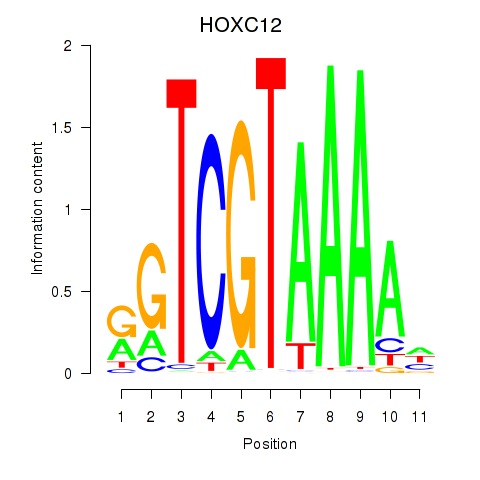
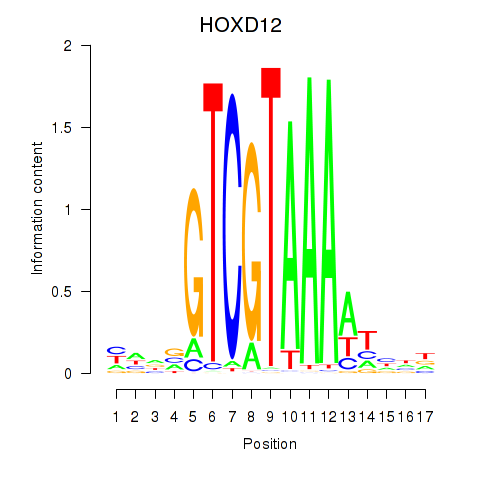
Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for HOXC12_HOXD12
Z-value: 0.67
Transcription factors associated with HOXC12_HOXD12
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC12
|
ENSG00000123407.3 | HOXC12 |
|
HOXD12
|
ENSG00000170178.5 | HOXD12 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD12 | hg19_v2_chr2_+_176964458_176964540 | 0.59 | 1.3e-01 | Click! |
| HOXC12 | hg19_v2_chr12_+_54348618_54348693 | 0.23 | 5.9e-01 | Click! |
Activity profile of HOXC12_HOXD12 motif
Sorted Z-values of HOXC12_HOXD12 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC12_HOXD12
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_156787425 | 0.95 |
ENST00000537611.2 |
ASIC5 |
acid-sensing (proton-gated) ion channel family member 5 |
| chr10_+_31608054 | 0.84 |
ENST00000320985.10 ENST00000361642.5 ENST00000560721.2 ENST00000558440.1 ENST00000424869.1 ENST00000542815.3 |
ZEB1 |
zinc finger E-box binding homeobox 1 |
| chr17_-_66951474 | 0.74 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr9_-_73029540 | 0.69 |
ENST00000377126.2 |
KLF9 |
Kruppel-like factor 9 |
| chr12_-_91573132 | 0.67 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr2_-_225811747 | 0.65 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr2_+_201450591 | 0.58 |
ENST00000374700.2 |
AOX1 |
aldehyde oxidase 1 |
| chr22_-_20307532 | 0.57 |
ENST00000405465.3 ENST00000248879.3 |
DGCR6L |
DiGeorge syndrome critical region gene 6-like |
| chr15_-_99789736 | 0.51 |
ENST00000560235.1 ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23 |
tetratricopeptide repeat domain 23 |
| chr4_-_70626314 | 0.46 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr7_+_138915102 | 0.45 |
ENST00000486663.1 |
UBN2 |
ubinuclein 2 |
| chr4_+_70894130 | 0.43 |
ENST00000526767.1 ENST00000530128.1 ENST00000381057.3 |
HTN3 |
histatin 3 |
| chr8_-_60031762 | 0.43 |
ENST00000361421.1 |
TOX |
thymocyte selection-associated high mobility group box |
| chr12_-_95945246 | 0.43 |
ENST00000258499.3 |
USP44 |
ubiquitin specific peptidase 44 |
| chr8_-_120605194 | 0.41 |
ENST00000522167.1 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr8_+_16884740 | 0.37 |
ENST00000318063.5 |
MICU3 |
mitochondrial calcium uptake family, member 3 |
| chr7_+_150065278 | 0.37 |
ENST00000519397.1 ENST00000479668.1 ENST00000540729.1 |
REPIN1 |
replication initiator 1 |
| chr5_+_169010638 | 0.37 |
ENST00000265295.4 ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1 |
spindle apparatus coiled-coil protein 1 |
| chr19_+_7011509 | 0.35 |
ENST00000377296.3 |
AC025278.1 |
Uncharacterized protein |
| chr3_+_148457585 | 0.33 |
ENST00000402260.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr8_+_17013515 | 0.33 |
ENST00000262096.8 |
ZDHHC2 |
zinc finger, DHHC-type containing 2 |
| chr5_-_146833222 | 0.33 |
ENST00000534907.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr7_-_27169801 | 0.32 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr1_+_114522049 | 0.32 |
ENST00000369551.1 ENST00000320334.4 |
OLFML3 |
olfactomedin-like 3 |
| chr8_-_13134045 | 0.31 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr7_+_154720173 | 0.29 |
ENST00000397551.2 |
PAXIP1-AS2 |
PAXIP1 antisense RNA 2 |
| chr5_-_146833485 | 0.29 |
ENST00000398514.3 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr14_+_21359558 | 0.28 |
ENST00000304639.3 |
RNASE3 |
ribonuclease, RNase A family, 3 |
| chr6_-_166796461 | 0.28 |
ENST00000360961.6 ENST00000341756.6 |
MPC1 |
mitochondrial pyruvate carrier 1 |
| chr17_+_29664830 | 0.28 |
ENST00000444181.2 ENST00000417592.2 |
NF1 |
neurofibromin 1 |
| chr3_-_18466026 | 0.27 |
ENST00000417717.2 |
SATB1 |
SATB homeobox 1 |
| chr5_-_76383133 | 0.26 |
ENST00000255198.2 |
ZBED3 |
zinc finger, BED-type containing 3 |
| chr7_+_90339169 | 0.26 |
ENST00000436577.2 |
CDK14 |
cyclin-dependent kinase 14 |
| chr5_-_146781153 | 0.25 |
ENST00000520473.1 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr1_+_202431859 | 0.25 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr20_-_30311703 | 0.25 |
ENST00000450273.1 ENST00000456404.1 ENST00000420488.1 ENST00000439267.1 |
BCL2L1 |
BCL2-like 1 |
| chr15_+_79166065 | 0.24 |
ENST00000559690.1 ENST00000559158.1 |
MORF4L1 |
mortality factor 4 like 1 |
| chr2_+_161993465 | 0.24 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr7_+_152456904 | 0.23 |
ENST00000537264.1 |
ACTR3B |
ARP3 actin-related protein 3 homolog B (yeast) |
| chr8_-_12668962 | 0.22 |
ENST00000534827.1 |
RP11-252C15.1 |
RP11-252C15.1 |
| chr12_-_31743901 | 0.22 |
ENST00000354285.4 |
DENND5B |
DENN/MADD domain containing 5B |
| chr20_+_12989596 | 0.22 |
ENST00000434210.1 ENST00000399002.2 |
SPTLC3 |
serine palmitoyltransferase, long chain base subunit 3 |
| chr12_+_120105558 | 0.22 |
ENST00000229328.5 ENST00000541640.1 |
PRKAB1 |
protein kinase, AMP-activated, beta 1 non-catalytic subunit |
| chr15_+_72410629 | 0.21 |
ENST00000340912.4 ENST00000544171.1 |
SENP8 |
SUMO/sentrin specific peptidase family member 8 |
| chr3_-_195310802 | 0.21 |
ENST00000421243.1 ENST00000453131.1 |
APOD |
apolipoprotein D |
| chr12_-_76879852 | 0.21 |
ENST00000548341.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr19_+_18668672 | 0.21 |
ENST00000601630.1 ENST00000599000.1 ENST00000595073.1 ENST00000596785.1 |
KXD1 |
KxDL motif containing 1 |
| chr9_+_2029019 | 0.21 |
ENST00000382194.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr2_+_55746746 | 0.21 |
ENST00000406691.3 ENST00000349456.4 ENST00000407816.3 ENST00000403007.3 |
CCDC104 |
coiled-coil domain containing 104 |
| chr19_-_14945933 | 0.21 |
ENST00000322301.3 |
OR7A5 |
olfactory receptor, family 7, subfamily A, member 5 |
| chr1_+_158975744 | 0.21 |
ENST00000426592.2 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr22_+_18893736 | 0.20 |
ENST00000331444.6 |
DGCR6 |
DiGeorge syndrome critical region gene 6 |
| chr5_-_176433693 | 0.20 |
ENST00000507513.1 ENST00000511320.1 |
UIMC1 |
ubiquitin interaction motif containing 1 |
| chr11_-_67275542 | 0.20 |
ENST00000531506.1 |
CDK2AP2 |
cyclin-dependent kinase 2 associated protein 2 |
| chr12_-_88535747 | 0.20 |
ENST00000309041.7 |
CEP290 |
centrosomal protein 290kDa |
| chr10_-_90712520 | 0.19 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr10_-_27529486 | 0.19 |
ENST00000375888.1 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chr2_-_73460334 | 0.19 |
ENST00000258083.2 |
PRADC1 |
protease-associated domain containing 1 |
| chr19_-_48752812 | 0.18 |
ENST00000359009.4 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr2_+_161993412 | 0.18 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr2_+_132160448 | 0.18 |
ENST00000437751.1 |
AC073869.19 |
long intergenic non-protein coding RNA 1120 |
| chr22_+_29168652 | 0.18 |
ENST00000249064.4 ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117 |
coiled-coil domain containing 117 |
| chr4_+_26585686 | 0.18 |
ENST00000505206.1 ENST00000511789.1 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr12_+_104697504 | 0.17 |
ENST00000527879.1 |
EID3 |
EP300 interacting inhibitor of differentiation 3 |
| chr11_+_20044600 | 0.17 |
ENST00000311043.8 |
NAV2 |
neuron navigator 2 |
| chr13_+_49551020 | 0.17 |
ENST00000541916.1 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr19_-_48753104 | 0.17 |
ENST00000447740.2 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr19_+_18668572 | 0.17 |
ENST00000540691.1 ENST00000539106.1 ENST00000222307.4 |
KXD1 |
KxDL motif containing 1 |
| chr1_+_42928945 | 0.17 |
ENST00000428554.2 |
CCDC30 |
coiled-coil domain containing 30 |
| chr12_-_53074182 | 0.17 |
ENST00000252244.3 |
KRT1 |
keratin 1 |
| chr1_-_246670519 | 0.17 |
ENST00000388985.4 ENST00000490107.1 |
SMYD3 |
SET and MYND domain containing 3 |
| chr13_-_24895566 | 0.17 |
ENST00000422229.2 |
AL359736.1 |
protein PCOTH isoform 1 |
| chr5_+_145718587 | 0.17 |
ENST00000230732.4 |
POU4F3 |
POU class 4 homeobox 3 |
| chr7_-_132261223 | 0.17 |
ENST00000423507.2 |
PLXNA4 |
plexin A4 |
| chr1_+_59775752 | 0.16 |
ENST00000371212.1 |
FGGY |
FGGY carbohydrate kinase domain containing |
| chr6_+_20534672 | 0.16 |
ENST00000274695.4 ENST00000378624.4 |
CDKAL1 |
CDK5 regulatory subunit associated protein 1-like 1 |
| chr8_-_93029865 | 0.16 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr15_+_26360970 | 0.16 |
ENST00000556159.1 ENST00000557523.1 |
LINC00929 |
long intergenic non-protein coding RNA 929 |
| chr12_-_13248562 | 0.16 |
ENST00000457134.2 ENST00000537302.1 |
GSG1 |
germ cell associated 1 |
| chr12_-_13248705 | 0.15 |
ENST00000396310.2 |
GSG1 |
germ cell associated 1 |
| chr4_+_146539415 | 0.15 |
ENST00000281317.5 |
MMAA |
methylmalonic aciduria (cobalamin deficiency) cblA type |
| chr2_-_152146385 | 0.15 |
ENST00000414946.1 ENST00000243346.5 |
NMI |
N-myc (and STAT) interactor |
| chr19_-_44905741 | 0.15 |
ENST00000591679.1 ENST00000544719.2 ENST00000330997.4 ENST00000585868.1 ENST00000588212.1 |
ZNF285 CTC-512J12.6 |
zinc finger protein 285 Uncharacterized protein |
| chr4_-_17513702 | 0.15 |
ENST00000428702.2 ENST00000508623.1 ENST00000513615.1 |
QDPR |
quinoid dihydropteridine reductase |
| chr1_-_47407097 | 0.15 |
ENST00000457840.2 |
CYP4A11 |
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr7_-_27183263 | 0.15 |
ENST00000222726.3 |
HOXA5 |
homeobox A5 |
| chr19_-_23869999 | 0.14 |
ENST00000601935.1 ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675 |
zinc finger protein 675 |
| chr16_-_12897642 | 0.14 |
ENST00000433677.2 ENST00000261660.4 ENST00000381774.4 |
CPPED1 |
calcineurin-like phosphoesterase domain containing 1 |
| chr4_-_114682224 | 0.14 |
ENST00000342666.5 ENST00000515496.1 ENST00000514328.1 ENST00000508738.1 ENST00000379773.2 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr12_+_59989918 | 0.14 |
ENST00000547379.1 ENST00000549465.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr5_+_135468516 | 0.14 |
ENST00000507118.1 ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5 |
SMAD family member 5 |
| chr12_-_13248732 | 0.14 |
ENST00000396302.3 |
GSG1 |
germ cell associated 1 |
| chr6_+_32812568 | 0.14 |
ENST00000414474.1 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr4_-_114682364 | 0.14 |
ENST00000511664.1 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr4_+_26585538 | 0.14 |
ENST00000264866.4 |
TBC1D19 |
TBC1 domain family, member 19 |
| chr1_-_53686261 | 0.14 |
ENST00000294360.4 |
C1orf123 |
chromosome 1 open reading frame 123 |
| chr19_+_7745708 | 0.14 |
ENST00000596148.1 ENST00000317378.5 ENST00000426877.2 |
TRAPPC5 |
trafficking protein particle complex 5 |
| chr5_-_176433582 | 0.13 |
ENST00000506128.1 |
UIMC1 |
ubiquitin interaction motif containing 1 |
| chr5_-_156569754 | 0.13 |
ENST00000420343.1 |
MED7 |
mediator complex subunit 7 |
| chr17_-_15496722 | 0.13 |
ENST00000472534.1 |
CDRT1 |
CMT1A duplicated region transcript 1 |
| chr2_+_190541153 | 0.13 |
ENST00000313581.4 ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR |
ankyrin and armadillo repeat containing |
| chr5_-_180670370 | 0.13 |
ENST00000502844.1 |
GNB2L1 |
guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 |
| chr17_+_17685422 | 0.13 |
ENST00000395774.1 |
RAI1 |
retinoic acid induced 1 |
| chr21_-_46012386 | 0.12 |
ENST00000400368.1 |
KRTAP10-6 |
keratin associated protein 10-6 |
| chr16_+_20818020 | 0.12 |
ENST00000564274.1 ENST00000563068.1 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr9_+_137979506 | 0.12 |
ENST00000539529.1 ENST00000392991.4 ENST00000371793.3 |
OLFM1 |
olfactomedin 1 |
| chr19_+_21106081 | 0.12 |
ENST00000300540.3 ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85 |
zinc finger protein 85 |
| chr19_+_13885252 | 0.12 |
ENST00000221576.4 |
C19orf53 |
chromosome 19 open reading frame 53 |
| chr7_-_132261253 | 0.12 |
ENST00000321063.4 |
PLXNA4 |
plexin A4 |
| chr10_+_124670121 | 0.12 |
ENST00000368894.1 |
FAM24A |
family with sequence similarity 24, member A |
| chr17_-_39637392 | 0.12 |
ENST00000246639.2 ENST00000393989.1 |
KRT35 |
keratin 35 |
| chr1_+_151735431 | 0.12 |
ENST00000321531.5 ENST00000315067.8 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr3_-_56502375 | 0.12 |
ENST00000288221.6 |
ERC2 |
ELKS/RAB6-interacting/CAST family member 2 |
| chr9_-_124976154 | 0.12 |
ENST00000482062.1 |
LHX6 |
LIM homeobox 6 |
| chr7_-_27702455 | 0.12 |
ENST00000265395.2 |
HIBADH |
3-hydroxyisobutyrate dehydrogenase |
| chr12_-_30848914 | 0.11 |
ENST00000256079.4 |
IPO8 |
importin 8 |
| chr9_-_124976185 | 0.11 |
ENST00000464484.2 |
LHX6 |
LIM homeobox 6 |
| chr3_+_179322573 | 0.11 |
ENST00000493866.1 ENST00000472629.1 ENST00000482604.1 |
NDUFB5 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 5, 16kDa |
| chr5_-_156569850 | 0.11 |
ENST00000524219.1 |
HAVCR2 |
hepatitis A virus cellular receptor 2 |
| chr19_+_21106028 | 0.11 |
ENST00000597314.1 ENST00000601924.1 |
ZNF85 |
zinc finger protein 85 |
| chr8_-_105479270 | 0.11 |
ENST00000521573.2 ENST00000351513.2 |
DPYS |
dihydropyrimidinase |
| chr7_-_76247617 | 0.11 |
ENST00000441393.1 |
POMZP3 |
POM121 and ZP3 fusion |
| chr17_-_73127826 | 0.11 |
ENST00000582170.1 ENST00000245552.2 |
NT5C |
5', 3'-nucleotidase, cytosolic |
| chr2_+_61372226 | 0.11 |
ENST00000426997.1 |
C2orf74 |
chromosome 2 open reading frame 74 |
| chr15_-_55657428 | 0.11 |
ENST00000568543.1 |
CCPG1 |
cell cycle progression 1 |
| chr1_-_15735925 | 0.11 |
ENST00000427824.1 |
RP3-467K16.4 |
RP3-467K16.4 |
| chr7_+_134331550 | 0.11 |
ENST00000344924.3 ENST00000418040.1 ENST00000393132.2 |
BPGM |
2,3-bisphosphoglycerate mutase |
| chr2_-_211179883 | 0.10 |
ENST00000352451.3 |
MYL1 |
myosin, light chain 1, alkali; skeletal, fast |
| chr1_+_36396677 | 0.10 |
ENST00000373191.4 ENST00000397828.2 |
AGO3 |
argonaute RISC catalytic component 3 |
| chr19_-_23869970 | 0.10 |
ENST00000601010.1 |
ZNF675 |
zinc finger protein 675 |
| chr1_+_171107241 | 0.10 |
ENST00000236166.3 |
FMO6P |
flavin containing monooxygenase 6 pseudogene |
| chr4_-_83765613 | 0.10 |
ENST00000503937.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr1_+_171060018 | 0.10 |
ENST00000367755.4 ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3 |
flavin containing monooxygenase 3 |
| chr17_+_45286706 | 0.10 |
ENST00000393450.1 ENST00000572303.1 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr2_-_152589670 | 0.10 |
ENST00000604864.1 ENST00000603639.1 |
NEB |
nebulin |
| chr6_-_110011718 | 0.09 |
ENST00000532976.1 |
AK9 |
adenylate kinase 9 |
| chr3_+_138327417 | 0.09 |
ENST00000338446.4 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr14_+_88471468 | 0.09 |
ENST00000267549.3 |
GPR65 |
G protein-coupled receptor 65 |
| chr3_+_138327542 | 0.09 |
ENST00000360570.3 ENST00000393035.2 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr5_-_39270725 | 0.09 |
ENST00000512138.1 ENST00000512982.1 ENST00000540520.1 |
FYB |
FYN binding protein |
| chrY_+_2709527 | 0.09 |
ENST00000250784.8 |
RPS4Y1 |
ribosomal protein S4, Y-linked 1 |
| chr6_-_112080256 | 0.09 |
ENST00000462856.2 ENST00000229471.4 |
FYN |
FYN oncogene related to SRC, FGR, YES |
| chr17_-_36904437 | 0.09 |
ENST00000585100.1 ENST00000360797.2 ENST00000578109.1 ENST00000579882.1 |
PCGF2 |
polycomb group ring finger 2 |
| chr1_+_219347203 | 0.09 |
ENST00000366927.3 |
LYPLAL1 |
lysophospholipase-like 1 |
| chr11_-_26593677 | 0.09 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chr7_+_141811539 | 0.09 |
ENST00000550469.2 ENST00000477922.3 |
RP11-1220K2.2 |
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr14_+_24702127 | 0.09 |
ENST00000557854.1 ENST00000348719.7 ENST00000559104.1 ENST00000456667.3 |
GMPR2 |
guanosine monophosphate reductase 2 |
| chr11_+_101785727 | 0.09 |
ENST00000263468.8 |
KIAA1377 |
KIAA1377 |
| chr6_-_36515177 | 0.09 |
ENST00000229812.7 |
STK38 |
serine/threonine kinase 38 |
| chr7_+_152456829 | 0.09 |
ENST00000377776.3 ENST00000256001.8 ENST00000397282.2 |
ACTR3B |
ARP3 actin-related protein 3 homolog B (yeast) |
| chr14_-_77889860 | 0.09 |
ENST00000555603.1 |
NOXRED1 |
NADP-dependent oxidoreductase domain containing 1 |
| chr3_-_157251383 | 0.09 |
ENST00000487753.1 ENST00000489602.1 ENST00000461299.1 ENST00000479987.1 |
VEPH1 |
ventricular zone expressed PH domain-containing 1 |
| chr2_-_85645545 | 0.09 |
ENST00000409275.1 |
CAPG |
capping protein (actin filament), gelsolin-like |
| chr1_-_156647189 | 0.08 |
ENST00000368223.3 |
NES |
nestin |
| chr12_-_122879969 | 0.08 |
ENST00000540304.1 |
CLIP1 |
CAP-GLY domain containing linker protein 1 |
| chr1_+_24104869 | 0.08 |
ENST00000246151.4 |
PITHD1 |
PITH (C-terminal proteasome-interacting domain of thioredoxin-like) domain containing 1 |
| chr1_-_147599549 | 0.08 |
ENST00000369228.5 |
NBPF24 |
neuroblastoma breakpoint family, member 24 |
| chr17_+_37856214 | 0.08 |
ENST00000445658.2 |
ERBB2 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 2 |
| chr1_-_148347506 | 0.08 |
ENST00000369189.3 |
NBPF20 |
neuroblastoma breakpoint family, member 20 |
| chr6_-_131321863 | 0.08 |
ENST00000528282.1 |
EPB41L2 |
erythrocyte membrane protein band 4.1-like 2 |
| chrX_+_153775821 | 0.08 |
ENST00000263518.6 ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr8_-_27850141 | 0.08 |
ENST00000524352.1 |
SCARA5 |
scavenger receptor class A, member 5 (putative) |
| chr18_+_9103957 | 0.08 |
ENST00000400033.1 |
NDUFV2 |
NADH dehydrogenase (ubiquinone) flavoprotein 2, 24kDa |
| chr6_+_26204825 | 0.08 |
ENST00000360441.4 |
HIST1H4E |
histone cluster 1, H4e |
| chr17_-_34890709 | 0.08 |
ENST00000544606.1 |
MYO19 |
myosin XIX |
| chr8_+_77318769 | 0.08 |
ENST00000518732.1 |
RP11-706J10.1 |
long intergenic non-protein coding RNA 1111 |
| chr8_+_107738343 | 0.08 |
ENST00000521592.1 |
OXR1 |
oxidation resistance 1 |
| chr2_+_108994466 | 0.08 |
ENST00000272452.2 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
| chr12_+_10658201 | 0.08 |
ENST00000322446.3 |
EIF2S3L |
Putative eukaryotic translation initiation factor 2 subunit 3-like protein |
| chr5_+_140767452 | 0.08 |
ENST00000519479.1 |
PCDHGB4 |
protocadherin gamma subfamily B, 4 |
| chr8_-_92053212 | 0.08 |
ENST00000285419.3 |
TMEM55A |
transmembrane protein 55A |
| chr11_+_33278811 | 0.08 |
ENST00000303296.4 ENST00000379016.3 |
HIPK3 |
homeodomain interacting protein kinase 3 |
| chr14_-_24701539 | 0.08 |
ENST00000534348.1 ENST00000524927.1 ENST00000250495.5 |
NEDD8-MDP1 NEDD8 |
NEDD8-MDP1 readthrough neural precursor cell expressed, developmentally down-regulated 8 |
| chr6_+_155537771 | 0.08 |
ENST00000275246.7 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
| chr9_+_91933726 | 0.08 |
ENST00000534113.2 |
SECISBP2 |
SECIS binding protein 2 |
| chr3_+_137906109 | 0.08 |
ENST00000481646.1 ENST00000469044.1 ENST00000491704.1 ENST00000461600.1 |
ARMC8 |
armadillo repeat containing 8 |
| chr6_+_57182400 | 0.08 |
ENST00000607273.1 |
PRIM2 |
primase, DNA, polypeptide 2 (58kDa) |
| chr13_+_53602894 | 0.08 |
ENST00000219022.2 |
OLFM4 |
olfactomedin 4 |
| chr12_-_110939870 | 0.08 |
ENST00000447578.2 ENST00000546588.1 ENST00000360579.7 ENST00000549970.1 ENST00000549578.1 |
VPS29 |
vacuolar protein sorting 29 homolog (S. cerevisiae) |
| chr1_-_202896310 | 0.07 |
ENST00000367261.3 |
KLHL12 |
kelch-like family member 12 |
| chr7_+_28452130 | 0.07 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr11_-_77705687 | 0.07 |
ENST00000529807.1 ENST00000527522.1 ENST00000534064.1 |
INTS4 |
integrator complex subunit 4 |
| chr19_-_22018966 | 0.07 |
ENST00000599906.1 ENST00000354959.4 |
ZNF43 |
zinc finger protein 43 |
| chr19_-_48759119 | 0.07 |
ENST00000522889.1 ENST00000520753.1 ENST00000519940.1 ENST00000519332.1 ENST00000521437.1 ENST00000520007.1 ENST00000521613.1 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr3_-_17783990 | 0.07 |
ENST00000429383.4 ENST00000446863.1 ENST00000414349.1 ENST00000428355.1 ENST00000425944.1 ENST00000445294.1 ENST00000444471.1 ENST00000415814.2 |
TBC1D5 |
TBC1 domain family, member 5 |
| chr2_+_39103103 | 0.07 |
ENST00000340556.6 ENST00000410014.1 ENST00000409665.1 ENST00000409077.2 ENST00000409131.2 |
MORN2 |
MORN repeat containing 2 |
| chr7_+_23338819 | 0.07 |
ENST00000466681.1 |
MALSU1 |
mitochondrial assembly of ribosomal large subunit 1 |
| chr11_-_19223523 | 0.07 |
ENST00000265968.3 |
CSRP3 |
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr2_+_170335924 | 0.07 |
ENST00000554017.1 ENST00000392663.2 ENST00000513963.1 |
BBS5 RP11-724O16.1 |
Bardet-Biedl syndrome 5 Bardet-Biedl syndrome 5 protein; Uncharacterized protein |
| chr20_-_17539456 | 0.07 |
ENST00000544874.1 ENST00000377868.2 |
BFSP1 |
beaded filament structural protein 1, filensin |
| chr19_+_13858593 | 0.07 |
ENST00000221554.8 |
CCDC130 |
coiled-coil domain containing 130 |
| chr22_+_23134974 | 0.07 |
ENST00000390314.2 |
IGLV2-11 |
immunoglobulin lambda variable 2-11 |
| chr4_+_169842707 | 0.07 |
ENST00000503290.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr7_+_138145076 | 0.07 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr19_-_48753064 | 0.07 |
ENST00000520153.1 ENST00000357778.5 ENST00000520015.1 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr7_+_100551239 | 0.07 |
ENST00000319509.7 |
MUC3A |
mucin 3A, cell surface associated |
| chr19_-_5680202 | 0.07 |
ENST00000590389.1 |
C19orf70 |
chromosome 19 open reading frame 70 |
| chr4_-_17513851 | 0.07 |
ENST00000281243.5 |
QDPR |
quinoid dihydropteridine reductase |
| chr11_-_32457075 | 0.06 |
ENST00000448076.3 |
WT1 |
Wilms tumor 1 |
| chr5_-_175612149 | 0.06 |
ENST00000515403.1 |
RP11-844P9.2 |
RP11-844P9.2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0046110 | xanthine metabolic process(GO:0046110) |
| 0.1 | 0.8 | GO:0045602 | negative regulation of endothelial cell differentiation(GO:0045602) |
| 0.1 | 0.7 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.3 | GO:0021793 | chemorepulsion of branchiomotor axon(GO:0021793) |
| 0.1 | 0.3 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.1 | 0.3 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.1 | 0.2 | GO:2000097 | regulation of smooth muscle cell-matrix adhesion(GO:2000097) |
| 0.1 | 0.2 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.1 | 0.4 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.3 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.7 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.3 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.1 | 0.3 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.2 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.7 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.7 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.2 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.9 | GO:0051764 | actin crosslink formation(GO:0051764) |
| 0.0 | 0.2 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.5 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.2 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.1 | GO:0006756 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.2 | GO:0060154 | cellular process regulating host cell cycle in response to virus(GO:0060154) |
| 0.0 | 0.1 | GO:0006574 | valine catabolic process(GO:0006574) |
| 0.0 | 0.3 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.2 | GO:2000035 | regulation of stem cell division(GO:2000035) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.0 | 0.2 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.2 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.4 | GO:2000394 | positive regulation of lamellipodium morphogenesis(GO:2000394) |
| 0.0 | 0.1 | GO:2001106 | regulation of Rho guanyl-nucleotide exchange factor activity(GO:2001106) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.5 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.3 | GO:1901725 | regulation of cardiac muscle cell action potential involved in regulation of contraction(GO:0098909) regulation of histone deacetylase activity(GO:1901725) regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1902514) |
| 0.0 | 0.2 | GO:0016926 | protein desumoylation(GO:0016926) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.4 | GO:0090266 | regulation of mitotic cell cycle spindle assembly checkpoint(GO:0090266) regulation of mitotic spindle checkpoint(GO:1903504) |
| 0.0 | 0.3 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:1904936 | cerebral cortex GABAergic interneuron migration(GO:0021853) interneuron migration(GO:1904936) |
| 0.0 | 0.2 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.0 | 0.1 | GO:0090071 | negative regulation of ribosome biogenesis(GO:0090071) |
| 0.0 | 0.1 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.1 | GO:0009615 | response to virus(GO:0009615) |
| 0.0 | 0.1 | GO:0044861 | protein transport into plasma membrane raft(GO:0044861) |
| 0.0 | 0.1 | GO:0072299 | negative regulation of metanephric glomerulus development(GO:0072299) negative regulation of metanephric glomerular mesangial cell proliferation(GO:0072302) |
| 0.0 | 0.1 | GO:0033088 | negative regulation of immature T cell proliferation in thymus(GO:0033088) |
| 0.0 | 0.3 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.2 | GO:0030916 | otic vesicle formation(GO:0030916) |
| 0.0 | 0.2 | GO:0046520 | sphingoid biosynthetic process(GO:0046520) |
| 0.0 | 0.4 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.2 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.0 | GO:0044209 | AMP salvage(GO:0044209) |
| 0.0 | 0.2 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.1 | GO:0001692 | histamine metabolic process(GO:0001692) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:0006269 | DNA replication, synthesis of RNA primer(GO:0006269) |
| 0.0 | 0.1 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
| 0.0 | 0.0 | GO:2000768 | glomerular parietal epithelial cell differentiation(GO:0072139) positive regulation of nephron tubule epithelial cell differentiation(GO:2000768) |
| 0.0 | 0.3 | GO:0043101 | purine-containing compound salvage(GO:0043101) |
| 0.0 | 0.2 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:2000332 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:2000389 | regulation of neutrophil extravasation(GO:2000389) |
| 0.0 | 0.1 | GO:0006561 | proline biosynthetic process(GO:0006561) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.7 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:1902560 | GMP reductase complex(GO:1902560) |
| 0.0 | 0.5 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.3 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.2 | GO:0017059 | serine C-palmitoyltransferase complex(GO:0017059) endoplasmic reticulum palmitoyltransferase complex(GO:0031211) |
| 0.0 | 0.4 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.2 | GO:0030915 | Smc5-Smc6 complex(GO:0030915) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.3 | GO:0002116 | semaphorin receptor complex(GO:0002116) |
| 0.0 | 0.3 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.3 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.4 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.9 | GO:0031941 | filamentous actin(GO:0031941) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.0 | GO:0014802 | terminal cisterna(GO:0014802) |
| 0.0 | 0.1 | GO:0044352 | pinosome(GO:0044352) macropinosome(GO:0044354) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.0 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0005658 | alpha DNA polymerase:primase complex(GO:0005658) |
| 0.0 | 0.3 | GO:0031235 | intrinsic component of the cytoplasmic side of the plasma membrane(GO:0031235) |
| 0.0 | 0.2 | GO:0036038 | MKS complex(GO:0036038) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.6 | GO:0004031 | aldehyde oxidase activity(GO:0004031) |
| 0.1 | 0.3 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.1 | 0.4 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.5 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 0.7 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.9 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.1 | 0.4 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.1 | 0.4 | GO:0043515 | kinetochore binding(GO:0043515) |
| 0.1 | 0.5 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.3 | GO:0016657 | GMP reductase activity(GO:0003920) oxidoreductase activity, acting on NAD(P)H, nitrogenous group as acceptor(GO:0016657) |
| 0.0 | 0.2 | GO:0004758 | serine C-palmitoyltransferase activity(GO:0004758) C-palmitoyltransferase activity(GO:0016454) |
| 0.0 | 0.2 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.1 | GO:0004619 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.5 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.2 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.2 | GO:0019784 | SUMO-specific protease activity(GO:0016929) NEDD8-specific protease activity(GO:0019784) |
| 0.0 | 0.1 | GO:0003896 | DNA primase activity(GO:0003896) |
| 0.0 | 0.2 | GO:0001162 | RNA polymerase II intronic transcription regulatory region sequence-specific DNA binding(GO:0001162) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.3 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.3 | GO:0017154 | semaphorin receptor activity(GO:0017154) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.2 | GO:0004679 | AMP-activated protein kinase activity(GO:0004679) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.0 | 0.1 | GO:0044729 | hemi-methylated DNA-binding(GO:0044729) |
| 0.0 | 0.1 | GO:0047631 | ADP-ribose diphosphatase activity(GO:0047631) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.1 | GO:0035368 | selenocysteine insertion sequence binding(GO:0035368) |
| 0.0 | 0.0 | GO:0002054 | nucleobase binding(GO:0002054) |
| 0.0 | 0.3 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0070888 | E-box binding(GO:0070888) |
| 0.0 | 0.1 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.3 | REACTOME PURINE SALVAGE | Genes involved in Purine salvage |
| 0.0 | 0.2 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.3 | REACTOME RAS ACTIVATION UOPN CA2 INFUX THROUGH NMDA RECEPTOR | Genes involved in Ras activation uopn Ca2+ infux through NMDA receptor |
| 0.0 | 0.2 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |