Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for HOXD1
Z-value: 0.53
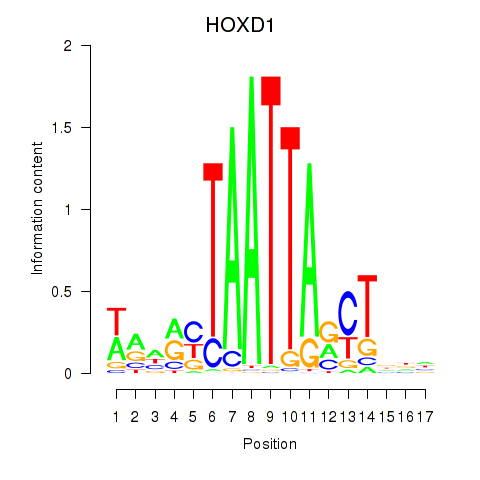
Transcription factors associated with HOXD1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD1
|
ENSG00000128645.11 | HOXD1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD1 | hg19_v2_chr2_+_177053307_177053402 | 0.52 | 1.9e-01 | Click! |
Activity profile of HOXD1 motif
Sorted Z-values of HOXD1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr3_-_151034734 | 0.64 |
ENST00000260843.4 |
GPR87 |
G protein-coupled receptor 87 |
| chr19_-_36001113 | 0.48 |
ENST00000434389.1 |
DMKN |
dermokine |
| chr11_-_102651343 | 0.46 |
ENST00000279441.4 ENST00000539681.1 |
MMP10 |
matrix metallopeptidase 10 (stromelysin 2) |
| chr5_+_66300446 | 0.44 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr3_+_111718036 | 0.36 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr3_+_111718173 | 0.35 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr3_+_111717600 | 0.35 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr4_+_144354644 | 0.32 |
ENST00000512843.1 |
GAB1 |
GRB2-associated binding protein 1 |
| chr3_+_111717511 | 0.32 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr4_-_74486217 | 0.31 |
ENST00000335049.5 ENST00000307439.5 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr4_-_25865159 | 0.30 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr11_+_65554493 | 0.22 |
ENST00000335987.3 |
OVOL1 |
ovo-like zinc finger 1 |
| chr9_-_5339873 | 0.21 |
ENST00000223862.1 ENST00000223858.4 |
RLN1 |
relaxin 1 |
| chr12_+_29376673 | 0.20 |
ENST00000547116.1 |
FAR2 |
fatty acyl CoA reductase 2 |
| chr20_-_50418972 | 0.20 |
ENST00000395997.3 |
SALL4 |
spalt-like transcription factor 4 |
| chr20_-_50418947 | 0.20 |
ENST00000371539.3 |
SALL4 |
spalt-like transcription factor 4 |
| chr12_+_29376592 | 0.20 |
ENST00000182377.4 |
FAR2 |
fatty acyl CoA reductase 2 |
| chr20_-_50419055 | 0.19 |
ENST00000217086.4 |
SALL4 |
spalt-like transcription factor 4 |
| chr3_-_191000172 | 0.18 |
ENST00000427544.2 |
UTS2B |
urotensin 2B |
| chr4_-_74486109 | 0.18 |
ENST00000395777.2 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr4_-_74486347 | 0.16 |
ENST00000342081.3 |
RASSF6 |
Ras association (RalGDS/AF-6) domain family member 6 |
| chr17_-_27418537 | 0.16 |
ENST00000408971.2 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr11_-_124190184 | 0.15 |
ENST00000357438.2 |
OR8D2 |
olfactory receptor, family 8, subfamily D, member 2 |
| chrX_+_107288239 | 0.15 |
ENST00000217957.5 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr12_+_10163231 | 0.14 |
ENST00000396502.1 ENST00000338896.5 |
CLEC12B |
C-type lectin domain family 12, member B |
| chr6_+_130339710 | 0.12 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr9_+_124329336 | 0.11 |
ENST00000394340.3 ENST00000436835.1 ENST00000259371.2 |
DAB2IP |
DAB2 interacting protein |
| chr8_-_49834299 | 0.10 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chrX_+_107288197 | 0.10 |
ENST00000415430.3 |
VSIG1 |
V-set and immunoglobulin domain containing 1 |
| chr8_-_49833978 | 0.09 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr2_-_99871570 | 0.08 |
ENST00000333017.2 ENST00000409679.1 ENST00000423306.1 |
LYG2 |
lysozyme G-like 2 |
| chr2_+_228736335 | 0.08 |
ENST00000440997.1 ENST00000545118.1 |
DAW1 |
dynein assembly factor with WDR repeat domains 1 |
| chr17_-_10372875 | 0.08 |
ENST00000255381.2 |
MYH4 |
myosin, heavy chain 4, skeletal muscle |
| chr14_+_56584414 | 0.08 |
ENST00000559044.1 |
PELI2 |
pellino E3 ubiquitin protein ligase family member 2 |
| chrX_+_114827818 | 0.07 |
ENST00000420625.2 |
PLS3 |
plastin 3 |
| chr11_+_60163775 | 0.07 |
ENST00000300187.6 ENST00000395005.2 |
MS4A14 |
membrane-spanning 4-domains, subfamily A, member 14 |
| chr1_+_117544366 | 0.06 |
ENST00000256652.4 ENST00000369470.1 |
CD101 |
CD101 molecule |
| chr1_-_201140673 | 0.06 |
ENST00000367333.2 |
TMEM9 |
transmembrane protein 9 |
| chr13_-_36050819 | 0.06 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr17_-_38956205 | 0.06 |
ENST00000306658.7 |
KRT28 |
keratin 28 |
| chr4_+_86748898 | 0.06 |
ENST00000509300.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr2_+_135596180 | 0.06 |
ENST00000283054.4 ENST00000392928.1 |
ACMSD |
aminocarboxymuconate semialdehyde decarboxylase |
| chr12_-_10022735 | 0.06 |
ENST00000228438.2 |
CLEC2B |
C-type lectin domain family 2, member B |
| chr3_-_126327398 | 0.05 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr3_+_129247479 | 0.05 |
ENST00000296271.3 |
RHO |
rhodopsin |
| chr12_-_22063787 | 0.05 |
ENST00000544039.1 |
ABCC9 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 9 |
| chr2_+_135596106 | 0.05 |
ENST00000356140.5 |
ACMSD |
aminocarboxymuconate semialdehyde decarboxylase |
| chr5_+_63802109 | 0.05 |
ENST00000334025.2 |
RGS7BP |
regulator of G-protein signaling 7 binding protein |
| chr15_+_58702742 | 0.05 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr1_-_92952433 | 0.05 |
ENST00000294702.5 |
GFI1 |
growth factor independent 1 transcription repressor |
| chr4_+_86749045 | 0.05 |
ENST00000514229.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr12_+_28410128 | 0.04 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr10_+_99205894 | 0.04 |
ENST00000370854.3 ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16 |
zinc finger, DHHC-type containing 16 |
| chr11_+_73498898 | 0.04 |
ENST00000535529.1 ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48 |
mitochondrial ribosomal protein L48 |
| chr19_-_3557570 | 0.04 |
ENST00000355415.2 |
MFSD12 |
major facilitator superfamily domain containing 12 |
| chr10_+_99205959 | 0.04 |
ENST00000352634.4 ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16 |
zinc finger, DHHC-type containing 16 |
| chr12_-_23737534 | 0.04 |
ENST00000396007.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr7_-_73256838 | 0.03 |
ENST00000297873.4 |
WBSCR27 |
Williams Beuren syndrome chromosome region 27 |
| chr11_+_118398178 | 0.03 |
ENST00000302783.4 ENST00000539546.1 |
TTC36 |
tetratricopeptide repeat domain 36 |
| chr11_-_71823715 | 0.03 |
ENST00000545944.1 ENST00000502597.2 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr12_-_52761262 | 0.03 |
ENST00000257901.3 |
KRT85 |
keratin 85 |
| chr11_-_71823266 | 0.03 |
ENST00000538919.1 ENST00000539395.1 ENST00000542531.1 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr10_-_72142345 | 0.03 |
ENST00000373224.1 ENST00000446961.1 ENST00000358141.2 ENST00000357631.2 |
LRRC20 |
leucine rich repeat containing 20 |
| chr11_-_71823796 | 0.03 |
ENST00000545680.1 ENST00000543587.1 ENST00000538393.1 ENST00000535234.1 ENST00000227618.4 ENST00000535503.1 |
ANAPC15 |
anaphase promoting complex subunit 15 |
| chr18_-_33709268 | 0.02 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chrX_+_43515467 | 0.02 |
ENST00000338702.3 ENST00000542639.1 |
MAOA |
monoamine oxidase A |
| chr14_+_32798547 | 0.02 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr4_-_139051839 | 0.01 |
ENST00000514600.1 ENST00000513895.1 ENST00000512536.1 |
LINC00616 |
long intergenic non-protein coding RNA 616 |
| chr4_-_138453559 | 0.01 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr18_-_44181442 | 0.01 |
ENST00000398722.4 |
LOXHD1 |
lipoxygenase homology domains 1 |
| chr11_+_71900703 | 0.01 |
ENST00000393681.2 |
FOLR1 |
folate receptor 1 (adult) |
| chr3_-_101039402 | 0.01 |
ENST00000193391.7 |
IMPG2 |
interphotoreceptor matrix proteoglycan 2 |
| chr12_-_118797475 | 0.01 |
ENST00000541786.1 ENST00000419821.2 ENST00000541878.1 |
TAOK3 |
TAO kinase 3 |
| chr15_-_55562479 | 0.01 |
ENST00000564609.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr10_-_99205607 | 0.01 |
ENST00000477692.2 ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1 |
exosome component 1 |
| chr5_-_96478466 | 0.01 |
ENST00000274382.4 |
LIX1 |
Lix1 homolog (chicken) |
| chr19_+_11071546 | 0.01 |
ENST00000358026.2 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr14_+_32798462 | 0.01 |
ENST00000280979.4 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr3_-_99569821 | 0.01 |
ENST00000487087.1 |
FILIP1L |
filamin A interacting protein 1-like |
| chr7_-_81399329 | 0.00 |
ENST00000453411.1 ENST00000444829.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chrX_+_77154935 | 0.00 |
ENST00000481445.1 |
COX7B |
cytochrome c oxidase subunit VIIb |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.2 | GO:1901994 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.4 | GO:0010166 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.6 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.6 | GO:0035589 | G-protein coupled purinergic nucleotide receptor signaling pathway(GO:0035589) |
| 0.0 | 0.1 | GO:0043553 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) negative regulation of phosphatidylinositol 3-kinase activity(GO:0043553) |
| 0.0 | 0.1 | GO:0019805 | quinolinate biosynthetic process(GO:0019805) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0002767 | immune response-inhibiting cell surface receptor signaling pathway(GO:0002767) |
| 0.0 | 0.2 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.4 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 0.1 | GO:0008282 | ATP-sensitive potassium channel complex(GO:0008282) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.0 | 0.6 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.0 | 0.1 | GO:0008281 | sulfonylurea receptor activity(GO:0008281) |
| 0.0 | 0.1 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME DEGRADATION OF THE EXTRACELLULAR MATRIX | Genes involved in Degradation of the extracellular matrix |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |