Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for HOXC13
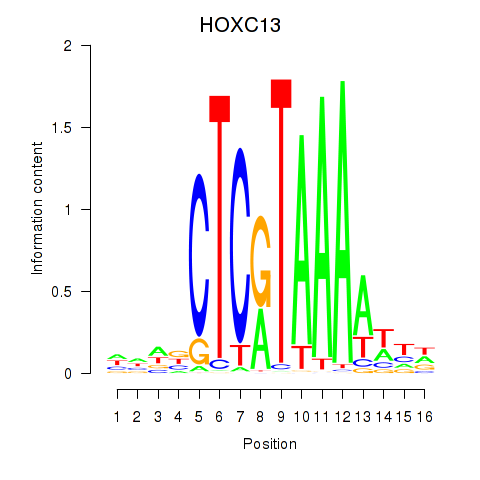
Z-value: 0.67
Transcription factors associated with HOXC13
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC13
|
ENSG00000123364.3 | HOXC13 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC13 | hg19_v2_chr12_+_54332535_54332636 | -0.80 | 1.8e-02 | Click! |
Activity profile of HOXC13 motif
Sorted Z-values of HOXC13 motif
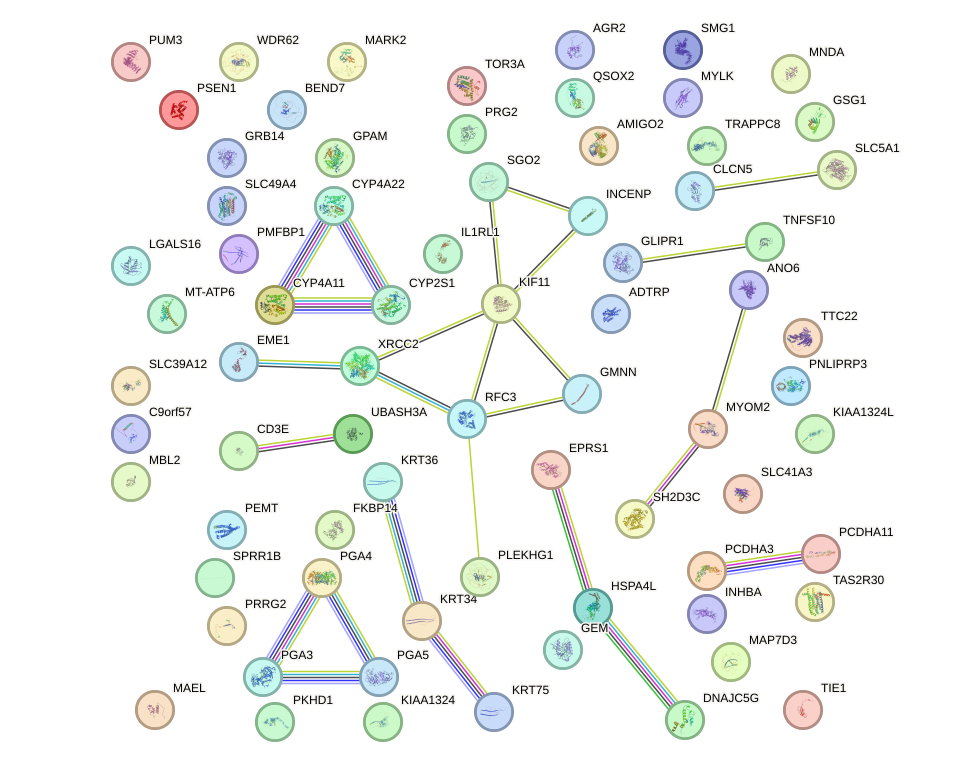
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC13
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr21_+_25801041 | 0.91 |
ENST00000453784.2 ENST00000423581.1 |
AP000476.1 |
AP000476.1 |
| chr18_+_61554932 | 0.83 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr1_+_153003671 | 0.70 |
ENST00000307098.4 |
SPRR1B |
small proline-rich protein 1B |
| chr17_-_39538550 | 0.48 |
ENST00000394001.1 |
KRT34 |
keratin 34 |
| chr3_-_172241250 | 0.44 |
ENST00000420541.2 ENST00000241261.2 |
TNFSF10 |
tumor necrosis factor (ligand) superfamily, member 10 |
| chr7_-_41742697 | 0.43 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr2_-_165424973 | 0.39 |
ENST00000543549.1 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr1_+_43766642 | 0.36 |
ENST00000372476.3 |
TIE1 |
tyrosine kinase with immunoglobulin-like and EGF-like domains 1 |
| chr12_-_52828147 | 0.36 |
ENST00000252245.5 |
KRT75 |
keratin 75 |
| chr10_+_118187379 | 0.35 |
ENST00000369230.3 |
PNLIPRP3 |
pancreatic lipase-related protein 3 |
| chr9_-_139137648 | 0.31 |
ENST00000358701.5 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
| chr8_+_101170563 | 0.30 |
ENST00000520508.1 ENST00000388798.2 |
SPAG1 |
sperm associated antigen 1 |
| chr1_-_169680745 | 0.30 |
ENST00000236147.4 |
SELL |
selectin L |
| chr17_-_39646116 | 0.28 |
ENST00000328119.6 |
KRT36 |
keratin 36 |
| chr12_-_47473425 | 0.27 |
ENST00000550413.1 |
AMIGO2 |
adhesion molecule with Ig-like domain 2 |
| chr9_+_40028620 | 0.27 |
ENST00000426179.1 |
AL353791.1 |
AL353791.1 |
| chr19_+_40146534 | 0.26 |
ENST00000392051.3 |
LGALS16 |
lectin, galactoside-binding, soluble, 16 |
| chr3_-_123339343 | 0.26 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr3_-_123339418 | 0.26 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chr3_-_112127981 | 0.25 |
ENST00000486726.2 |
RP11-231E6.1 |
RP11-231E6.1 |
| chr19_+_50084561 | 0.22 |
ENST00000246794.5 |
PRRG2 |
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr11_+_61891445 | 0.22 |
ENST00000394818.3 ENST00000533896.1 ENST00000278849.4 |
INCENP |
inner centromere protein antigens 135/155kDa |
| chr10_-_113943447 | 0.22 |
ENST00000369425.1 ENST00000348367.4 ENST00000423155.1 |
GPAM |
glycerol-3-phosphate acyltransferase, mitochondrial |
| chrX_+_139791917 | 0.22 |
ENST00000607004.1 ENST00000370535.3 |
LINC00632 |
long intergenic non-protein coding RNA 632 |
| chr5_+_140248518 | 0.21 |
ENST00000398640.2 |
PCDHA11 |
protocadherin alpha 11 |
| chrX_+_49687216 | 0.21 |
ENST00000376088.3 |
CLCN5 |
chloride channel, voltage-sensitive 5 |
| chr4_-_128887069 | 0.21 |
ENST00000541133.1 ENST00000296468.3 ENST00000513559.1 |
MFSD8 |
major facilitator superfamily domain containing 8 |
| chr1_+_179050512 | 0.20 |
ENST00000367627.3 |
TOR3A |
torsin family 3, member A |
| chr6_-_11807277 | 0.19 |
ENST00000379415.2 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr7_-_16872932 | 0.19 |
ENST00000419572.2 ENST00000412973.1 |
AGR2 |
anterior gradient 2 |
| chr11_+_118175132 | 0.18 |
ENST00000361763.4 |
CD3E |
CD3e molecule, epsilon (CD3-TCR complex) |
| chr1_-_55266926 | 0.18 |
ENST00000371276.4 |
TTC22 |
tetratricopeptide repeat domain 22 |
| chr13_+_34392200 | 0.18 |
ENST00000434425.1 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
| chr21_+_39628655 | 0.17 |
ENST00000398925.1 ENST00000398928.1 ENST00000328656.4 ENST00000443341.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr5_+_140180635 | 0.17 |
ENST00000522353.2 ENST00000532566.2 |
PCDHA3 |
protocadherin alpha 3 |
| chr9_-_2844058 | 0.17 |
ENST00000397885.2 |
KIAA0020 |
KIAA0020 |
| chr15_+_77224045 | 0.17 |
ENST00000320963.5 ENST00000394883.3 |
RCN2 |
reticulocalbin 2, EF-hand calcium binding domain |
| chr15_+_77223960 | 0.16 |
ENST00000394885.3 |
RCN2 |
reticulocalbin 2, EF-hand calcium binding domain |
| chr18_+_61223393 | 0.16 |
ENST00000269491.1 ENST00000382768.1 |
SERPINB12 |
serpin peptidase inhibitor, clade B (ovalbumin), member 12 |
| chr13_+_34392185 | 0.16 |
ENST00000380071.3 |
RFC3 |
replication factor C (activator 1) 3, 38kDa |
| chr6_-_32784687 | 0.16 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr4_+_95679072 | 0.16 |
ENST00000515059.1 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr17_+_19091325 | 0.16 |
ENST00000584923.1 |
SNORD3A |
small nucleolar RNA, C/D box 3A |
| chr18_-_74839891 | 0.15 |
ENST00000581878.1 |
MBP |
myelin basic protein |
| chr3_+_122513642 | 0.15 |
ENST00000261038.5 |
DIRC2 |
disrupted in renal carcinoma 2 |
| chr2_+_201390843 | 0.15 |
ENST00000357799.4 ENST00000409203.3 |
SGOL2 |
shugoshin-like 2 (S. pombe) |
| chr12_-_11287243 | 0.15 |
ENST00000539585.1 |
TAS2R30 |
taste receptor, type 2, member 30 |
| chr8_-_95274536 | 0.15 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr11_-_104480019 | 0.14 |
ENST00000536529.1 ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1 |
RP11-886D15.1 |
| chr2_+_102953608 | 0.14 |
ENST00000311734.2 ENST00000409584.1 |
IL1RL1 |
interleukin 1 receptor-like 1 |
| chr20_-_18810797 | 0.14 |
ENST00000278779.4 |
C20orf78 |
chromosome 20 open reading frame 78 |
| chrX_-_38186775 | 0.14 |
ENST00000339363.3 ENST00000309513.3 ENST00000338898.3 ENST00000342811.3 ENST00000378505.2 |
RPGR |
retinitis pigmentosa GTPase regulator |
| chr1_-_85097431 | 0.14 |
ENST00000327308.3 |
C1orf180 |
chromosome 1 open reading frame 180 |
| chr1_+_109656532 | 0.13 |
ENST00000531664.1 ENST00000534476.1 |
KIAA1324 |
KIAA1324 |
| chr4_+_106631966 | 0.13 |
ENST00000360505.5 ENST00000510865.1 ENST00000509336.1 |
GSTCD |
glutathione S-transferase, C-terminal domain containing |
| chr10_+_18240814 | 0.12 |
ENST00000377374.4 |
SLC39A12 |
solute carrier family 39 (zinc transporter), member 12 |
| chrX_+_78003204 | 0.12 |
ENST00000435339.3 ENST00000514744.1 |
LPAR4 |
lysophosphatidic acid receptor 4 |
| chrX_-_135338503 | 0.12 |
ENST00000370663.5 |
MAP7D3 |
MAP7 domain containing 3 |
| chr10_+_94352956 | 0.12 |
ENST00000260731.3 |
KIF11 |
kinesin family member 11 |
| chr14_+_73634537 | 0.12 |
ENST00000406768.1 |
PSEN1 |
presenilin 1 |
| chr12_+_45609893 | 0.12 |
ENST00000320560.8 |
ANO6 |
anoctamin 6 |
| chr10_+_18240753 | 0.11 |
ENST00000377369.2 |
SLC39A12 |
solute carrier family 39 (zinc transporter), member 12 |
| chr14_+_97925151 | 0.11 |
ENST00000554862.1 ENST00000554260.1 ENST00000499910.2 |
CTD-2506J14.1 |
CTD-2506J14.1 |
| chr1_-_47407136 | 0.11 |
ENST00000462347.1 ENST00000371905.1 ENST00000310638.4 |
CYP4A11 |
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr17_+_48450575 | 0.11 |
ENST00000338165.4 ENST00000393271.2 ENST00000511519.2 |
EME1 |
essential meiotic structure-specific endonuclease 1 |
| chr4_+_128702969 | 0.11 |
ENST00000508776.1 ENST00000439123.2 |
HSPA4L |
heat shock 70kDa protein 4-like |
| chr6_-_51952367 | 0.11 |
ENST00000340994.4 |
PKHD1 |
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr1_+_158815588 | 0.11 |
ENST00000438394.1 |
MNDA |
myeloid cell nuclear differentiation antigen |
| chr6_+_151042224 | 0.11 |
ENST00000358517.2 |
PLEKHG1 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 1 |
| chr1_-_198990166 | 0.11 |
ENST00000427439.1 |
RP11-16L9.3 |
RP11-16L9.3 |
| chr19_+_21264980 | 0.11 |
ENST00000596053.1 ENST00000597086.1 ENST00000596143.1 ENST00000596367.1 ENST00000601416.1 |
ZNF714 |
zinc finger protein 714 |
| chr21_+_43823983 | 0.10 |
ENST00000291535.6 ENST00000450356.1 ENST00000319294.6 ENST00000398367.1 |
UBASH3A |
ubiquitin associated and SH3 domain containing A |
| chr17_-_17480779 | 0.10 |
ENST00000395782.1 |
PEMT |
phosphatidylethanolamine N-methyltransferase |
| chr9_-_88715044 | 0.10 |
ENST00000388711.3 ENST00000466178.1 |
GOLM1 |
golgi membrane protein 1 |
| chr2_+_27498289 | 0.10 |
ENST00000296097.3 ENST00000420191.1 |
DNAJC5G |
DnaJ (Hsp40) homolog, subfamily C, member 5 gamma |
| chr11_+_63606373 | 0.10 |
ENST00000402010.2 ENST00000315032.8 ENST00000377809.4 ENST00000413835.2 ENST00000377810.3 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr16_+_81348528 | 0.10 |
ENST00000568107.2 |
GAN |
gigaxonin |
| chr7_-_152373216 | 0.10 |
ENST00000359321.1 |
XRCC2 |
X-ray repair complementing defective repair in Chinese hamster cells 2 |
| chr1_-_47407097 | 0.10 |
ENST00000457840.2 |
CYP4A11 |
cytochrome P450, family 4, subfamily A, polypeptide 11 |
| chr6_-_51952418 | 0.10 |
ENST00000371117.3 |
PKHD1 |
polycystic kidney and hepatic disease 1 (autosomal recessive) |
| chr2_+_192109911 | 0.10 |
ENST00000418908.1 ENST00000339514.4 ENST00000392318.3 |
MYO1B |
myosin IB |
| chr16_-_18908196 | 0.10 |
ENST00000565324.1 ENST00000561947.1 |
SMG1 |
SMG1 phosphatidylinositol 3-kinase-related kinase |
| chr3_-_125775629 | 0.10 |
ENST00000383598.2 |
SLC41A3 |
solute carrier family 41, member 3 |
| chr10_-_6019984 | 0.09 |
ENST00000525219.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr15_+_69373184 | 0.09 |
ENST00000558147.1 ENST00000440444.1 |
LINC00277 |
long intergenic non-protein coding RNA 277 |
| chr8_+_95565947 | 0.09 |
ENST00000523011.1 |
RP11-267M23.4 |
RP11-267M23.4 |
| chr4_+_8321882 | 0.09 |
ENST00000509453.1 ENST00000503186.1 |
RP11-774O3.2 RP11-774O3.1 |
RP11-774O3.2 RP11-774O3.1 |
| chr17_+_35732955 | 0.09 |
ENST00000300618.4 |
C17orf78 |
chromosome 17 open reading frame 78 |
| chr1_-_150208363 | 0.09 |
ENST00000436748.2 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chrM_+_8527 | 0.09 |
ENST00000361899.2 |
MT-ATP6 |
mitochondrially encoded ATP synthase 6 |
| chr10_+_18240834 | 0.09 |
ENST00000377371.3 ENST00000539911.1 |
SLC39A12 |
solute carrier family 39 (zinc transporter), member 12 |
| chr1_+_47603109 | 0.09 |
ENST00000371890.3 ENST00000294337.3 ENST00000371891.3 |
CYP4A22 |
cytochrome P450, family 4, subfamily A, polypeptide 22 |
| chr1_-_220220000 | 0.09 |
ENST00000366923.3 |
EPRS |
glutamyl-prolyl-tRNA synthetase |
| chr10_-_13544945 | 0.08 |
ENST00000378605.3 ENST00000341083.3 |
BEND7 |
BEN domain containing 7 |
| chr16_-_72206034 | 0.08 |
ENST00000537465.1 ENST00000237353.10 |
PMFBP1 |
polyamine modulated factor 1 binding protein 1 |
| chr11_+_60995849 | 0.08 |
ENST00000537932.1 |
PGA4 |
pepsinogen 4, group I (pepsinogen A) |
| chr19_+_41698927 | 0.08 |
ENST00000310054.4 |
CYP2S1 |
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr1_+_166958346 | 0.08 |
ENST00000367872.4 |
MAEL |
maelstrom spermatogenic transposon silencer |
| chr8_+_1993173 | 0.08 |
ENST00000523438.1 |
MYOM2 |
myomesin 2 |
| chr1_-_150208320 | 0.08 |
ENST00000534220.1 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr9_-_74675521 | 0.08 |
ENST00000377024.3 |
C9orf57 |
chromosome 9 open reading frame 57 |
| chr3_+_57875738 | 0.08 |
ENST00000417128.1 ENST00000438794.1 |
SLMAP |
sarcolemma associated protein |
| chr18_-_10748498 | 0.08 |
ENST00000579949.1 |
PIEZO2 |
piezo-type mechanosensitive ion channel component 2 |
| chr1_-_150208412 | 0.08 |
ENST00000532744.1 ENST00000369114.5 ENST00000369115.2 ENST00000369116.4 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr9_-_88714421 | 0.08 |
ENST00000388712.3 |
GOLM1 |
golgi membrane protein 1 |
| chr9_-_130533615 | 0.08 |
ENST00000373277.4 |
SH2D3C |
SH2 domain containing 3C |
| chr12_-_13248598 | 0.08 |
ENST00000337630.6 ENST00000545699.1 |
GSG1 |
germ cell associated 1 |
| chr3_-_98241760 | 0.08 |
ENST00000507874.1 ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1 |
claudin domain containing 1 |
| chr22_+_32439019 | 0.08 |
ENST00000266088.4 |
SLC5A1 |
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr12_+_75874580 | 0.08 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr3_+_57875711 | 0.08 |
ENST00000442599.2 |
SLMAP |
sarcolemma associated protein |
| chr17_+_35732916 | 0.08 |
ENST00000586700.1 |
C17orf78 |
chromosome 17 open reading frame 78 |
| chr7_-_30066233 | 0.08 |
ENST00000222803.5 |
FKBP14 |
FK506 binding protein 14, 22 kDa |
| chr7_-_86595190 | 0.07 |
ENST00000398276.2 ENST00000416314.1 ENST00000425689.1 |
KIAA1324L |
KIAA1324-like |
| chr2_-_177502254 | 0.07 |
ENST00000339037.3 |
AC017048.3 |
long intergenic non-protein coding RNA 1116 |
| chr19_+_36545833 | 0.07 |
ENST00000401500.2 ENST00000270301.7 ENST00000427823.2 |
WDR62 |
WD repeat domain 62 |
| chr11_-_114430570 | 0.07 |
ENST00000251921.2 |
NXPE1 |
neurexophilin and PC-esterase domain family, member 1 |
| chr1_-_150208498 | 0.07 |
ENST00000314136.8 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr9_-_37034028 | 0.07 |
ENST00000520281.1 ENST00000446742.1 ENST00000522003.1 ENST00000523145.1 ENST00000414447.1 ENST00000377847.2 ENST00000377853.2 ENST00000377852.2 ENST00000523241.1 ENST00000520154.1 ENST00000358127.4 |
PAX5 |
paired box 5 |
| chr8_+_1993152 | 0.07 |
ENST00000262113.4 |
MYOM2 |
myomesin 2 |
| chr6_+_88182643 | 0.07 |
ENST00000369556.3 ENST00000544441.1 ENST00000369552.4 ENST00000369557.5 |
SLC35A1 |
solute carrier family 35 (CMP-sialic acid transporter), member A1 |
| chr1_-_150208291 | 0.07 |
ENST00000533654.1 |
ANP32E |
acidic (leucine-rich) nuclear phosphoprotein 32 family, member E |
| chr5_-_79287060 | 0.07 |
ENST00000512560.1 ENST00000509852.1 ENST00000512528.1 |
MTX3 |
metaxin 3 |
| chr3_+_111630451 | 0.07 |
ENST00000495180.1 |
PHLDB2 |
pleckstrin homology-like domain, family B, member 2 |
| chrY_+_23698778 | 0.07 |
ENST00000303902.5 |
RBMY1A1 |
RNA binding motif protein, Y-linked, family 1, member A1 |
| chr14_+_23727694 | 0.07 |
ENST00000399905.1 ENST00000470456.1 |
C14orf164 |
chromosome 14 open reading frame 164 |
| chr12_-_13248562 | 0.07 |
ENST00000457134.2 ENST00000537302.1 |
GSG1 |
germ cell associated 1 |
| chr12_-_13248705 | 0.07 |
ENST00000396310.2 |
GSG1 |
germ cell associated 1 |
| chrX_+_133733457 | 0.06 |
ENST00000440614.1 |
RP11-308B5.2 |
RP11-308B5.2 |
| chr2_-_223520770 | 0.06 |
ENST00000536361.1 |
FARSB |
phenylalanyl-tRNA synthetase, beta subunit |
| chrX_-_100129320 | 0.06 |
ENST00000372966.3 |
NOX1 |
NADPH oxidase 1 |
| chr8_-_17555164 | 0.06 |
ENST00000297488.6 |
MTUS1 |
microtubule associated tumor suppressor 1 |
| chr5_-_180076580 | 0.06 |
ENST00000502649.1 |
FLT4 |
fms-related tyrosine kinase 4 |
| chr14_-_92333873 | 0.06 |
ENST00000435962.2 |
TC2N |
tandem C2 domains, nuclear |
| chr10_+_24738355 | 0.06 |
ENST00000307544.6 |
KIAA1217 |
KIAA1217 |
| chr9_-_32526299 | 0.06 |
ENST00000379882.1 ENST00000379883.2 |
DDX58 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr4_+_88720698 | 0.06 |
ENST00000226284.5 |
IBSP |
integrin-binding sialoprotein |
| chr12_+_4758264 | 0.06 |
ENST00000266544.5 |
NDUFA9 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
| chr11_-_85376121 | 0.06 |
ENST00000527447.1 |
CREBZF |
CREB/ATF bZIP transcription factor |
| chr4_-_155533787 | 0.06 |
ENST00000407946.1 ENST00000405164.1 ENST00000336098.3 ENST00000393846.2 ENST00000404648.3 ENST00000443553.1 |
FGG |
fibrinogen gamma chain |
| chr15_+_71185148 | 0.06 |
ENST00000443425.2 ENST00000560755.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr4_+_122722466 | 0.06 |
ENST00000243498.5 ENST00000379663.3 ENST00000509800.1 |
EXOSC9 |
exosome component 9 |
| chr9_-_5304432 | 0.06 |
ENST00000416837.1 ENST00000308420.3 |
RLN2 |
relaxin 2 |
| chr14_-_50319758 | 0.06 |
ENST00000298310.5 |
NEMF |
nuclear export mediator factor |
| chr1_+_90460661 | 0.05 |
ENST00000340281.4 ENST00000361911.5 ENST00000370447.3 ENST00000455342.2 |
ZNF326 |
zinc finger protein 326 |
| chr8_+_133879193 | 0.05 |
ENST00000377869.1 ENST00000220616.4 |
TG |
thyroglobulin |
| chr5_+_42756903 | 0.05 |
ENST00000361970.5 ENST00000388827.4 |
CCDC152 |
coiled-coil domain containing 152 |
| chr15_+_71184931 | 0.05 |
ENST00000560369.1 ENST00000260382.5 |
LRRC49 |
leucine rich repeat containing 49 |
| chr12_+_102514019 | 0.05 |
ENST00000537257.1 ENST00000358383.5 ENST00000392911.2 |
PARPBP |
PARP1 binding protein |
| chr12_-_7904201 | 0.05 |
ENST00000354629.5 |
CLEC4C |
C-type lectin domain family 4, member C |
| chr3_+_101498269 | 0.05 |
ENST00000491511.2 |
NXPE3 |
neurexophilin and PC-esterase domain family, member 3 |
| chr19_-_11456935 | 0.05 |
ENST00000590788.1 ENST00000586590.1 ENST00000589555.1 ENST00000586956.1 ENST00000593256.2 ENST00000447337.1 ENST00000591677.1 ENST00000586701.1 ENST00000589655.1 |
TMEM205 RAB3D |
transmembrane protein 205 RAB3D, member RAS oncogene family |
| chrX_+_113818545 | 0.05 |
ENST00000371951.1 ENST00000276198.1 ENST00000371950.3 |
HTR2C |
5-hydroxytryptamine (serotonin) receptor 2C, G protein-coupled |
| chr9_+_103947311 | 0.05 |
ENST00000395056.2 |
LPPR1 |
Lipid phosphate phosphatase-related protein type 1 |
| chr17_+_28268623 | 0.05 |
ENST00000394835.3 ENST00000320856.5 ENST00000394832.2 ENST00000378738.3 |
EFCAB5 |
EF-hand calcium binding domain 5 |
| chr19_-_6737576 | 0.05 |
ENST00000601716.1 ENST00000264080.7 |
GPR108 |
G protein-coupled receptor 108 |
| chr19_+_11350278 | 0.05 |
ENST00000252453.8 |
C19orf80 |
chromosome 19 open reading frame 80 |
| chr15_-_41836441 | 0.05 |
ENST00000567866.1 ENST00000561603.1 ENST00000304330.4 ENST00000566863.1 |
RPAP1 |
RNA polymerase II associated protein 1 |
| chr19_-_35719609 | 0.05 |
ENST00000324675.3 |
FAM187B |
family with sequence similarity 187, member B |
| chr5_-_180076613 | 0.05 |
ENST00000261937.6 ENST00000393347.3 |
FLT4 |
fms-related tyrosine kinase 4 |
| chr8_+_118147498 | 0.04 |
ENST00000519688.1 ENST00000456015.2 |
SLC30A8 |
solute carrier family 30 (zinc transporter), member 8 |
| chr11_+_3011093 | 0.04 |
ENST00000332881.2 |
AC131971.1 |
HCG1782999; PRO0943; Uncharacterized protein |
| chr16_+_67906919 | 0.04 |
ENST00000358933.5 |
EDC4 |
enhancer of mRNA decapping 4 |
| chr19_+_21265028 | 0.04 |
ENST00000291770.7 |
ZNF714 |
zinc finger protein 714 |
| chr14_-_50319482 | 0.04 |
ENST00000546046.1 ENST00000555970.1 ENST00000554626.1 ENST00000545773.1 ENST00000556672.1 |
NEMF |
nuclear export mediator factor |
| chr18_-_52989217 | 0.04 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr4_+_54243917 | 0.04 |
ENST00000507166.1 |
FIP1L1 |
factor interacting with PAPOLA and CPSF1 |
| chr19_-_11456905 | 0.04 |
ENST00000588560.1 ENST00000592952.1 |
TMEM205 |
transmembrane protein 205 |
| chr19_-_11456722 | 0.04 |
ENST00000354882.5 |
TMEM205 |
transmembrane protein 205 |
| chr4_-_69536346 | 0.04 |
ENST00000338206.5 |
UGT2B15 |
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr12_-_4647606 | 0.04 |
ENST00000261250.3 ENST00000541014.1 ENST00000545746.1 ENST00000542080.1 |
C12orf4 |
chromosome 12 open reading frame 4 |
| chr3_-_179169181 | 0.04 |
ENST00000497513.1 |
GNB4 |
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr12_-_9360966 | 0.04 |
ENST00000261336.2 |
PZP |
pregnancy-zone protein |
| chr4_-_148605265 | 0.04 |
ENST00000541232.1 ENST00000322396.6 |
PRMT10 |
protein arginine methyltransferase 10 (putative) |
| chr19_-_11456872 | 0.04 |
ENST00000586218.1 |
TMEM205 |
transmembrane protein 205 |
| chr9_-_21351377 | 0.04 |
ENST00000380210.1 |
IFNA6 |
interferon, alpha 6 |
| chrX_-_100129128 | 0.04 |
ENST00000372960.4 ENST00000372964.1 ENST00000217885.5 |
NOX1 |
NADPH oxidase 1 |
| chr19_-_23869999 | 0.04 |
ENST00000601935.1 ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675 |
zinc finger protein 675 |
| chr12_+_45609862 | 0.04 |
ENST00000423947.3 ENST00000435642.1 |
ANO6 |
anoctamin 6 |
| chr4_-_164395014 | 0.04 |
ENST00000280605.3 |
TKTL2 |
transketolase-like 2 |
| chr3_-_46037299 | 0.04 |
ENST00000296137.2 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
| chr7_+_23749894 | 0.04 |
ENST00000433467.2 |
STK31 |
serine/threonine kinase 31 |
| chr1_-_33502441 | 0.04 |
ENST00000548033.1 ENST00000487289.1 ENST00000373449.2 ENST00000480134.1 ENST00000467905.1 |
AK2 |
adenylate kinase 2 |
| chr6_+_29141311 | 0.04 |
ENST00000377167.2 |
OR2J2 |
olfactory receptor, family 2, subfamily J, member 2 |
| chr17_-_42092313 | 0.04 |
ENST00000587529.1 ENST00000206380.3 ENST00000542039.1 |
TMEM101 |
transmembrane protein 101 |
| chr11_+_63449045 | 0.04 |
ENST00000354497.4 |
RTN3 |
reticulon 3 |
| chr19_+_16999654 | 0.04 |
ENST00000248076.3 |
F2RL3 |
coagulation factor II (thrombin) receptor-like 3 |
| chr19_-_11457082 | 0.04 |
ENST00000587948.1 |
TMEM205 |
transmembrane protein 205 |
| chr19_+_29456034 | 0.04 |
ENST00000589921.1 |
LINC00906 |
long intergenic non-protein coding RNA 906 |
| chr4_-_71532339 | 0.04 |
ENST00000254801.4 |
IGJ |
immunoglobulin J polypeptide, linker protein for immunoglobulin alpha and mu polypeptides |
| chr13_-_103451307 | 0.03 |
ENST00000376004.4 |
KDELC1 |
KDEL (Lys-Asp-Glu-Leu) containing 1 |
| chr4_+_70861647 | 0.03 |
ENST00000246895.4 ENST00000381060.2 |
STATH |
statherin |
| chr7_-_4877677 | 0.03 |
ENST00000538469.1 |
RADIL |
Ras association and DIL domains |
| chr3_-_52864680 | 0.03 |
ENST00000406595.1 ENST00000485816.1 ENST00000434759.3 ENST00000346281.5 ENST00000266041.4 |
ITIH4 |
inter-alpha-trypsin inhibitor heavy chain family, member 4 |
| chr19_-_6279932 | 0.03 |
ENST00000252674.7 |
MLLT1 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 1 |
| chr7_+_64838712 | 0.03 |
ENST00000328747.7 ENST00000431504.1 ENST00000357512.2 |
ZNF92 |
zinc finger protein 92 |
| chr21_-_27423339 | 0.03 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr20_+_36946029 | 0.03 |
ENST00000417318.1 |
BPI |
bactericidal/permeability-increasing protein |
| chr15_+_71228826 | 0.03 |
ENST00000558456.1 ENST00000560158.2 ENST00000558808.1 ENST00000559806.1 ENST00000559069.1 |
LRRC49 |
leucine rich repeat containing 49 |
| chr1_-_86174065 | 0.03 |
ENST00000370574.3 ENST00000431532.2 |
ZNHIT6 |
zinc finger, HIT-type containing 6 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.5 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.2 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.3 | GO:1900264 | regulation of DNA-directed DNA polymerase activity(GO:1900262) positive regulation of DNA-directed DNA polymerase activity(GO:1900264) |
| 0.0 | 0.2 | GO:0097045 | phosphatidylserine exposure on blood platelet(GO:0097045) |
| 0.0 | 0.2 | GO:0002669 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.2 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.1 | GO:0003095 | pressure natriuresis(GO:0003095) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0006433 | glutamyl-tRNA aminoacylation(GO:0006424) prolyl-tRNA aminoacylation(GO:0006433) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.2 | GO:1903899 | lung goblet cell differentiation(GO:0060480) positive regulation of PERK-mediated unfolded protein response(GO:1903899) |
| 0.0 | 0.1 | GO:0045726 | positive regulation of integrin biosynthetic process(GO:0045726) |
| 0.0 | 0.2 | GO:0051563 | smooth endoplasmic reticulum calcium ion homeostasis(GO:0051563) |
| 0.0 | 0.2 | GO:1904207 | regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904207) positive regulation of chemokine (C-C motif) ligand 2 secretion(GO:1904209) |
| 0.0 | 0.2 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.9 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.0 | 0.4 | GO:0097296 | activation of cysteine-type endopeptidase activity involved in apoptotic signaling pathway(GO:0097296) |
| 0.0 | 0.2 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.3 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 | 0.1 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:0034473 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.3 | GO:0070234 | positive regulation of T cell apoptotic process(GO:0070234) |
| 0.0 | 1.8 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.0 | GO:0048611 | ectodermal digestive tract development(GO:0007439) embryonic ectodermal digestive tract development(GO:0048611) |
| 0.0 | 0.1 | GO:0097267 | omega-hydroxylase P450 pathway(GO:0097267) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.2 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.0 | 0.1 | GO:0050747 | sphingomyelin biosynthetic process(GO:0006686) positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 0.3 | GO:0005663 | DNA replication factor C complex(GO:0005663) |
| 0.0 | 0.1 | GO:0030849 | autosome(GO:0030849) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0000801 | central element(GO:0000801) |
| 0.0 | 0.4 | GO:0000812 | Swr1 complex(GO:0000812) |
| 0.0 | 0.1 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.2 | GO:0033269 | internode region of axon(GO:0033269) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.1 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.2 | GO:1990712 | HFE-transferrin receptor complex(GO:1990712) |
| 0.0 | 0.1 | GO:0070765 | gamma-secretase complex(GO:0070765) |
| 0.0 | 0.7 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0097427 | microtubule bundle(GO:0097427) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.2 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.1 | 0.3 | GO:0030395 | lactose binding(GO:0030395) |
| 0.1 | 0.3 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.1 | 0.5 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.3 | GO:0033170 | DNA clamp loader activity(GO:0003689) protein-DNA loading ATPase activity(GO:0033170) |
| 0.0 | 0.3 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.1 | GO:0080101 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.1 | GO:0018685 | alkane 1-monooxygenase activity(GO:0018685) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.1 | GO:0004827 | glutamate-tRNA ligase activity(GO:0004818) proline-tRNA ligase activity(GO:0004827) |
| 0.0 | 0.1 | GO:0030171 | voltage-gated proton channel activity(GO:0030171) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.3 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.2 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
| 0.0 | 0.1 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.0 | 0.4 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.0 | 0.4 | GO:0004806 | triglyceride lipase activity(GO:0004806) |
| 0.0 | 0.1 | GO:0016713 | oxidoreductase activity, acting on paired donors, with incorporation or reduction of molecular oxygen, reduced iron-sulfur protein as one donor, and incorporation of one atom of oxygen(GO:0016713) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0008821 | crossover junction endodeoxyribonuclease activity(GO:0008821) |
| 0.0 | 0.4 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.3 | REACTOME POL SWITCHING | Genes involved in Polymerase switching |
| 0.0 | 0.4 | REACTOME EXTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Extrinsic Pathway for Apoptosis |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |