENST00000419594.1 |
HEPH |
hephaestin |
| chr10_+_60759378 | 0.20 |
ENST00000432535.1 |
LINC00844 |
long intergenic non-protein coding RNA 844 |
| chrX_+_105937068 | 0.19 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr1_+_153003671 | 0.18 |
ENST00000307098.4 |
SPRR1B |
small proline-rich protein 1B |
| chr1_-_9129895 | 0.17 |
ENST00000473209.1 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr2_-_216878305 | 0.17 |
ENST00000263268.6 |
MREG |
melanoregulin |
| chr6_+_72922505 | 0.16 |
ENST00000401910.3 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr6_+_72922590 | 0.15 |
ENST00000523963.1 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr20_+_58179582 | 0.15 |
ENST00000371015.1 ENST00000395639.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr18_-_52989217 | 0.15 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr8_+_52730143 | 0.14 |
ENST00000415643.1 |
AC090186.1 |
Uncharacterized protein |
| chr8_+_31497271 | 0.14 |
ENST00000520407.1 |
NRG1 |
neuregulin 1 |
| chr7_+_22766766 | 0.14 |
ENST00000426291.1 ENST00000401651.1 ENST00000258743.5 ENST00000420258.2 ENST00000407492.1 ENST00000401630.3 ENST00000406575.1 |
IL6 |
interleukin 6 (interferon, beta 2) |
| chr2_-_152382500 | 0.13 |
ENST00000434685.1 |
NEB |
nebulin |
| chr21_-_43771226 | 0.13 |
ENST00000291526.4 |
TFF2 |
trefoil factor 2 |
| chr12_-_52779433 | 0.13 |
ENST00000257951.3 |
KRT84 |
keratin 84 |
| chr3_+_63428982 | 0.12 |
ENST00000479198.1 ENST00000460711.1 ENST00000465156.1 |
SYNPR |
synaptoporin |
| chr8_+_42873548 | 0.12 |
ENST00000533338.1 ENST00000534420.1 |
HOOK3 RP11-598P20.5 |
hook microtubule-tethering protein 3 Uncharacterized protein |
| chr10_-_96829246 | 0.12 |
ENST00000371270.3 ENST00000535898.1 ENST00000539050.1 |
CYP2C8 |
cytochrome P450, family 2, subfamily C, polypeptide 8 |
| chrX_+_65382381 | 0.12 |
ENST00000519389.1 |
HEPH |
hephaestin |
| chr3_+_173116225 | 0.12 |
ENST00000457714.1 |
NLGN1 |
neuroligin 1 |
| chr13_+_78315466 | 0.11 |
ENST00000314070.5 ENST00000462234.1 |
SLAIN1 |
SLAIN motif family, member 1 |
| chr3_-_138048682 | 0.11 |
ENST00000383180.2 |
NME9 |
NME/NM23 family member 9 |
| chr3_-_151034734 | 0.11 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chrX_-_101771645 | 0.11 |
ENST00000289373.4 |
TMSB15A |
thymosin beta 15a |
| chr4_+_156824840 | 0.11 |
ENST00000536354.2 |
TDO2 |
tryptophan 2,3-dioxygenase |
| chr6_-_27114577 | 0.11 |
ENST00000356950.1 ENST00000396891.4 |
HIST1H2BK |
histone cluster 1, H2bk |
| chr9_+_12775011 | 0.11 |
ENST00000319264.3 |
LURAP1L |
leucine rich adaptor protein 1-like |
| chr18_-_52989525 | 0.11 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr8_-_124553437 | 0.11 |
ENST00000517956.1 ENST00000443022.2 |
FBXO32 |
F-box protein 32 |
| chr1_+_244515930 | 0.10 |
ENST00000366537.1 ENST00000308105.4 |
C1orf100 |
chromosome 1 open reading frame 100 |
| chr8_+_38831683 | 0.10 |
ENST00000302495.4 |
HTRA4 |
HtrA serine peptidase 4 |
| chr6_-_27775694 | 0.10 |
ENST00000377401.2 |
HIST1H2BL |
histone cluster 1, H2bl |
| chr7_+_35756092 | 0.10 |
ENST00000458087.3 |
AC018647.3 |
AC018647.3 |
| chr6_+_132891461 | 0.10 |
ENST00000275198.1 |
TAAR6 |
trace amine associated receptor 6 |
| chr7_+_16793160 | 0.10 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr7_+_35756186 | 0.10 |
ENST00000430518.1 |
AC018647.3 |
AC018647.3 |
| chr3_-_185270342 | 0.10 |
ENST00000424591.2 |
LIPH |
lipase, member H |
| chr18_-_53069419 | 0.09 |
ENST00000570177.2 |
TCF4 |
transcription factor 4 |
| chr14_+_21525981 | 0.09 |
ENST00000308227.2 |
RNASE8 |
ribonuclease, RNase A family, 8 |
| chr6_+_46761118 | 0.09 |
ENST00000230588.4 |
MEP1A |
meprin A, alpha (PABA peptide hydrolase) |
| chrX_+_16668278 | 0.09 |
ENST00000380200.3 |
S100G |
S100 calcium binding protein G |
| chr3_+_63428752 | 0.09 |
ENST00000295894.5 |
SYNPR |
synaptoporin |
| chr1_+_151735431 | 0.09 |
ENST00000321531.5 ENST00000315067.8 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr7_+_26331541 | 0.09 |
ENST00000416246.1 ENST00000338523.4 ENST00000412416.1 |
SNX10 |
sorting nexin 10 |
| chr12_+_70219052 | 0.09 |
ENST00000552032.2 ENST00000547771.2 |
MYRFL |
myelin regulatory factor-like |
| chr16_+_57653854 | 0.08 |
ENST00000568908.1 ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr11_-_119999611 | 0.08 |
ENST00000529044.1 |
TRIM29 |
tripartite motif containing 29 |
| chr11_-_119999539 | 0.08 |
ENST00000541857.1 |
TRIM29 |
tripartite motif containing 29 |
| chr4_+_71337834 | 0.08 |
ENST00000304887.5 |
MUC7 |
mucin 7, secreted |
| chr4_+_146539415 | 0.08 |
ENST00000281317.5 |
MMAA |
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr13_-_20077417 | 0.08 |
ENST00000382978.1 ENST00000400230.2 ENST00000255310.6 |
TPTE2 |
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr16_+_57653989 | 0.08 |
ENST00000567835.1 ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr6_+_122793058 | 0.08 |
ENST00000392491.2 |
PKIB |
protein kinase (cAMP-dependent, catalytic) inhibitor beta |
| chrX_-_33229636 | 0.08 |
ENST00000357033.4 |
DMD |
dystrophin |
| chr1_+_160370344 | 0.08 |
ENST00000368061.2 |
VANGL2 |
VANGL planar cell polarity protein 2 |
| chr9_-_73736511 | 0.08 |
ENST00000377110.3 ENST00000377111.2 |
TRPM3 |
transient receptor potential cation channel, subfamily M, member 3 |
| chr9_-_114521783 | 0.08 |
ENST00000394779.3 ENST00000394777.4 |
C9orf84 |
chromosome 9 open reading frame 84 |
| chr7_-_75401513 | 0.08 |
ENST00000005180.4 |
CCL26 |
chemokine (C-C motif) ligand 26 |
| chr18_+_61223393 | 0.08 |
ENST00000269491.1 ENST00000382768.1 |
SERPINB12 |
serpin peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr10_-_13523073 | 0.08 |
ENST00000440282.1 |
BEND7 |
BEN domain containing 7 |
| chr1_-_100643765 | 0.08 |
ENST00000370137.1 ENST00000370138.1 ENST00000342895.3 |
LRRC39 |
leucine rich repeat containing 39 |
| chr3_-_38835501 | 0.08 |
ENST00000449082.2 |
SCN10A |
sodium channel, voltage-gated, type X, alpha subunit |
| chr2_+_158114051 | 0.08 |
ENST00000259056.4 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr6_-_27782548 | 0.08 |
ENST00000333151.3 |
HIST1H2AJ |
histone cluster 1, H2aj |
| chr7_-_41742697 | 0.08 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr7_+_138915102 | 0.08 |
ENST00000486663.1 |
UBN2 |
ubinuclein 2 |
| chr17_-_56621665 | 0.07 |
ENST00000321691.3 |
C17orf47 |
chromosome 17 open reading frame 47 |
| chr5_-_43412418 | 0.07 |
ENST00000537013.1 ENST00000361115.4 |
CCL28 |
chemokine (C-C motif) ligand 28 |
| chr12_-_15038779 | 0.07 |
ENST00000228938.5 ENST00000539261.1 |
MGP |
matrix Gla protein |
| chr14_-_61748550 | 0.07 |
ENST00000555868.1 |
TMEM30B |
transmembrane protein 30B |
| chr19_-_54676846 | 0.07 |
ENST00000301187.4 |
TMC4 |
transmembrane channel-like 4 |
| chr6_-_132967142 | 0.07 |
ENST00000275216.1 |
TAAR1 |
trace amine associated receptor 1 |
| chr8_-_95220775 | 0.07 |
ENST00000441892.2 ENST00000521491.1 ENST00000027335.3 |
CDH17 |
cadherin 17, LI cadherin (liver-intestine) |
| chr11_-_132813566 | 0.07 |
ENST00000331898.7 |
OPCML |
opioid binding protein/cell adhesion molecule-like |
| chr12_-_49418407 | 0.07 |
ENST00000526209.1 |
KMT2D |
lysine (K)-specific methyltransferase 2D |
| chr14_-_25479811 | 0.07 |
ENST00000550887.1 |
STXBP6 |
syntaxin binding protein 6 (amisyn) |
| chr9_-_95640218 | 0.07 |
ENST00000395506.3 ENST00000375495.3 ENST00000332591.6 |
ZNF484 |
zinc finger protein 484 |
| chr10_+_85933494 | 0.07 |
ENST00000372126.3 |
C10orf99 |
chromosome 10 open reading frame 99 |
| chr18_-_53253323 | 0.07 |
ENST00000540999.1 ENST00000563888.2 |
TCF4 |
transcription factor 4 |
| chr1_+_162351503 | 0.07 |
ENST00000458626.2 |
C1orf226 |
chromosome 1 open reading frame 226 |
| chr18_-_53253112 | 0.07 |
ENST00000568673.1 ENST00000562847.1 ENST00000568147.1 |
TCF4 |
transcription factor 4 |
| chr5_+_149546334 | 0.06 |
ENST00000231656.8 |
CDX1 |
caudal type homeobox 1 |
| chr4_+_48018781 | 0.06 |
ENST00000295461.5 |
NIPAL1 |
NIPA-like domain containing 1 |
| chr9_-_19786926 | 0.06 |
ENST00000341998.2 ENST00000286344.3 |
SLC24A2 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2 |
| chr9_+_2717502 | 0.06 |
ENST00000382082.3 |
KCNV2 |
potassium channel, subfamily V, member 2 |
| chr14_-_55369525 | 0.06 |
ENST00000543643.2 ENST00000536224.2 ENST00000395514.1 ENST00000491895.2 |
GCH1 |
GTP cyclohydrolase 1 |
| chr17_+_32646055 | 0.06 |
ENST00000394620.1 |
CCL8 |
chemokine (C-C motif) ligand 8 |
| chr4_-_156787425 | 0.06 |
ENST00000537611.2 |
ASIC5 |
acid-sensing (proton-gated) ion channel family member 5 |
| chrX_-_117119243 | 0.06 |
ENST00000539496.1 ENST00000469946.1 |
KLHL13 |
kelch-like family member 13 |
| chr8_-_114449112 | 0.06 |
ENST00000455883.2 ENST00000352409.3 ENST00000297405.5 |
CSMD3 |
CUB and Sushi multiple domains 3 |
| chr7_-_50633078 | 0.06 |
ENST00000444124.2 |
DDC |
dopa decarboxylase (aromatic L-amino acid decarboxylase) |
| chrX_-_11284095 | 0.06 |
ENST00000303025.6 ENST00000534860.1 |
ARHGAP6 |
Rho GTPase activating protein 6 |
| chr3_+_148447887 | 0.06 |
ENST00000475347.1 ENST00000474935.1 ENST00000461609.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr5_-_137475071 | 0.06 |
ENST00000265191.2 |
NME5 |
NME/NM23 family member 5 |
| chrX_-_100604184 | 0.06 |
ENST00000372902.3 |
TIMM8A |
translocase of inner mitochondrial membrane 8 homolog A (yeast) |
| chr11_+_63606373 | 0.06 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr6_-_111888474 | 0.06 |
ENST00000368735.1 |
TRAF3IP2 |
TRAF3 interacting protein 2 |
| chr12_-_11548496 | 0.06 |
ENST00000389362.4 ENST00000565533.1 ENST00000546254.1 |
PRB2 PRB1 |
proline-rich protein BstNI subfamily 2 proline-rich protein BstNI subfamily 1 |
| chr10_-_62332357 | 0.06 |
ENST00000503366.1 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr4_+_88754113 | 0.06 |
ENST00000560249.1 ENST00000540395.1 ENST00000511670.1 ENST00000361056.3 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr9_+_82188077 | 0.05 |
ENST00000425506.1 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr14_-_72458326 | 0.05 |
ENST00000542853.1 |
AC005477.1 |
AC005477.1 |
| chr12_-_81992111 | 0.05 |
ENST00000443686.3 ENST00000407050.4 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr20_+_52105495 | 0.05 |
ENST00000439873.2 |
AL354993.1 |
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr8_-_49834299 | 0.05 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr2_-_105030466 | 0.05 |
ENST00000449772.1 |
AC068535.3 |
AC068535.3 |
| chr6_+_27775899 | 0.05 |
ENST00000358739.3 |
HIST1H2AI |
histone cluster 1, H2ai |
| chr4_+_88754069 | 0.05 |
ENST00000395102.4 ENST00000497649.2 |
MEPE |
matrix extracellular phosphoglycoprotein |
| chr6_+_131958436 | 0.05 |
ENST00000357639.3 ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3 |
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr12_-_18243119 | 0.05 |
ENST00000538724.1 ENST00000229002.2 |
RERGL |
RERG/RAS-like |
| chr12_+_131438443 | 0.05 |
ENST00000261654.5 |
GPR133 |
G protein-coupled receptor 133 |
| chrX_-_133792480 | 0.05 |
ENST00000359237.4 |
PLAC1 |
placenta-specific 1 |
| chr6_+_21593972 | 0.05 |
ENST00000244745.1 ENST00000543472.1 |
SOX4 |
SRY (sex determining region Y)-box 4 |
| chr3_+_189507432 | 0.05 |
ENST00000354600.5 |
TP63 |
tumor protein p63 |
| chr9_+_21967137 | 0.05 |
ENST00000441769.2 |
C9orf53 |
chromosome 9 open reading frame 53 |
| chr9_-_21202204 | 0.05 |
ENST00000239347.3 |
IFNA7 |
interferon, alpha 7 |
| chr3_-_164796269 | 0.05 |
ENST00000264382.3 |
SI |
sucrase-isomaltase (alpha-glucosidase) |
| chr7_+_80275953 | 0.05 |
ENST00000538969.1 ENST00000544133.1 ENST00000433696.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr6_+_26156551 | 0.05 |
ENST00000304218.3 |
HIST1H1E |
histone cluster 1, H1e |
| chr1_+_62439037 | 0.05 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr20_-_1974692 | 0.05 |
ENST00000217305.2 ENST00000539905.1 |
PDYN |
prodynorphin |
| chr4_+_169013666 | 0.05 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chr9_-_124990680 | 0.05 |
ENST00000541397.2 ENST00000560485.1 |
LHX6 |
LIM homeobox 6 |
| chr16_-_80926457 | 0.05 |
ENST00000563626.1 ENST00000562231.1 |
RP11-314O13.1 |
RP11-314O13.1 |
| chr7_+_23719749 | 0.05 |
ENST00000409192.3 ENST00000344962.4 ENST00000409653.1 ENST00000409994.3 |
FAM221A |
family with sequence similarity 221, member A |
| chr3_+_189507460 | 0.05 |
ENST00000434928.1 |
TP63 |
tumor protein p63 |
| chr4_-_138453606 | 0.05 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr6_+_111408698 | 0.05 |
ENST00000368851.5 |
SLC16A10 |
solute carrier family 16 (aromatic amino acid transporter), member 10 |
| chr1_+_211432700 | 0.05 |
ENST00000452621.2 |
RCOR3 |
REST corepressor 3 |
| chr18_+_61554932 | 0.05 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr4_-_110723335 | 0.05 |
ENST00000394634.2 |
CFI |
complement factor I |
| chr11_+_64879317 | 0.05 |
ENST00000526809.1 ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2 |
transmembrane 7 superfamily member 2 |
| chr4_-_110723194 | 0.05 |
ENST00000394635.3 |
CFI |
complement factor I |
| chr12_-_11508520 | 0.05 |
ENST00000545626.1 ENST00000500254.2 |
PRB1 |
proline-rich protein BstNI subfamily 1 |
| chr11_+_118401706 | 0.05 |
ENST00000411589.2 ENST00000442938.2 ENST00000359862.4 |
TMEM25 |
transmembrane protein 25 |
| chr2_+_210444142 | 0.05 |
ENST00000360351.4 ENST00000361559.4 |
MAP2 |
microtubule-associated protein 2 |
| chr6_-_99797522 | 0.05 |
ENST00000389677.5 |
FAXC |
failed axon connections homolog (Drosophila) |
| chr4_-_110723134 | 0.05 |
ENST00000510800.1 ENST00000512148.1 |
CFI |
complement factor I |
| chr16_+_23652773 | 0.04 |
ENST00000563998.1 ENST00000568589.1 ENST00000568272.1 |
DCTN5 |
dynactin 5 (p25) |
| chr11_+_69924639 | 0.04 |
ENST00000538023.1 ENST00000398543.2 |
ANO1 |
anoctamin 1, calcium activated chloride channel |
| chr15_-_76304731 | 0.04 |
ENST00000394907.3 |
NRG4 |
neuregulin 4 |
| chrX_+_24483338 | 0.04 |
ENST00000379162.4 ENST00000441463.2 |
PDK3 |
pyruvate dehydrogenase kinase, isozyme 3 |
| chr8_+_77318769 | 0.04 |
ENST00000518732.1 |
RP11-706J10.1 |
long intergenic non-protein coding RNA 1111 |
| chr8_+_86089619 | 0.04 |
ENST00000256117.5 ENST00000416274.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr16_-_4852915 | 0.04 |
ENST00000322048.7 |
ROGDI |
rogdi homolog (Drosophila) |
| chr8_+_77593448 | 0.04 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr3_+_159557637 | 0.04 |
ENST00000445224.2 |
SCHIP1 |
schwannomin interacting protein 1 |
| chr8_+_86089460 | 0.04 |
ENST00000418930.2 |
E2F5 |
E2F transcription factor 5, p130-binding |
| chr19_-_54676884 | 0.04 |
ENST00000376591.4 |
TMC4 |
transmembrane channel-like 4 |
| chr18_+_34124507 | 0.04 |
ENST00000591635.1 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr12_-_81763184 | 0.04 |
ENST00000548670.1 ENST00000541570.2 ENST00000553058.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr7_-_50860565 | 0.04 |
ENST00000403097.1 |
GRB10 |
growth factor receptor-bound protein 10 |
| chr7_-_69062391 | 0.04 |
ENST00000436600.2 |
RP5-942I16.1 |
RP5-942I16.1 |
| chr10_+_35456444 | 0.04 |
ENST00000361599.4 |
CREM |
cAMP responsive element modulator |
| chr22_-_30642782 | 0.04 |
ENST00000249075.3 |
LIF |
leukemia inhibitory factor |
| chr9_-_133814455 | 0.04 |
ENST00000448616.1 |
FIBCD1 |
fibrinogen C domain containing 1 |
| chr17_-_39684550 | 0.04 |
ENST00000455635.1 ENST00000361566.3 |
KRT19 |
keratin 19 |
| chr11_+_124481361 | 0.04 |
ENST00000284288.2 |
PANX3 |
pannexin 3 |
| chr2_-_136288113 | 0.04 |
ENST00000401392.1 |
ZRANB3 |
zinc finger, RAN-binding domain containing 3 |
| chr3_-_57326704 | 0.04 |
ENST00000487349.1 ENST00000389601.3 |
ASB14 |
ankyrin repeat and SOCS box containing 14 |
| chr17_-_39526052 | 0.04 |
ENST00000251646.3 |
KRT33B |
keratin 33B |
| chr2_+_169923577 | 0.04 |
ENST00000432060.2 |
DHRS9 |
dehydrogenase/reductase (SDR family) member 9 |
| chr7_+_80275752 | 0.04 |
ENST00000419819.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr3_-_178969403 | 0.04 |
ENST00000314235.5 ENST00000392685.2 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr7_-_16872932 | 0.04 |
ENST00000419572.2 ENST00000412973.1 |
AGR2 |
anterior gradient 2 |
| chr1_-_9129631 | 0.04 |
ENST00000377414.3 |
SLC2A5 |
solute carrier family 2 (facilitated glucose/fructose transporter), member 5 |
| chr3_-_18480260 | 0.04 |
ENST00000454909.2 |
SATB1 |
SATB homeobox 1 |
| chr12_-_8814669 | 0.04 |
ENST00000535411.1 ENST00000540087.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr2_+_105050794 | 0.04 |
ENST00000429464.1 ENST00000414442.1 ENST00000447380.1 |
AC013402.2 |
long intergenic non-protein coding RNA 1102 |
| chr3_+_89156674 | 0.04 |
ENST00000336596.2 |
EPHA3 |
EPH receptor A3 |
| chr1_+_197382957 | 0.04 |
ENST00000367397.1 |
CRB1 |
crumbs homolog 1 (Drosophila) |
| chr11_+_118401899 | 0.04 |
ENST00000528373.1 ENST00000544878.1 ENST00000354284.4 ENST00000533137.1 ENST00000532762.1 ENST00000526973.1 ENST00000354064.7 ENST00000533102.1 ENST00000313236.5 ENST00000527267.1 ENST00000524725.1 ENST00000533689.1 |
TMEM25 |
transmembrane protein 25 |
| chr16_+_55600580 | 0.04 |
ENST00000457326.2 |
CAPNS2 |
calpain, small subunit 2 |
| chr8_-_66701319 | 0.04 |
ENST00000379419.4 |
PDE7A |
phosphodiesterase 7A |
| chr12_-_81763127 | 0.04 |
ENST00000541017.1 |
PPFIA2 |
protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 2 |
| chr3_-_178984759 | 0.04 |
ENST00000349697.2 ENST00000497599.1 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr3_-_51813009 | 0.04 |
ENST00000398780.3 |
IQCF6 |
IQ motif containing F6 |
| chr11_-_102826434 | 0.04 |
ENST00000340273.4 ENST00000260302.3 |
MMP13 |
matrix metallopeptidase 13 (collagenase 3) |
| chr4_-_174451370 | 0.04 |
ENST00000359562.4 |
HAND2 |
heart and neural crest derivatives expressed 2 |
| chr2_-_69180083 | 0.04 |
ENST00000328895.4 |
GKN2 |
gastrokine 2 |
| chr1_+_66458072 | 0.04 |
ENST00000423207.2 |
PDE4B |
phosphodiesterase 4B, cAMP-specific |
| chr18_-_74728998 | 0.04 |
ENST00000359645.3 ENST00000397875.3 ENST00000397869.3 ENST00000578193.1 ENST00000578873.1 ENST00000397866.4 ENST00000528160.1 ENST00000527041.1 ENST00000526111.1 ENST00000397865.5 ENST00000382582.3 |
MBP |
myelin basic protein |
| chr7_+_138943265 | 0.04 |
ENST00000483726.1 |
UBN2 |
ubinuclein 2 |
| chr9_+_103947311 | 0.04 |
ENST00000395056.2 |
LPPR1 |
Lipid phosphate phosphatase-related protein type 1 |
| chr7_+_80275621 | 0.04 |
ENST00000426978.1 ENST00000432207.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr1_+_26485511 | 0.04 |
ENST00000374268.3 |
FAM110D |
family with sequence similarity 110, member D |
| chr7_+_47834880 | 0.04 |
ENST00000258776.4 |
C7orf69 |
chromosome 7 open reading frame 69 |
| chr6_-_136847610 | 0.04 |
ENST00000454590.1 ENST00000432797.2 |
MAP7 |
microtubule-associated protein 7 |
| chr6_-_26247259 | 0.04 |
ENST00000244537.4 |
HIST1H4G |
histone cluster 1, H4g |
| chr6_+_27114861 | 0.04 |
ENST00000377459.1 |
HIST1H2AH |
histone cluster 1, H2ah |
| chr14_+_75536335 | 0.04 |
ENST00000554763.1 ENST00000439583.2 ENST00000526130.1 ENST00000525046.1 |
ZC2HC1C |
zinc finger, C2HC-type containing 1C |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.1 | GO:0099545 | trans-synaptic signaling by trans-synaptic complex(GO:0099545) |
| 0.0 | 0.1 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.9 | GO:0006825 | copper ion transport(GO:0006825) |
| 0.0 | 0.1 | GO:0061341 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.1 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.0 | 0.1 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.0 | 0.1 | GO:0097156 | fasciculation of motor neuron axon(GO:0097156) |
| 0.0 | 0.2 | GO:0098704 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.1 | GO:1900827 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.0 | GO:0010621 | negative regulation of transcription by transcription factor localization(GO:0010621) |
| 0.0 | 0.1 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:2000334 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0061760 | antifungal innate immune response(GO:0061760) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.3 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0002933 | lipid hydroxylation(GO:0002933) |
| 0.0 | 0.1 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.0 | GO:0002636 | positive regulation of germinal center formation(GO:0002636) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.1 | GO:0035910 | N-terminal peptidyl-lysine acetylation(GO:0018076) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.0 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.0 | 0.1 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.0 | 0.1 | GO:1903899 | positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0021853 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.0 | GO:1990641 | response to iron ion starvation(GO:1990641) |
| 0.0 | 0.0 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.2 | GO:0006183 | GTP biosynthetic process(GO:0006183) |
| 0.0 | 0.0 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.0 | GO:0036292 | DNA rewinding(GO:0036292) |
| 0.0 | 0.0 | GO:1903978 | negative regulation of macrophage colony-stimulating factor signaling pathway(GO:1902227) negative regulation of response to macrophage colony-stimulating factor(GO:1903970) negative regulation of cellular response to macrophage colony-stimulating factor stimulus(GO:1903973) regulation of microglial cell activation(GO:1903978) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.1 | GO:0097679 | other organism cytoplasm(GO:0097679) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.1 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0004322 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.1 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.1 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.0 | 0.4 | GO:0008061 | chitin binding(GO:0008061) |
| 0.0 | 0.2 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.1 | GO:0034875 | oxidoreductase activity, acting on CH or CH2 groups, quinone or similar compound as acceptor(GO:0033695) caffeine oxidase activity(GO:0034875) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.1 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0035673 | oligopeptide transmembrane transporter activity(GO:0035673) |
| 0.0 | 0.1 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.0 | 0.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.1 | GO:0031728 | CCR3 chemokine receptor binding(GO:0031728) |
| 0.0 | 0.0 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0005138 | interleukin-6 receptor binding(GO:0005138) |
| 0.0 | 0.1 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.1 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.1 | GO:0004522 | ribonuclease A activity(GO:0004522) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.0 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.0 | GO:0003947 | (N-acetylneuraminyl)-galactosylglucosylceramide N-acetylgalactosaminyltransferase activity(GO:0003947) |
| 0.0 | 0.0 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0008273 | calcium, potassium:sodium antiporter activity(GO:0008273) |
| 0.0 | 0.0 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME METAL ION SLC TRANSPORTERS | Genes involved in Metal ion SLC transporters |
| 0.0 | 0.3 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |