Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for HOXC9
Z-value: 0.95
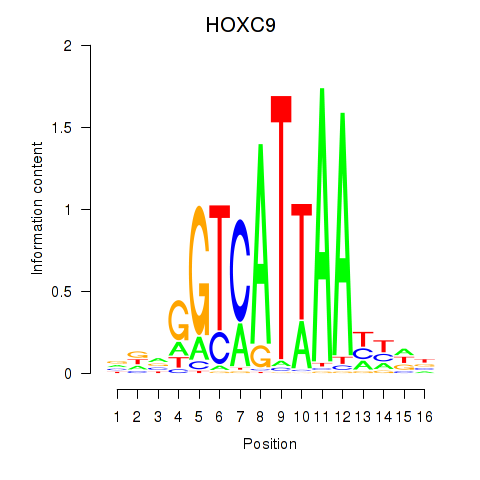
Transcription factors associated with HOXC9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXC9
|
ENSG00000180806.4 | HOXC9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXC9 | hg19_v2_chr12_+_54393880_54393962 | 0.52 | 1.8e-01 | Click! |
Activity profile of HOXC9 motif
Sorted Z-values of HOXC9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXC9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91546926 | 3.39 |
ENST00000550758.1 |
DCN |
decorin |
| chr1_+_163039143 | 2.45 |
ENST00000531057.1 ENST00000527809.1 ENST00000367908.4 |
RGS4 |
regulator of G-protein signaling 4 |
| chr4_-_70626314 | 2.11 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr6_-_52859046 | 1.33 |
ENST00000457564.1 ENST00000541324.1 ENST00000370960.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr4_+_70916119 | 1.31 |
ENST00000246896.3 ENST00000511674.1 |
HTN1 |
histatin 1 |
| chr2_-_225811747 | 1.21 |
ENST00000409592.3 |
DOCK10 |
dedicator of cytokinesis 10 |
| chr5_-_111093167 | 1.18 |
ENST00000446294.2 ENST00000419114.2 |
NREP |
neuronal regeneration related protein |
| chr4_-_70626430 | 1.02 |
ENST00000310613.3 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr8_+_16884740 | 1.00 |
ENST00000318063.5 |
MICU3 |
mitochondrial calcium uptake family, member 3 |
| chr7_+_138915102 | 0.95 |
ENST00000486663.1 |
UBN2 |
ubinuclein 2 |
| chr17_-_66951474 | 0.95 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr1_+_221051699 | 0.92 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr4_-_156787425 | 0.92 |
ENST00000537611.2 |
ASIC5 |
acid-sensing (proton-gated) ion channel family member 5 |
| chr5_+_156696362 | 0.89 |
ENST00000377576.3 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr10_-_13344341 | 0.89 |
ENST00000396920.3 |
PHYH |
phytanoyl-CoA 2-hydroxylase |
| chr4_+_70894130 | 0.86 |
ENST00000526767.1 ENST00000530128.1 ENST00000381057.3 |
HTN3 |
histatin 3 |
| chr22_-_32651326 | 0.81 |
ENST00000266086.4 |
SLC5A4 |
solute carrier family 5 (glucose activated ion channel), member 4 |
| chr11_+_19798964 | 0.78 |
ENST00000527559.2 |
NAV2 |
neuron navigator 2 |
| chr13_-_33760216 | 0.76 |
ENST00000255486.4 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr12_-_63328817 | 0.61 |
ENST00000228705.6 |
PPM1H |
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr14_+_21359558 | 0.61 |
ENST00000304639.3 |
RNASE3 |
ribonuclease, RNase A family, 3 |
| chr16_-_12897642 | 0.59 |
ENST00000433677.2 ENST00000261660.4 ENST00000381774.4 |
CPPED1 |
calcineurin-like phosphoesterase domain containing 1 |
| chr2_+_158114051 | 0.56 |
ENST00000259056.4 |
GALNT5 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 5 (GalNAc-T5) |
| chr19_-_58864848 | 0.55 |
ENST00000263100.3 |
A1BG |
alpha-1-B glycoprotein |
| chr3_+_69812701 | 0.55 |
ENST00000472437.1 |
MITF |
microphthalmia-associated transcription factor |
| chr7_+_7606497 | 0.55 |
ENST00000340080.4 ENST00000405785.1 ENST00000433635.1 |
MIOS |
missing oocyte, meiosis regulator, homolog (Drosophila) |
| chr5_-_10761206 | 0.55 |
ENST00000432074.2 ENST00000230895.6 |
DAP |
death-associated protein |
| chr6_-_88876058 | 0.54 |
ENST00000369501.2 |
CNR1 |
cannabinoid receptor 1 (brain) |
| chr12_-_104234966 | 0.54 |
ENST00000392876.3 |
NT5DC3 |
5'-nucleotidase domain containing 3 |
| chr2_-_224467093 | 0.52 |
ENST00000305409.2 |
SCG2 |
secretogranin II |
| chr11_+_5710919 | 0.51 |
ENST00000379965.3 ENST00000425490.1 |
TRIM22 |
tripartite motif containing 22 |
| chr2_+_210444142 | 0.51 |
ENST00000360351.4 ENST00000361559.4 |
MAP2 |
microtubule-associated protein 2 |
| chr8_-_93029865 | 0.48 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr7_+_141811539 | 0.48 |
ENST00000550469.2 ENST00000477922.3 |
RP11-1220K2.2 |
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr2_+_109237717 | 0.48 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr11_+_65266507 | 0.47 |
ENST00000544868.1 |
MALAT1 |
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr12_-_15374343 | 0.46 |
ENST00000256953.2 ENST00000546331.1 |
RERG |
RAS-like, estrogen-regulated, growth inhibitor |
| chr3_+_138066539 | 0.46 |
ENST00000289104.4 |
MRAS |
muscle RAS oncogene homolog |
| chr5_-_139930713 | 0.45 |
ENST00000602657.1 |
SRA1 |
steroid receptor RNA activator 1 |
| chr2_+_62132800 | 0.45 |
ENST00000538736.1 |
COMMD1 |
copper metabolism (Murr1) domain containing 1 |
| chr10_+_45495898 | 0.44 |
ENST00000298299.3 |
ZNF22 |
zinc finger protein 22 |
| chr11_-_26593779 | 0.43 |
ENST00000529533.1 |
MUC15 |
mucin 15, cell surface associated |
| chr11_-_26593677 | 0.43 |
ENST00000527569.1 |
MUC15 |
mucin 15, cell surface associated |
| chr10_-_27529486 | 0.42 |
ENST00000375888.1 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chr1_-_204183071 | 0.42 |
ENST00000308302.3 |
GOLT1A |
golgi transport 1A |
| chr2_+_109271481 | 0.42 |
ENST00000542845.1 ENST00000393314.2 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr2_+_62132781 | 0.39 |
ENST00000311832.5 |
COMMD1 |
copper metabolism (Murr1) domain containing 1 |
| chr4_-_119759795 | 0.37 |
ENST00000419654.2 |
SEC24D |
SEC24 family member D |
| chr19_-_48752812 | 0.36 |
ENST00000359009.4 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr6_-_52859968 | 0.36 |
ENST00000370959.1 |
GSTA4 |
glutathione S-transferase alpha 4 |
| chr2_+_161993465 | 0.34 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr19_-_48753104 | 0.34 |
ENST00000447740.2 |
CARD8 |
caspase recruitment domain family, member 8 |
| chr2_+_162016827 | 0.33 |
ENST00000429217.1 ENST00000406287.1 ENST00000402568.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr12_-_31743901 | 0.33 |
ENST00000354285.4 |
DENND5B |
DENN/MADD domain containing 5B |
| chr2_+_109204743 | 0.33 |
ENST00000332345.6 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr21_-_31859755 | 0.32 |
ENST00000334055.3 |
KRTAP19-2 |
keratin associated protein 19-2 |
| chr9_+_131549610 | 0.32 |
ENST00000223865.8 |
TBC1D13 |
TBC1 domain family, member 13 |
| chr16_-_66583701 | 0.31 |
ENST00000527800.1 ENST00000525974.1 ENST00000563369.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr6_+_83903061 | 0.30 |
ENST00000369724.4 ENST00000539997.1 |
RWDD2A |
RWD domain containing 2A |
| chr15_-_20193370 | 0.30 |
ENST00000558565.2 |
IGHV3OR15-7 |
immunoglobulin heavy variable 3/OR15-7 (pseudogene) |
| chr10_-_127505167 | 0.29 |
ENST00000368786.1 |
UROS |
uroporphyrinogen III synthase |
| chr16_-_66584059 | 0.29 |
ENST00000417693.3 ENST00000544898.1 ENST00000569718.1 ENST00000527284.1 ENST00000299697.7 ENST00000451102.2 |
TK2 |
thymidine kinase 2, mitochondrial |
| chr6_+_32812568 | 0.29 |
ENST00000414474.1 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr2_+_162016804 | 0.29 |
ENST00000392749.2 ENST00000440506.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr20_+_57414743 | 0.29 |
ENST00000313949.7 |
GNAS |
GNAS complex locus |
| chr12_-_10605929 | 0.28 |
ENST00000347831.5 ENST00000359151.3 |
KLRC1 |
killer cell lectin-like receptor subfamily C, member 1 |
| chr3_+_185046676 | 0.27 |
ENST00000428617.1 ENST00000443863.1 |
MAP3K13 |
mitogen-activated protein kinase kinase kinase 13 |
| chr12_-_10573149 | 0.27 |
ENST00000381904.2 ENST00000381903.2 ENST00000396439.2 |
KLRC3 |
killer cell lectin-like receptor subfamily C, member 3 |
| chr20_-_33732952 | 0.27 |
ENST00000541621.1 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr2_+_161993412 | 0.27 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr13_-_24007815 | 0.27 |
ENST00000382298.3 |
SACS |
spastic ataxia of Charlevoix-Saguenay (sacsin) |
| chr2_+_162016916 | 0.27 |
ENST00000405852.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr9_+_2029019 | 0.26 |
ENST00000382194.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr8_+_7693993 | 0.26 |
ENST00000314265.2 |
DEFB104A |
defensin, beta 104A |
| chr1_-_147599549 | 0.26 |
ENST00000369228.5 |
NBPF24 |
neuroblastoma breakpoint family, member 24 |
| chr1_-_148347506 | 0.25 |
ENST00000369189.3 |
NBPF20 |
neuroblastoma breakpoint family, member 20 |
| chr2_-_111230393 | 0.25 |
ENST00000447537.2 ENST00000413601.2 |
LIMS3L LIMS3L |
LIM and senescent cell antigen-like-containing domain protein 3; Uncharacterized protein; cDNA FLJ59124, highly similar to Particularly interesting newCys-His protein; cDNA, FLJ79109, highly similar to Particularly interesting newCys-His protein LIM and senescent cell antigen-like domains 3-like |
| chr13_+_49551020 | 0.25 |
ENST00000541916.1 |
FNDC3A |
fibronectin type III domain containing 3A |
| chr6_+_28249299 | 0.25 |
ENST00000405948.2 |
PGBD1 |
piggyBac transposable element derived 1 |
| chr11_-_26593649 | 0.24 |
ENST00000455601.2 |
MUC15 |
mucin 15, cell surface associated |
| chr2_+_109204909 | 0.24 |
ENST00000393310.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr17_-_46690839 | 0.24 |
ENST00000498634.2 |
HOXB8 |
homeobox B8 |
| chr1_-_622053 | 0.24 |
ENST00000332831.2 |
OR4F16 |
olfactory receptor, family 4, subfamily F, member 16 |
| chr8_-_116504448 | 0.24 |
ENST00000518018.1 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr6_+_28249332 | 0.23 |
ENST00000259883.3 |
PGBD1 |
piggyBac transposable element derived 1 |
| chrX_+_16668278 | 0.23 |
ENST00000380200.3 |
S100G |
S100 calcium binding protein G |
| chr7_+_80275621 | 0.23 |
ENST00000426978.1 ENST00000432207.1 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr12_+_25348186 | 0.22 |
ENST00000555711.1 ENST00000556885.1 ENST00000554266.1 ENST00000556351.1 ENST00000556927.1 ENST00000556402.1 ENST00000553788.1 |
LYRM5 |
LYR motif containing 5 |
| chr12_+_104235229 | 0.22 |
ENST00000551650.1 |
RP11-650K20.3 |
Uncharacterized protein |
| chr12_+_75874580 | 0.22 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr12_+_25348139 | 0.22 |
ENST00000557540.2 ENST00000381356.4 |
LYRM5 |
LYR motif containing 5 |
| chr20_+_57414795 | 0.22 |
ENST00000371098.2 ENST00000371075.3 |
GNAS |
GNAS complex locus |
| chrX_+_120181457 | 0.22 |
ENST00000328078.1 |
GLUD2 |
glutamate dehydrogenase 2 |
| chr7_+_80275752 | 0.22 |
ENST00000419819.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr10_-_115614127 | 0.21 |
ENST00000369305.1 |
DCLRE1A |
DNA cross-link repair 1A |
| chr18_-_12656715 | 0.21 |
ENST00000462226.1 ENST00000497844.2 ENST00000309836.5 ENST00000453447.2 |
SPIRE1 |
spire-type actin nucleation factor 1 |
| chr5_+_98109322 | 0.21 |
ENST00000513185.1 |
RGMB |
repulsive guidance molecule family member b |
| chr2_-_207629997 | 0.21 |
ENST00000454776.2 |
MDH1B |
malate dehydrogenase 1B, NAD (soluble) |
| chr22_+_32149927 | 0.21 |
ENST00000437411.1 ENST00000535622.1 ENST00000536766.1 ENST00000400242.3 ENST00000266091.3 ENST00000400249.2 ENST00000400246.1 ENST00000382105.2 |
DEPDC5 |
DEP domain containing 5 |
| chr19_-_54850417 | 0.21 |
ENST00000291759.4 |
LILRA4 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 4 |
| chrX_+_153775821 | 0.21 |
ENST00000263518.6 ENST00000470142.1 ENST00000393549.2 ENST00000455588.2 ENST00000369602.3 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr5_-_125930929 | 0.21 |
ENST00000553117.1 ENST00000447989.2 ENST00000409134.3 |
ALDH7A1 |
aldehyde dehydrogenase 7 family, member A1 |
| chr18_-_32870148 | 0.20 |
ENST00000589178.1 ENST00000333206.5 ENST00000592278.1 ENST00000592211.1 ENST00000420878.3 ENST00000383091.2 ENST00000586922.2 |
ZSCAN30 RP11-158H5.7 |
zinc finger and SCAN domain containing 30 RP11-158H5.7 |
| chrX_+_43515467 | 0.20 |
ENST00000338702.3 ENST00000542639.1 |
MAOA |
monoamine oxidase A |
| chr12_-_90049828 | 0.20 |
ENST00000261173.2 ENST00000348959.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr6_-_83903600 | 0.20 |
ENST00000506587.1 ENST00000507554.1 |
PGM3 |
phosphoglucomutase 3 |
| chr5_-_176433693 | 0.20 |
ENST00000507513.1 ENST00000511320.1 |
UIMC1 |
ubiquitin interaction motif containing 1 |
| chr14_+_57671888 | 0.20 |
ENST00000391612.1 |
AL391152.1 |
AL391152.1 |
| chrX_-_16887963 | 0.19 |
ENST00000380084.4 |
RBBP7 |
retinoblastoma binding protein 7 |
| chr5_-_145562147 | 0.19 |
ENST00000545646.1 ENST00000274562.9 ENST00000510191.1 ENST00000394434.2 |
LARS |
leucyl-tRNA synthetase |
| chr2_+_110656005 | 0.19 |
ENST00000437679.2 |
LIMS3 |
LIM and senescent cell antigen-like domains 3 |
| chr6_-_111804905 | 0.19 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr12_-_90049878 | 0.19 |
ENST00000359142.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr6_-_26189304 | 0.18 |
ENST00000340756.2 |
HIST1H4D |
histone cluster 1, H4d |
| chr12_-_13248732 | 0.18 |
ENST00000396302.3 |
GSG1 |
germ cell associated 1 |
| chr5_+_135468516 | 0.18 |
ENST00000507118.1 ENST00000511116.1 ENST00000545279.1 ENST00000545620.1 |
SMAD5 |
SMAD family member 5 |
| chr1_+_192127578 | 0.18 |
ENST00000367460.3 |
RGS18 |
regulator of G-protein signaling 18 |
| chr10_-_31146615 | 0.18 |
ENST00000444692.2 |
ZNF438 |
zinc finger protein 438 |
| chr1_+_212965170 | 0.18 |
ENST00000532324.1 ENST00000366974.4 ENST00000530441.1 ENST00000526641.1 ENST00000531963.1 ENST00000366973.4 ENST00000526997.1 ENST00000488246.2 |
TATDN3 |
TatD DNase domain containing 3 |
| chr21_-_31869451 | 0.17 |
ENST00000334058.2 |
KRTAP19-4 |
keratin associated protein 19-4 |
| chr2_-_69664586 | 0.17 |
ENST00000303698.3 ENST00000394305.1 ENST00000410022.2 |
NFU1 |
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr9_+_131549483 | 0.17 |
ENST00000372648.5 ENST00000539497.1 |
TBC1D13 |
TBC1 domain family, member 13 |
| chr13_+_21714711 | 0.17 |
ENST00000607003.1 ENST00000492245.1 |
SAP18 |
Sin3A-associated protein, 18kDa |
| chr12_+_58176525 | 0.17 |
ENST00000543727.1 ENST00000540550.1 ENST00000323833.8 ENST00000350762.5 ENST00000550559.1 ENST00000548851.1 ENST00000434359.1 ENST00000457189.1 |
TSFM |
Ts translation elongation factor, mitochondrial |
| chr5_-_58295712 | 0.16 |
ENST00000317118.8 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr16_+_22501658 | 0.16 |
ENST00000415833.2 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr1_-_158656488 | 0.16 |
ENST00000368147.4 |
SPTA1 |
spectrin, alpha, erythrocytic 1 (elliptocytosis 2) |
| chr1_+_26798955 | 0.16 |
ENST00000361427.5 |
HMGN2 |
high mobility group nucleosomal binding domain 2 |
| chr7_-_156685890 | 0.16 |
ENST00000353442.5 |
LMBR1 |
limb development membrane protein 1 |
| chr1_-_212965104 | 0.16 |
ENST00000422588.2 ENST00000366975.6 ENST00000366977.3 ENST00000366976.1 |
NSL1 |
NSL1, MIS12 kinetochore complex component |
| chr7_+_80275953 | 0.16 |
ENST00000538969.1 ENST00000544133.1 ENST00000433696.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr11_-_62911693 | 0.16 |
ENST00000417740.1 ENST00000326192.5 |
SLC22A24 |
solute carrier family 22, member 24 |
| chr5_-_148442584 | 0.16 |
ENST00000394358.2 ENST00000512049.1 |
SH3TC2 |
SH3 domain and tetratricopeptide repeats 2 |
| chr20_+_56964169 | 0.16 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr10_+_134150835 | 0.16 |
ENST00000432555.2 |
LRRC27 |
leucine rich repeat containing 27 |
| chr13_+_36050881 | 0.15 |
ENST00000537702.1 |
NBEA |
neurobeachin |
| chr19_-_20748541 | 0.15 |
ENST00000427401.4 ENST00000594419.1 |
ZNF737 |
zinc finger protein 737 |
| chr6_-_146057144 | 0.15 |
ENST00000367519.3 |
EPM2A |
epilepsy, progressive myoclonus type 2A, Lafora disease (laforin) |
| chr2_-_230096756 | 0.15 |
ENST00000354069.6 |
PID1 |
phosphotyrosine interaction domain containing 1 |
| chr5_+_176731572 | 0.15 |
ENST00000503853.1 |
PRELID1 |
PRELI domain containing 1 |
| chr13_+_108921977 | 0.15 |
ENST00000430559.1 ENST00000375887.4 |
TNFSF13B |
tumor necrosis factor (ligand) superfamily, member 13b |
| chr8_-_105479270 | 0.15 |
ENST00000521573.2 ENST00000351513.2 |
DPYS |
dihydropyrimidinase |
| chr3_+_63953415 | 0.14 |
ENST00000484332.1 |
ATXN7 |
ataxin 7 |
| chrX_-_13835147 | 0.14 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr5_-_148033726 | 0.14 |
ENST00000354217.2 ENST00000314512.6 ENST00000362016.2 |
HTR4 |
5-hydroxytryptamine (serotonin) receptor 4, G protein-coupled |
| chr11_+_114310102 | 0.14 |
ENST00000265881.5 |
REXO2 |
RNA exonuclease 2 |
| chr2_+_108994633 | 0.13 |
ENST00000409309.3 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
| chr2_+_108994466 | 0.13 |
ENST00000272452.2 |
SULT1C4 |
sulfotransferase family, cytosolic, 1C, member 4 |
| chr16_-_66764119 | 0.13 |
ENST00000569320.1 |
DYNC1LI2 |
dynein, cytoplasmic 1, light intermediate chain 2 |
| chr8_-_62602327 | 0.13 |
ENST00000445642.3 ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH |
aspartate beta-hydroxylase |
| chr9_+_82187630 | 0.13 |
ENST00000265284.6 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr14_+_52327350 | 0.13 |
ENST00000555472.1 ENST00000556766.1 |
GNG2 |
guanine nucleotide binding protein (G protein), gamma 2 |
| chr16_+_25228242 | 0.13 |
ENST00000219660.5 |
AQP8 |
aquaporin 8 |
| chr17_+_45286706 | 0.13 |
ENST00000393450.1 ENST00000572303.1 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr12_-_11091862 | 0.13 |
ENST00000537503.1 |
TAS2R14 |
taste receptor, type 2, member 14 |
| chr17_-_9862772 | 0.13 |
ENST00000580865.1 ENST00000583882.1 |
GAS7 |
growth arrest-specific 7 |
| chr12_-_95945246 | 0.13 |
ENST00000258499.3 |
USP44 |
ubiquitin specific peptidase 44 |
| chr18_+_32558208 | 0.13 |
ENST00000436190.2 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr9_-_15472730 | 0.13 |
ENST00000481862.1 |
PSIP1 |
PC4 and SFRS1 interacting protein 1 |
| chr5_-_42811986 | 0.13 |
ENST00000511224.1 ENST00000507920.1 ENST00000510965.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr3_+_138340049 | 0.12 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr1_+_35734562 | 0.12 |
ENST00000314607.6 ENST00000373297.2 |
ZMYM4 |
zinc finger, MYM-type 4 |
| chr2_-_242212227 | 0.12 |
ENST00000427007.1 ENST00000458564.1 ENST00000452065.1 ENST00000427183.2 ENST00000426343.1 ENST00000422080.1 ENST00000449504.1 ENST00000449864.1 ENST00000391975.1 |
HDLBP |
high density lipoprotein binding protein |
| chr6_+_47749718 | 0.12 |
ENST00000489301.2 ENST00000371211.2 ENST00000393699.2 |
OPN5 |
opsin 5 |
| chr9_+_80912059 | 0.12 |
ENST00000347159.2 ENST00000376588.3 |
PSAT1 |
phosphoserine aminotransferase 1 |
| chr19_+_21106081 | 0.12 |
ENST00000300540.3 ENST00000595854.1 ENST00000601284.1 ENST00000328178.8 ENST00000599885.1 ENST00000596476.1 ENST00000345030.6 |
ZNF85 |
zinc finger protein 85 |
| chrX_-_15332665 | 0.12 |
ENST00000537676.1 ENST00000344384.4 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr10_+_74927875 | 0.12 |
ENST00000242505.6 |
FAM149B1 |
family with sequence similarity 149, member B1 |
| chr17_+_42264556 | 0.12 |
ENST00000319511.6 ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr18_-_59274139 | 0.11 |
ENST00000586949.1 |
RP11-879F14.2 |
RP11-879F14.2 |
| chr2_-_207630033 | 0.11 |
ENST00000449792.1 |
MDH1B |
malate dehydrogenase 1B, NAD (soluble) |
| chr19_-_23869999 | 0.11 |
ENST00000601935.1 ENST00000359788.4 ENST00000600313.1 ENST00000596211.1 ENST00000599168.1 |
ZNF675 |
zinc finger protein 675 |
| chr5_-_68665084 | 0.11 |
ENST00000509462.1 |
TAF9 |
TAF9 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 32kDa |
| chr4_-_52883786 | 0.11 |
ENST00000343457.3 |
LRRC66 |
leucine rich repeat containing 66 |
| chr1_+_63989004 | 0.11 |
ENST00000371088.4 |
EFCAB7 |
EF-hand calcium binding domain 7 |
| chr9_+_82187487 | 0.11 |
ENST00000435650.1 ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr6_+_158733692 | 0.11 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chr4_-_103748880 | 0.11 |
ENST00000453744.2 ENST00000349311.8 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr19_+_3506261 | 0.11 |
ENST00000441788.2 |
FZR1 |
fizzy/cell division cycle 20 related 1 (Drosophila) |
| chr12_-_27167233 | 0.11 |
ENST00000535819.1 ENST00000543803.1 ENST00000535423.1 ENST00000539741.1 ENST00000343028.4 ENST00000545600.1 ENST00000543088.1 |
TM7SF3 |
transmembrane 7 superfamily member 3 |
| chr1_+_151735431 | 0.11 |
ENST00000321531.5 ENST00000315067.8 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr15_+_45544426 | 0.11 |
ENST00000347644.3 ENST00000560438.1 |
SLC28A2 |
solute carrier family 28 (concentrative nucleoside transporter), member 2 |
| chr11_-_55703876 | 0.10 |
ENST00000301532.3 |
OR5I1 |
olfactory receptor, family 5, subfamily I, member 1 |
| chrX_+_13671225 | 0.10 |
ENST00000545566.1 ENST00000544987.1 ENST00000314720.4 |
TCEANC |
transcription elongation factor A (SII) N-terminal and central domain containing |
| chr14_-_45252031 | 0.10 |
ENST00000556405.1 |
RP11-398E10.1 |
RP11-398E10.1 |
| chr10_+_115614370 | 0.10 |
ENST00000369301.3 |
NHLRC2 |
NHL repeat containing 2 |
| chr16_-_56553882 | 0.10 |
ENST00000568104.1 |
BBS2 |
Bardet-Biedl syndrome 2 |
| chr7_-_156685841 | 0.10 |
ENST00000354505.4 ENST00000540390.1 |
LMBR1 |
limb development membrane protein 1 |
| chr4_-_103749179 | 0.10 |
ENST00000502690.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr17_+_33914276 | 0.10 |
ENST00000592545.1 ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr2_-_109605663 | 0.10 |
ENST00000409271.1 ENST00000258443.2 ENST00000376651.1 |
EDAR |
ectodysplasin A receptor |
| chr11_+_114310164 | 0.09 |
ENST00000544196.1 ENST00000539754.1 ENST00000539275.1 |
REXO2 |
RNA exonuclease 2 |
| chr8_-_93978357 | 0.09 |
ENST00000522925.1 ENST00000522903.1 ENST00000537541.1 ENST00000518748.1 ENST00000519069.1 ENST00000521988.1 |
TRIQK |
triple QxxK/R motif containing |
| chr9_+_137979506 | 0.09 |
ENST00000539529.1 ENST00000392991.4 ENST00000371793.3 |
OLFM1 |
olfactomedin 1 |
| chr5_-_148033693 | 0.09 |
ENST00000377888.3 ENST00000360693.3 |
HTR4 |
5-hydroxytryptamine (serotonin) receptor 4, G protein-coupled |
| chr19_+_21106028 | 0.09 |
ENST00000597314.1 ENST00000601924.1 |
ZNF85 |
zinc finger protein 85 |
| chr2_-_70475701 | 0.09 |
ENST00000282574.4 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr5_-_16738451 | 0.09 |
ENST00000274203.9 ENST00000515803.1 |
MYO10 |
myosin X |
| chr9_-_179018 | 0.09 |
ENST00000431099.2 ENST00000382447.4 ENST00000382389.1 ENST00000377447.3 ENST00000314367.10 ENST00000356521.4 ENST00000382393.1 ENST00000377400.4 |
CBWD1 |
COBW domain containing 1 |
| chr14_+_55494323 | 0.09 |
ENST00000339298.2 |
SOCS4 |
suppressor of cytokine signaling 4 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.2 | 3.4 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.2 | 0.9 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.2 | 0.6 | GO:0046125 | thymidine metabolic process(GO:0046104) pyrimidine deoxyribonucleoside metabolic process(GO:0046125) |
| 0.1 | 0.5 | GO:0099553 | trans-synaptic signaling by lipid, modulating synaptic transmission(GO:0099552) trans-synaptic signaling by endocannabinoid, modulating synaptic transmission(GO:0099553) |
| 0.1 | 3.4 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.1 | 0.8 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.1 | 0.9 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.3 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.1 | 0.8 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.1 | 0.3 | GO:0036509 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 0.9 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.1 | 1.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.1 | 0.6 | GO:0072564 | lipoprotein particle mediated signaling(GO:0055095) low-density lipoprotein particle mediated signaling(GO:0055096) blood microparticle formation(GO:0072564) regulation of blood microparticle formation(GO:2000332) positive regulation of blood microparticle formation(GO:2000334) |
| 0.1 | 1.0 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.1 | 0.5 | GO:0040032 | post-embryonic body morphogenesis(GO:0040032) |
| 0.1 | 0.2 | GO:0031456 | glycine betaine biosynthetic process from choline(GO:0019285) glycine betaine metabolic process(GO:0031455) glycine betaine biosynthetic process(GO:0031456) |
| 0.1 | 0.2 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.1 | 0.4 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.9 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.1 | 2.4 | GO:0000188 | inactivation of MAPK activity(GO:0000188) |
| 0.0 | 0.1 | GO:0031296 | B cell costimulation(GO:0031296) |
| 0.0 | 0.1 | GO:0006208 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) beta-alanine metabolic process(GO:0019482) thymine metabolic process(GO:0019859) uracil metabolic process(GO:0019860) |
| 0.0 | 0.4 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 2.2 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.6 | GO:0002227 | innate immune response in mucosa(GO:0002227) |
| 0.0 | 1.7 | GO:1901687 | glutathione derivative metabolic process(GO:1901685) glutathione derivative biosynthetic process(GO:1901687) |
| 0.0 | 0.1 | GO:0003147 | neural crest cell migration involved in heart formation(GO:0003147) anterior neural tube closure(GO:0061713) |
| 0.0 | 0.7 | GO:0050713 | negative regulation of interleukin-1 beta secretion(GO:0050713) |
| 0.0 | 0.2 | GO:0019255 | glucose 1-phosphate metabolic process(GO:0019255) |
| 0.0 | 0.2 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.2 | GO:2001170 | negative regulation of mitochondrial DNA replication(GO:0090298) negative regulation of mitochondrial DNA metabolic process(GO:1901859) negative regulation of ATP biosynthetic process(GO:2001170) |
| 0.0 | 0.5 | GO:0048245 | eosinophil chemotaxis(GO:0048245) |
| 0.0 | 0.9 | GO:0006851 | mitochondrial calcium ion transport(GO:0006851) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0070537 | histone H2A K63-linked deubiquitination(GO:0070537) |
| 0.0 | 0.3 | GO:1904628 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 1.5 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.2 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.0 | 0.2 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.2 | GO:0001880 | Mullerian duct regression(GO:0001880) |
| 0.0 | 0.8 | GO:1904659 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.3 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 1.3 | GO:0034198 | cellular response to amino acid starvation(GO:0034198) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0042816 | vitamin B6 metabolic process(GO:0042816) |
| 0.0 | 0.2 | GO:0042420 | dopamine catabolic process(GO:0042420) |
| 0.0 | 0.1 | GO:0038108 | negative regulation of appetite by leptin-mediated signaling pathway(GO:0038108) |
| 0.0 | 0.1 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.0 | 0.2 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.4 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 1.4 | GO:0016266 | O-glycan processing(GO:0016266) |
| 0.0 | 0.2 | GO:0008104 | protein localization(GO:0008104) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.2 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
| 0.0 | 0.2 | GO:0032287 | peripheral nervous system myelin maintenance(GO:0032287) |
| 0.0 | 0.3 | GO:0090084 | negative regulation of inclusion body assembly(GO:0090084) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:0051610 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.1 | GO:2000660 | negative regulation of interleukin-1-mediated signaling pathway(GO:2000660) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0097068 | response to thyroxine(GO:0097068) response to L-phenylalanine derivative(GO:1904386) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.6 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0000492 | box C/D snoRNP assembly(GO:0000492) |
| 0.0 | 0.2 | GO:0060009 | Sertoli cell development(GO:0060009) |
| 0.0 | 0.1 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.1 | GO:0003190 | atrioventricular valve formation(GO:0003190) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 3.4 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.1 | 0.5 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.8 | GO:0031462 | Cul2-RING ubiquitin ligase complex(GO:0031462) |
| 0.0 | 0.5 | GO:0061700 | GATOR2 complex(GO:0061700) |
| 0.0 | 0.2 | GO:0035859 | Seh1-associated complex(GO:0035859) Iml1 complex(GO:1990130) |
| 0.0 | 0.7 | GO:0072559 | NLRP3 inflammasome complex(GO:0072559) |
| 0.0 | 0.8 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.3 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.5 | GO:0008385 | IkappaB kinase complex(GO:0008385) |
| 0.0 | 0.2 | GO:0000444 | MIS12/MIND type complex(GO:0000444) |
| 0.0 | 0.2 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.3 | GO:0070852 | cell body fiber(GO:0070852) |
| 0.0 | 0.2 | GO:0032437 | cuticular plate(GO:0032437) |
| 0.0 | 0.4 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.2 | GO:0070531 | BRCA1-A complex(GO:0070531) |
| 0.0 | 0.6 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.4 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.4 | GO:0032591 | dendritic spine membrane(GO:0032591) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0070761 | PCAF complex(GO:0000125) pre-snoRNP complex(GO:0070761) |
| 0.0 | 0.9 | GO:0031907 | peroxisomal matrix(GO:0005782) microbody lumen(GO:0031907) |
| 0.0 | 0.3 | GO:0071564 | npBAF complex(GO:0071564) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.5 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.2 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.2 | 0.6 | GO:0004137 | deoxycytidine kinase activity(GO:0004137) thymidine kinase activity(GO:0004797) |
| 0.2 | 0.9 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 3.4 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.1 | 0.8 | GO:0032089 | NACHT domain binding(GO:0032089) |
| 0.1 | 0.5 | GO:0004949 | cannabinoid receptor activity(GO:0004949) |
| 0.1 | 1.0 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.1 | 0.6 | GO:0070892 | lipoteichoic acid receptor activity(GO:0070892) |
| 0.1 | 0.3 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.1 | 0.8 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.1 | 2.3 | GO:0001965 | G-protein alpha-subunit binding(GO:0001965) |
| 0.1 | 0.2 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.1 | 0.3 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.2 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0004157 | dihydropyrimidinase activity(GO:0004157) |
| 0.0 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.5 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 3.3 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.2 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.1 | GO:0061714 | folic acid receptor activity(GO:0061714) |
| 0.0 | 0.5 | GO:0070513 | death domain binding(GO:0070513) |
| 0.0 | 0.8 | GO:0070300 | phosphatidic acid binding(GO:0070300) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.8 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 1.7 | GO:0004364 | glutathione transferase activity(GO:0004364) |
| 0.0 | 0.2 | GO:0004614 | phosphoglucomutase activity(GO:0004614) |
| 0.0 | 0.1 | GO:0043337 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
| 0.0 | 0.5 | GO:0008253 | 5'-nucleotidase activity(GO:0008253) |
| 0.0 | 0.1 | GO:0097100 | supercoiled DNA binding(GO:0097100) |
| 0.0 | 0.2 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.5 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0005415 | nucleoside:sodium symporter activity(GO:0005415) |
| 0.0 | 0.1 | GO:0008483 | transaminase activity(GO:0008483) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.7 | GO:0042056 | chemoattractant activity(GO:0042056) |
| 0.0 | 0.1 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.6 | GO:0001530 | lipopolysaccharide binding(GO:0001530) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.4 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.4 | GO:0070530 | K63-linked polyubiquitin binding(GO:0070530) |
| 0.0 | 0.1 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.2 | GO:0031698 | beta-2 adrenergic receptor binding(GO:0031698) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.4 | GO:0005388 | calcium-transporting ATPase activity(GO:0005388) |
| 0.0 | 0.2 | GO:0030618 | transforming growth factor beta receptor, pathway-specific cytoplasmic mediator activity(GO:0030618) |
| 0.0 | 0.3 | GO:0070628 | proteasome binding(GO:0070628) |
| 0.0 | 0.0 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.0 | 0.1 | GO:0030229 | very-low-density lipoprotein particle receptor activity(GO:0030229) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.5 | GO:0030742 | GTP-dependent protein binding(GO:0030742) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.1 | 3.3 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 1.2 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.5 | PID NCADHERIN PATHWAY | N-cadherin signaling events |
| 0.0 | 0.6 | PID NFKAPPAB CANONICAL PATHWAY | Canonical NF-kappaB pathway |
| 0.0 | 0.9 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 1.0 | PID ILK PATHWAY | Integrin-linked kinase signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 3.4 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.1 | 3.4 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.1 | 1.7 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 3.1 | REACTOME G ALPHA Z SIGNALLING EVENTS | Genes involved in G alpha (z) signalling events |
| 0.0 | 1.5 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 1.7 | REACTOME GLUTATHIONE CONJUGATION | Genes involved in Glutathione conjugation |
| 0.0 | 1.0 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.6 | REACTOME G1 S SPECIFIC TRANSCRIPTION | Genes involved in G1/S-Specific Transcription |
| 0.0 | 0.7 | REACTOME ABC FAMILY PROTEINS MEDIATED TRANSPORT | Genes involved in ABC-family proteins mediated transport |
| 0.0 | 0.2 | REACTOME IKK COMPLEX RECRUITMENT MEDIATED BY RIP1 | Genes involved in IKK complex recruitment mediated by RIP1 |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.6 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.5 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF SUBSTRATES IN N GLYCAN BIOSYTHESIS | Genes involved in Synthesis of substrates in N-glycan biosythesis |
| 0.0 | 0.2 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.0 | 0.1 | REACTOME REGULATION OF ORNITHINE DECARBOXYLASE ODC | Genes involved in Regulation of ornithine decarboxylase (ODC) |
| 0.0 | 0.3 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |