Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for HOXD10
Z-value: 0.85
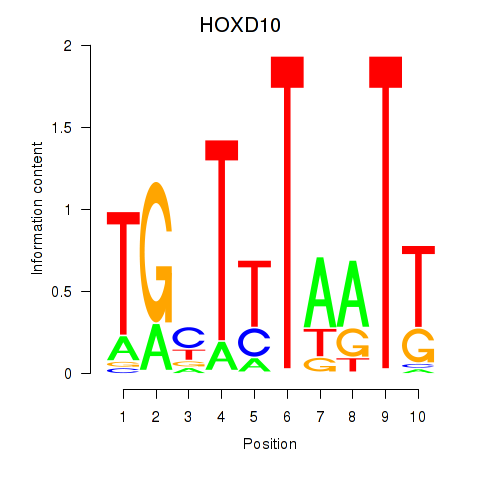
Transcription factors associated with HOXD10
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HOXD10
|
ENSG00000128710.5 | HOXD10 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HOXD10 | hg19_v2_chr2_+_176981307_176981307 | 0.39 | 3.4e-01 | Click! |
Activity profile of HOXD10 motif
Sorted Z-values of HOXD10 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HOXD10
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_40198527 | 1.33 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr18_+_29027696 | 1.11 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr2_-_161056762 | 0.88 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr2_-_161056802 | 0.78 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr1_-_242612779 | 0.76 |
ENST00000427495.1 |
PLD5 |
phospholipase D family, member 5 |
| chr19_-_6690723 | 0.75 |
ENST00000601008.1 |
C3 |
complement component 3 |
| chr18_+_61445007 | 0.74 |
ENST00000447428.1 ENST00000546027.1 |
SERPINB7 |
serpin peptidase inhibitor, clade B (ovalbumin), member 7 |
| chr1_+_62439037 | 0.72 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr4_+_77356248 | 0.68 |
ENST00000296043.6 |
SHROOM3 |
shroom family member 3 |
| chr2_-_165424973 | 0.66 |
ENST00000543549.1 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr19_-_51456321 | 0.63 |
ENST00000391809.2 |
KLK5 |
kallikrein-related peptidase 5 |
| chr17_+_48610074 | 0.61 |
ENST00000503690.1 ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3 |
epsin 3 |
| chr5_+_140227048 | 0.58 |
ENST00000532602.1 |
PCDHA9 |
protocadherin alpha 9 |
| chr5_+_140254884 | 0.56 |
ENST00000398631.2 |
PCDHA12 |
protocadherin alpha 12 |
| chr4_-_77328458 | 0.55 |
ENST00000388914.3 ENST00000434846.2 |
CCDC158 |
coiled-coil domain containing 158 |
| chr2_+_68961934 | 0.55 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr7_-_22233442 | 0.54 |
ENST00000401957.2 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr2_+_68961905 | 0.54 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr6_+_36097992 | 0.54 |
ENST00000211287.4 |
MAPK13 |
mitogen-activated protein kinase 13 |
| chrX_-_102565858 | 0.53 |
ENST00000449185.1 ENST00000536889.1 |
BEX2 |
brain expressed X-linked 2 |
| chr2_+_207804278 | 0.48 |
ENST00000272852.3 |
CPO |
carboxypeptidase O |
| chr12_+_20963647 | 0.45 |
ENST00000381545.3 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr11_+_63137251 | 0.45 |
ENST00000310969.4 ENST00000279178.3 |
SLC22A9 |
solute carrier family 22 (organic anion transporter), member 9 |
| chr12_+_20963632 | 0.44 |
ENST00000540853.1 ENST00000261196.2 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr1_+_186798073 | 0.43 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr12_-_12674032 | 0.43 |
ENST00000298573.4 |
DUSP16 |
dual specificity phosphatase 16 |
| chr2_+_68962014 | 0.38 |
ENST00000467265.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chrX_-_102565932 | 0.38 |
ENST00000372674.1 ENST00000372677.3 |
BEX2 |
brain expressed X-linked 2 |
| chr8_-_66701319 | 0.37 |
ENST00000379419.4 |
PDE7A |
phosphodiesterase 7A |
| chr16_+_30751953 | 0.36 |
ENST00000483578.1 |
RP11-2C24.4 |
RP11-2C24.4 |
| chr12_-_56882136 | 0.35 |
ENST00000311966.4 |
GLS2 |
glutaminase 2 (liver, mitochondrial) |
| chr3_+_46412345 | 0.35 |
ENST00000292303.4 |
CCR5 |
chemokine (C-C motif) receptor 5 (gene/pseudogene) |
| chr1_+_62417957 | 0.31 |
ENST00000307297.7 ENST00000543708.1 |
INADL |
InaD-like (Drosophila) |
| chr11_-_79151695 | 0.31 |
ENST00000278550.7 |
TENM4 |
teneurin transmembrane protein 4 |
| chr6_-_132967142 | 0.30 |
ENST00000275216.1 |
TAAR1 |
trace amine associated receptor 1 |
| chr2_-_165698662 | 0.30 |
ENST00000194871.6 ENST00000445474.2 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr18_-_19994830 | 0.29 |
ENST00000525417.1 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
| chr17_-_38938786 | 0.27 |
ENST00000301656.3 |
KRT27 |
keratin 27 |
| chr2_-_89521942 | 0.26 |
ENST00000482769.1 |
IGKV2-28 |
immunoglobulin kappa variable 2-28 |
| chr15_+_26360970 | 0.26 |
ENST00000556159.1 ENST00000557523.1 |
LINC00929 |
long intergenic non-protein coding RNA 929 |
| chr20_+_52105495 | 0.26 |
ENST00000439873.2 |
AL354993.1 |
Cell growth-inhibiting protein 7; HCG1784586; Uncharacterized protein |
| chr14_+_76776957 | 0.25 |
ENST00000512784.1 |
ESRRB |
estrogen-related receptor beta |
| chr1_-_153113927 | 0.25 |
ENST00000368752.4 |
SPRR2B |
small proline-rich protein 2B |
| chr10_+_6625605 | 0.25 |
ENST00000414894.1 ENST00000449648.1 |
PRKCQ-AS1 |
PRKCQ antisense RNA 1 |
| chr9_+_71944241 | 0.25 |
ENST00000257515.8 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr8_-_95274536 | 0.24 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr4_-_90758227 | 0.24 |
ENST00000506691.1 ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr12_+_41831485 | 0.23 |
ENST00000539469.2 ENST00000298919.7 |
PDZRN4 |
PDZ domain containing ring finger 4 |
| chr14_-_72458326 | 0.23 |
ENST00000542853.1 |
AC005477.1 |
AC005477.1 |
| chr4_-_90756769 | 0.22 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr18_+_55816546 | 0.22 |
ENST00000435432.2 ENST00000357895.5 ENST00000586263.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr6_-_29324054 | 0.22 |
ENST00000543825.1 |
OR5V1 |
olfactory receptor, family 5, subfamily V, member 1 |
| chr4_-_76944621 | 0.22 |
ENST00000306602.1 |
CXCL10 |
chemokine (C-X-C motif) ligand 10 |
| chr18_-_19997878 | 0.22 |
ENST00000391403.2 |
CTAGE1 |
cutaneous T-cell lymphoma-associated antigen 1 |
| chr22_-_29107919 | 0.22 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr13_+_78109884 | 0.22 |
ENST00000377246.3 ENST00000349847.3 |
SCEL |
sciellin |
| chr8_+_124084899 | 0.22 |
ENST00000287380.1 ENST00000309336.3 ENST00000519418.1 ENST00000327098.5 ENST00000522420.1 ENST00000521676.1 ENST00000378080.2 |
TBC1D31 |
TBC1 domain family, member 31 |
| chr3_+_140981456 | 0.21 |
ENST00000504264.1 |
ACPL2 |
acid phosphatase-like 2 |
| chr3_+_57881966 | 0.21 |
ENST00000495364.1 |
SLMAP |
sarcolemma associated protein |
| chr13_+_78109804 | 0.21 |
ENST00000535157.1 |
SCEL |
sciellin |
| chr6_+_121756809 | 0.20 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr18_-_44561988 | 0.20 |
ENST00000332567.4 |
TCEB3B |
transcription elongation factor B polypeptide 3B (elongin A2) |
| chr4_-_89951028 | 0.20 |
ENST00000506913.1 |
FAM13A |
family with sequence similarity 13, member A |
| chr17_-_4463856 | 0.20 |
ENST00000574584.1 ENST00000381550.3 ENST00000301395.3 |
GGT6 |
gamma-glutamyltransferase 6 |
| chr3_+_136676707 | 0.20 |
ENST00000329582.4 |
IL20RB |
interleukin 20 receptor beta |
| chr11_-_104769141 | 0.20 |
ENST00000508062.1 ENST00000422698.2 |
CASP12 |
caspase 12 (gene/pseudogene) |
| chr7_-_111032971 | 0.20 |
ENST00000450877.1 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr1_-_155942086 | 0.19 |
ENST00000368315.4 |
ARHGEF2 |
Rho/Rac guanine nucleotide exchange factor (GEF) 2 |
| chr2_+_90211643 | 0.19 |
ENST00000390277.2 |
IGKV3D-11 |
immunoglobulin kappa variable 3D-11 |
| chr10_-_120925054 | 0.19 |
ENST00000419372.1 ENST00000369131.4 ENST00000330036.6 ENST00000355697.2 |
SFXN4 |
sideroflexin 4 |
| chr12_-_25055177 | 0.19 |
ENST00000538118.1 |
BCAT1 |
branched chain amino-acid transaminase 1, cytosolic |
| chr4_-_72649763 | 0.19 |
ENST00000513476.1 |
GC |
group-specific component (vitamin D binding protein) |
| chr6_-_32557610 | 0.19 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chr4_+_100432161 | 0.19 |
ENST00000326581.4 ENST00000514652.1 |
C4orf17 |
chromosome 4 open reading frame 17 |
| chr16_-_30122717 | 0.19 |
ENST00000566613.1 |
GDPD3 |
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr3_-_27498235 | 0.19 |
ENST00000295736.5 ENST00000428386.1 ENST00000428179.1 |
SLC4A7 |
solute carrier family 4, sodium bicarbonate cotransporter, member 7 |
| chr16_-_20556492 | 0.18 |
ENST00000568098.1 |
ACSM2B |
acyl-CoA synthetase medium-chain family member 2B |
| chrX_+_37639302 | 0.18 |
ENST00000545017.1 ENST00000536160.1 |
CYBB |
cytochrome b-245, beta polypeptide |
| chr22_+_29469012 | 0.18 |
ENST00000400335.4 ENST00000400338.2 |
KREMEN1 |
kringle containing transmembrane protein 1 |
| chr19_-_53770972 | 0.17 |
ENST00000311170.4 |
VN1R4 |
vomeronasal 1 receptor 4 |
| chr2_+_89998789 | 0.17 |
ENST00000453166.2 |
IGKV2D-28 |
immunoglobulin kappa variable 2D-28 |
| chr13_+_32313658 | 0.17 |
ENST00000380314.1 ENST00000298386.2 |
RXFP2 |
relaxin/insulin-like family peptide receptor 2 |
| chr18_-_61329118 | 0.17 |
ENST00000332821.8 ENST00000283752.5 |
SERPINB3 |
serpin peptidase inhibitor, clade B (ovalbumin), member 3 |
| chr4_-_47983519 | 0.17 |
ENST00000358519.4 ENST00000544810.1 ENST00000402813.3 |
CNGA1 |
cyclic nucleotide gated channel alpha 1 |
| chr13_-_47012325 | 0.17 |
ENST00000409879.2 |
KIAA0226L |
KIAA0226-like |
| chr1_-_149459549 | 0.16 |
ENST00000369175.3 |
FAM72C |
family with sequence similarity 72, member C |
| chr3_+_57882024 | 0.16 |
ENST00000494088.1 |
SLMAP |
sarcolemma associated protein |
| chr10_-_52008313 | 0.16 |
ENST00000329428.6 ENST00000395526.4 ENST00000447815.1 |
ASAH2 |
N-acylsphingosine amidohydrolase (non-lysosomal ceramidase) 2 |
| chr9_+_125376948 | 0.16 |
ENST00000297913.2 |
OR1Q1 |
olfactory receptor, family 1, subfamily Q, member 1 |
| chr7_+_129847688 | 0.16 |
ENST00000297819.3 |
SSMEM1 |
serine-rich single-pass membrane protein 1 |
| chr21_-_31588365 | 0.16 |
ENST00000399899.1 |
CLDN8 |
claudin 8 |
| chr4_+_155484155 | 0.16 |
ENST00000509493.1 |
FGB |
fibrinogen beta chain |
| chr4_-_84035868 | 0.15 |
ENST00000426923.2 ENST00000509973.1 |
PLAC8 |
placenta-specific 8 |
| chr1_+_152975488 | 0.15 |
ENST00000542696.1 |
SPRR3 |
small proline-rich protein 3 |
| chr3_-_185538849 | 0.15 |
ENST00000421047.2 |
IGF2BP2 |
insulin-like growth factor 2 mRNA binding protein 2 |
| chr12_-_91451758 | 0.15 |
ENST00000266719.3 |
KERA |
keratocan |
| chr2_+_118572226 | 0.15 |
ENST00000263239.2 |
DDX18 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 18 |
| chr4_+_76481258 | 0.15 |
ENST00000311623.4 ENST00000435974.2 |
C4orf26 |
chromosome 4 open reading frame 26 |
| chr20_-_17539456 | 0.15 |
ENST00000544874.1 ENST00000377868.2 |
BFSP1 |
beaded filament structural protein 1, filensin |
| chr11_-_104827425 | 0.15 |
ENST00000393150.3 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr2_-_99870744 | 0.15 |
ENST00000409238.1 ENST00000423800.1 |
LYG2 |
lysozyme G-like 2 |
| chr5_-_137475071 | 0.15 |
ENST00000265191.2 |
NME5 |
NME/NM23 family member 5 |
| chr10_+_5488564 | 0.14 |
ENST00000449083.1 ENST00000380359.3 |
NET1 |
neuroepithelial cell transforming 1 |
| chr6_+_26365443 | 0.14 |
ENST00000527422.1 ENST00000356386.2 ENST00000396934.3 ENST00000377708.2 ENST00000396948.1 ENST00000508906.2 |
BTN3A2 |
butyrophilin, subfamily 3, member A2 |
| chr3_+_158288942 | 0.14 |
ENST00000491767.1 ENST00000355893.5 |
MLF1 |
myeloid leukemia factor 1 |
| chr18_-_3845293 | 0.14 |
ENST00000400145.2 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr2_+_1418154 | 0.14 |
ENST00000423320.1 ENST00000382198.1 |
TPO |
thyroid peroxidase |
| chr2_+_102456277 | 0.13 |
ENST00000421882.1 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr3_-_126327398 | 0.13 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr10_+_695888 | 0.13 |
ENST00000441152.2 |
PRR26 |
proline rich 26 |
| chr11_+_122753391 | 0.13 |
ENST00000307257.6 ENST00000227349.2 |
C11orf63 |
chromosome 11 open reading frame 63 |
| chr4_+_15341442 | 0.13 |
ENST00000397700.2 ENST00000295297.4 |
C1QTNF7 |
C1q and tumor necrosis factor related protein 7 |
| chr4_-_123377880 | 0.13 |
ENST00000226730.4 |
IL2 |
interleukin 2 |
| chr12_-_71148413 | 0.13 |
ENST00000440835.2 ENST00000549308.1 ENST00000550661.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr12_-_100656134 | 0.13 |
ENST00000548313.1 |
DEPDC4 |
DEP domain containing 4 |
| chr1_-_111743285 | 0.12 |
ENST00000357640.4 |
DENND2D |
DENN/MADD domain containing 2D |
| chr12_-_16759711 | 0.12 |
ENST00000447609.1 |
LMO3 |
LIM domain only 3 (rhombotin-like 2) |
| chr8_-_124279627 | 0.12 |
ENST00000357082.4 |
ZHX1-C8ORF76 |
ZHX1-C8ORF76 readthrough |
| chr4_-_68749745 | 0.12 |
ENST00000283916.6 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr17_-_10452929 | 0.12 |
ENST00000532183.2 ENST00000397183.2 ENST00000420805.1 |
MYH2 |
myosin, heavy chain 2, skeletal muscle, adult |
| chr10_+_96522361 | 0.12 |
ENST00000371321.3 |
CYP2C19 |
cytochrome P450, family 2, subfamily C, polypeptide 19 |
| chr12_-_71148357 | 0.12 |
ENST00000378778.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr4_-_68749699 | 0.12 |
ENST00000545541.1 |
TMPRSS11D |
transmembrane protease, serine 11D |
| chr7_+_141490017 | 0.11 |
ENST00000247883.4 |
TAS2R5 |
taste receptor, type 2, member 5 |
| chr1_-_156399184 | 0.11 |
ENST00000368243.1 ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61 |
chromosome 1 open reading frame 61 |
| chr4_-_69434245 | 0.11 |
ENST00000317746.2 |
UGT2B17 |
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr9_+_103189405 | 0.11 |
ENST00000395067.2 |
MSANTD3 |
Myb/SANT-like DNA-binding domain containing 3 |
| chr6_+_37897735 | 0.11 |
ENST00000373389.5 |
ZFAND3 |
zinc finger, AN1-type domain 3 |
| chr4_-_153332886 | 0.11 |
ENST00000603841.1 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr6_-_142409936 | 0.11 |
ENST00000258042.1 |
NMBR |
neuromedin B receptor |
| chr2_+_90273679 | 0.11 |
ENST00000423080.2 |
IGKV3D-7 |
immunoglobulin kappa variable 3D-7 |
| chr10_-_62332357 | 0.11 |
ENST00000503366.1 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr6_-_20212630 | 0.11 |
ENST00000324607.7 ENST00000541730.1 ENST00000536798.1 |
MBOAT1 |
membrane bound O-acyltransferase domain containing 1 |
| chr8_-_70016408 | 0.11 |
ENST00000518540.1 |
RP11-600K15.1 |
RP11-600K15.1 |
| chr11_-_58980342 | 0.11 |
ENST00000361050.3 |
MPEG1 |
macrophage expressed 1 |
| chr3_+_63428982 | 0.11 |
ENST00000479198.1 ENST00000460711.1 ENST00000465156.1 |
SYNPR |
synaptoporin |
| chr8_-_28347737 | 0.11 |
ENST00000517673.1 ENST00000518734.1 ENST00000346498.2 ENST00000380254.2 |
FBXO16 |
F-box protein 16 |
| chr1_+_244214577 | 0.11 |
ENST00000358704.4 |
ZBTB18 |
zinc finger and BTB domain containing 18 |
| chr21_-_27423339 | 0.11 |
ENST00000415997.1 |
APP |
amyloid beta (A4) precursor protein |
| chr11_-_27528301 | 0.11 |
ENST00000524596.1 ENST00000278193.2 |
LIN7C |
lin-7 homolog C (C. elegans) |
| chr6_+_96025341 | 0.10 |
ENST00000369293.1 ENST00000358812.4 |
MANEA |
mannosidase, endo-alpha |
| chr17_+_66624280 | 0.10 |
ENST00000585484.1 |
RP11-118B18.1 |
RP11-118B18.1 |
| chr10_+_116697946 | 0.10 |
ENST00000298746.3 |
TRUB1 |
TruB pseudouridine (psi) synthase family member 1 |
| chr19_+_49588677 | 0.10 |
ENST00000598984.1 ENST00000598441.1 |
SNRNP70 |
small nuclear ribonucleoprotein 70kDa (U1) |
| chr6_-_170101749 | 0.10 |
ENST00000448612.1 |
WDR27 |
WD repeat domain 27 |
| chr5_+_140174429 | 0.10 |
ENST00000520672.2 ENST00000378132.1 ENST00000526136.1 |
PCDHA2 |
protocadherin alpha 2 |
| chr3_+_158288960 | 0.10 |
ENST00000484955.1 ENST00000359117.5 ENST00000498592.1 ENST00000477042.1 ENST00000471745.1 ENST00000469452.1 |
MLF1 |
myeloid leukemia factor 1 |
| chr1_+_109256067 | 0.10 |
ENST00000271311.2 |
FNDC7 |
fibronectin type III domain containing 7 |
| chr12_+_118814344 | 0.10 |
ENST00000397564.2 |
SUDS3 |
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr11_+_55594695 | 0.10 |
ENST00000378397.1 |
OR5L2 |
olfactory receptor, family 5, subfamily L, member 2 |
| chr12_+_118814185 | 0.10 |
ENST00000543473.1 |
SUDS3 |
suppressor of defective silencing 3 homolog (S. cerevisiae) |
| chr6_+_27791862 | 0.10 |
ENST00000355057.1 |
HIST1H4J |
histone cluster 1, H4j |
| chr2_-_74875432 | 0.10 |
ENST00000536235.1 ENST00000421985.1 |
M1AP |
meiosis 1 associated protein |
| chrX_-_138914394 | 0.10 |
ENST00000327569.3 ENST00000361648.2 ENST00000370543.1 ENST00000359686.2 |
ATP11C |
ATPase, class VI, type 11C |
| chr17_+_40925454 | 0.10 |
ENST00000253794.2 ENST00000590339.1 ENST00000589520.1 |
VPS25 |
vacuolar protein sorting 25 homolog (S. cerevisiae) |
| chr6_-_32498046 | 0.09 |
ENST00000374975.3 |
HLA-DRB5 |
major histocompatibility complex, class II, DR beta 5 |
| chr6_+_131958436 | 0.09 |
ENST00000357639.3 ENST00000543135.1 ENST00000427148.2 ENST00000358229.5 |
ENPP3 |
ectonucleotide pyrophosphatase/phosphodiesterase 3 |
| chr3_+_196466710 | 0.09 |
ENST00000327134.3 |
PAK2 |
p21 protein (Cdc42/Rac)-activated kinase 2 |
| chr3_-_120461353 | 0.09 |
ENST00000483733.1 |
RABL3 |
RAB, member of RAS oncogene family-like 3 |
| chr1_+_180199393 | 0.09 |
ENST00000263726.2 |
LHX4 |
LIM homeobox 4 |
| chr2_+_157330081 | 0.09 |
ENST00000409674.1 |
GPD2 |
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr9_-_104145795 | 0.09 |
ENST00000259407.2 |
BAAT |
bile acid CoA: amino acid N-acyltransferase (glycine N-choloyltransferase) |
| chr16_-_71610985 | 0.09 |
ENST00000355962.4 |
TAT |
tyrosine aminotransferase |
| chr7_-_38305279 | 0.09 |
ENST00000443402.2 |
TRGC1 |
T cell receptor gamma constant 1 |
| chr2_+_183982238 | 0.09 |
ENST00000442895.2 ENST00000446612.1 ENST00000409798.1 |
NUP35 |
nucleoporin 35kDa |
| chr17_-_17480779 | 0.09 |
ENST00000395782.1 |
PEMT |
phosphatidylethanolamine N-methyltransferase |
| chr3_-_172859017 | 0.09 |
ENST00000351008.3 |
SPATA16 |
spermatogenesis associated 16 |
| chr2_-_90538397 | 0.09 |
ENST00000443397.3 |
RP11-685N3.1 |
Uncharacterized protein |
| chr11_+_7110165 | 0.09 |
ENST00000306904.5 |
RBMXL2 |
RNA binding motif protein, X-linked-like 2 |
| chr10_+_120967072 | 0.09 |
ENST00000392870.2 |
GRK5 |
G protein-coupled receptor kinase 5 |
| chr16_-_15180257 | 0.09 |
ENST00000540462.1 |
RRN3 |
RRN3 RNA polymerase I transcription factor homolog (S. cerevisiae) |
| chr3_+_130569429 | 0.09 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr9_-_47314222 | 0.09 |
ENST00000420228.1 ENST00000438517.1 ENST00000414020.1 |
AL953854.2 |
AL953854.2 |
| chrX_+_149887090 | 0.08 |
ENST00000538506.1 |
MTMR1 |
myotubularin related protein 1 |
| chr2_+_234590556 | 0.08 |
ENST00000373426.3 |
UGT1A7 |
UDP glucuronosyltransferase 1 family, polypeptide A7 |
| chr16_+_78056412 | 0.08 |
ENST00000299642.4 ENST00000575655.1 |
CLEC3A |
C-type lectin domain family 3, member A |
| chr9_+_4839762 | 0.08 |
ENST00000448872.2 ENST00000441844.1 |
RCL1 |
RNA terminal phosphate cyclase-like 1 |
| chr2_+_87565634 | 0.08 |
ENST00000421835.2 |
IGKV3OR2-268 |
immunoglobulin kappa variable 3/OR2-268 (non-functional) |
| chr12_-_13248732 | 0.08 |
ENST00000396302.3 |
GSG1 |
germ cell associated 1 |
| chr17_+_27369918 | 0.08 |
ENST00000323372.4 |
PIPOX |
pipecolic acid oxidase |
| chr6_+_123100853 | 0.08 |
ENST00000356535.4 |
FABP7 |
fatty acid binding protein 7, brain |
| chr2_-_105030466 | 0.08 |
ENST00000449772.1 |
AC068535.3 |
AC068535.3 |
| chr3_+_35721106 | 0.08 |
ENST00000474696.1 ENST00000412048.1 ENST00000396482.2 ENST00000432682.1 |
ARPP21 |
cAMP-regulated phosphoprotein, 21kDa |
| chr18_-_22932080 | 0.08 |
ENST00000584787.1 ENST00000361524.3 ENST00000538137.2 |
ZNF521 |
zinc finger protein 521 |
| chr8_+_26150628 | 0.08 |
ENST00000523925.1 ENST00000315985.7 |
PPP2R2A |
protein phosphatase 2, regulatory subunit B, alpha |
| chr3_-_194119083 | 0.08 |
ENST00000401815.1 |
GP5 |
glycoprotein V (platelet) |
| chr3_-_128206759 | 0.08 |
ENST00000430265.2 |
GATA2 |
GATA binding protein 2 |
| chr14_-_82089405 | 0.08 |
ENST00000554211.1 |
RP11-799P8.1 |
RP11-799P8.1 |
| chr14_+_31046959 | 0.08 |
ENST00000547532.1 ENST00000555429.1 |
G2E3 |
G2/M-phase specific E3 ubiquitin protein ligase |
| chr6_+_31021225 | 0.08 |
ENST00000565192.1 ENST00000562344.1 |
HCG22 |
HLA complex group 22 |
| chr1_+_40713573 | 0.08 |
ENST00000372766.3 |
TMCO2 |
transmembrane and coiled-coil domains 2 |
| chr12_-_13248705 | 0.07 |
ENST00000396310.2 |
GSG1 |
germ cell associated 1 |
| chr1_-_242162375 | 0.07 |
ENST00000357246.3 |
MAP1LC3C |
microtubule-associated protein 1 light chain 3 gamma |
| chr15_+_76352178 | 0.07 |
ENST00000388942.3 |
C15orf27 |
chromosome 15 open reading frame 27 |
| chr9_+_27109133 | 0.07 |
ENST00000519097.1 ENST00000380036.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr12_+_48876275 | 0.07 |
ENST00000314014.2 |
C12orf54 |
chromosome 12 open reading frame 54 |
| chr3_-_167191814 | 0.07 |
ENST00000466903.1 ENST00000264677.4 |
SERPINI2 |
serpin peptidase inhibitor, clade I (pancpin), member 2 |
| chr2_+_135011731 | 0.07 |
ENST00000281923.2 |
MGAT5 |
mannosyl (alpha-1,6-)-glycoprotein beta-1,6-N-acetyl-glucosaminyltransferase |
| chr12_-_13248562 | 0.07 |
ENST00000457134.2 ENST00000537302.1 |
GSG1 |
germ cell associated 1 |
| chrX_+_37639264 | 0.07 |
ENST00000378588.4 |
CYBB |
cytochrome b-245, beta polypeptide |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.7 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.2 | 0.7 | GO:0090362 | positive regulation of platelet-derived growth factor production(GO:0090362) |
| 0.2 | 1.7 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.1 | 0.4 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.6 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.3 | GO:0032289 | central nervous system myelin formation(GO:0032289) |
| 0.1 | 0.5 | GO:0032227 | negative regulation of synaptic transmission, dopaminergic(GO:0032227) negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.1 | 0.4 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.1 | 0.3 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.1 | 0.2 | GO:1903925 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.1 | 0.4 | GO:0006543 | glutamine catabolic process(GO:0006543) |
| 0.1 | 0.2 | GO:0010645 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.2 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.1 | 0.2 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.4 | GO:1905150 | regulation of voltage-gated sodium channel activity(GO:1905150) |
| 0.1 | 0.4 | GO:0039663 | fusion of virus membrane with host plasma membrane(GO:0019064) membrane fusion involved in viral entry into host cell(GO:0039663) multi-organism membrane fusion(GO:0044800) |
| 0.1 | 0.3 | GO:1904844 | response to L-glutamine(GO:1904844) cellular response to L-glutamine(GO:1904845) |
| 0.0 | 0.2 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.7 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.2 | GO:0035425 | autocrine signaling(GO:0035425) |
| 0.0 | 0.2 | GO:0071802 | negative regulation of podosome assembly(GO:0071802) |
| 0.0 | 0.0 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.0 | 0.2 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.2 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.2 | GO:0009082 | branched-chain amino acid biosynthetic process(GO:0009082) leucine biosynthetic process(GO:0009098) valine biosynthetic process(GO:0009099) |
| 0.0 | 0.1 | GO:0046013 | regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.0 | 0.1 | GO:0021526 | medial motor column neuron differentiation(GO:0021526) |
| 0.0 | 0.1 | GO:0072660 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.0 | 0.1 | GO:0010725 | regulation of primitive erythrocyte differentiation(GO:0010725) eosinophil fate commitment(GO:0035854) |
| 0.0 | 0.1 | GO:0050717 | positive regulation of interleukin-1 alpha secretion(GO:0050717) |
| 0.0 | 0.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.2 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.9 | GO:0043252 | sodium-independent organic anion transport(GO:0043252) |
| 0.0 | 0.1 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.1 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 0.1 | GO:2000638 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.2 | GO:1901509 | regulation of endothelial tube morphogenesis(GO:1901509) |
| 0.0 | 0.1 | GO:1905205 | positive regulation of connective tissue replacement(GO:1905205) |
| 0.0 | 0.1 | GO:0014005 | microglia differentiation(GO:0014004) microglia development(GO:0014005) cellular response to norepinephrine stimulus(GO:0071874) |
| 0.0 | 0.1 | GO:2000324 | positive regulation of glucocorticoid receptor signaling pathway(GO:2000324) |
| 0.0 | 1.3 | GO:0034260 | negative regulation of GTPase activity(GO:0034260) |
| 0.0 | 0.1 | GO:0043328 | protein targeting to vacuole involved in ubiquitin-dependent protein catabolic process via the multivesicular body sorting pathway(GO:0043328) |
| 0.0 | 0.3 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0009253 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0006127 | glycerophosphate shuttle(GO:0006127) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.1 | GO:1990481 | mRNA pseudouridine synthesis(GO:1990481) |
| 0.0 | 0.1 | GO:0016098 | monoterpenoid metabolic process(GO:0016098) |
| 0.0 | 0.0 | GO:0061534 | gamma-aminobutyric acid secretion, neurotransmission(GO:0061534) |
| 0.0 | 0.1 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.1 | GO:0007060 | male meiosis chromosome segregation(GO:0007060) |
| 0.0 | 0.1 | GO:0019477 | L-lysine catabolic process to acetyl-CoA(GO:0019474) L-lysine catabolic process(GO:0019477) L-lysine metabolic process(GO:0046440) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 0.0 | GO:0042040 | molybdenum incorporation into molybdenum-molybdopterin complex(GO:0018315) metal incorporation into metallo-molybdopterin complex(GO:0042040) glycine receptor clustering(GO:0072579) |
| 0.0 | 0.0 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.0 | GO:0034128 | negative regulation of MyD88-independent toll-like receptor signaling pathway(GO:0034128) |
| 0.0 | 0.1 | GO:0009804 | coumarin metabolic process(GO:0009804) |
| 0.0 | 0.1 | GO:0051342 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.7 | GO:0046627 | negative regulation of insulin receptor signaling pathway(GO:0046627) |
| 0.0 | 1.7 | GO:0070268 | cornification(GO:0070268) |
| 0.0 | 0.2 | GO:0097264 | self proteolysis(GO:0097264) |
| 0.0 | 0.1 | GO:0001180 | transcription initiation from RNA polymerase I promoter for nuclear large rRNA transcript(GO:0001180) |
| 0.0 | 0.1 | GO:1903361 | protein localization to basolateral plasma membrane(GO:1903361) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.1 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.0 | 0.0 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.1 | GO:0006228 | UTP biosynthetic process(GO:0006228) |
| 0.0 | 0.1 | GO:0006572 | tyrosine catabolic process(GO:0006572) |
| 0.0 | 0.2 | GO:0061303 | cornea development in camera-type eye(GO:0061303) |
| 0.0 | 0.1 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.0 | 0.1 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.0 | 0.2 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.1 | GO:0000480 | endonucleolytic cleavage in 5'-ETS of tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000480) |
| 0.0 | 0.1 | GO:0001778 | plasma membrane repair(GO:0001778) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.7 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.1 | 0.6 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.2 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.4 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 1.9 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0005826 | actomyosin contractile ring(GO:0005826) |
| 0.0 | 1.3 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.3 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.1 | GO:0009331 | glycerol-3-phosphate dehydrogenase complex(GO:0009331) |
| 0.0 | 0.3 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.3 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.1 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.0 | GO:0046696 | lipopolysaccharide receptor complex(GO:0046696) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.1 | 0.4 | GO:0004359 | glutaminase activity(GO:0004359) |
| 0.1 | 0.2 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.1 | 1.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.3 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.9 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.4 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.2 | GO:0052654 | branched-chain-amino-acid transaminase activity(GO:0004084) L-leucine transaminase activity(GO:0052654) L-valine transaminase activity(GO:0052655) L-isoleucine transaminase activity(GO:0052656) |
| 0.0 | 0.2 | GO:0005499 | vitamin D binding(GO:0005499) |
| 0.0 | 0.2 | GO:0017040 | ceramidase activity(GO:0017040) |
| 0.0 | 0.1 | GO:0080130 | L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.4 | GO:0019957 | C-C chemokine binding(GO:0019957) |
| 0.0 | 0.1 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.2 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.1 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.0 | 0.1 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.4 | GO:0017017 | MAP kinase tyrosine/serine/threonine phosphatase activity(GO:0017017) |
| 0.0 | 0.4 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0004447 | iodide peroxidase activity(GO:0004447) |
| 0.0 | 0.1 | GO:0052591 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |
| 0.0 | 0.5 | GO:0004707 | MAP kinase activity(GO:0004707) |
| 0.0 | 0.1 | GO:0003963 | RNA-3'-phosphate cyclase activity(GO:0003963) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.0 | 0.1 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
| 0.0 | 0.2 | GO:0004321 | fatty-acyl-CoA synthase activity(GO:0004321) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 1.9 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.0 | GO:0061598 | molybdopterin adenylyltransferase activity(GO:0061598) molybdopterin molybdotransferase activity(GO:0061599) |
| 0.0 | 0.2 | GO:0043855 | intracellular cyclic nucleotide activated cation channel activity(GO:0005221) cyclic nucleotide-gated ion channel activity(GO:0043855) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0016647 | oxidoreductase activity, acting on the CH-NH group of donors, oxygen as acceptor(GO:0016647) |
| 0.0 | 0.0 | GO:0017130 | poly(C) RNA binding(GO:0017130) |
| 0.0 | 0.2 | GO:0070990 | snRNP binding(GO:0070990) |
| 0.0 | 0.3 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 0.2 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0051425 | PTB domain binding(GO:0051425) |
| 0.0 | 0.1 | GO:0033192 | calmodulin-dependent protein phosphatase activity(GO:0033192) |
| 0.0 | 0.2 | GO:0004559 | alpha-mannosidase activity(GO:0004559) |
| 0.0 | 0.2 | GO:0016500 | protein-hormone receptor activity(GO:0016500) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.0 | 0.7 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | REACTOME BINDING AND ENTRY OF HIV VIRION | Genes involved in Binding and entry of HIV virion |
| 0.1 | 1.0 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.1 | 1.1 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.7 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.5 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.7 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 3.2 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.2 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.2 | REACTOME GAP JUNCTION DEGRADATION | Genes involved in Gap junction degradation |
| 0.0 | 0.2 | REACTOME THE NLRP3 INFLAMMASOME | Genes involved in The NLRP3 inflammasome |
| 0.0 | 0.2 | REACTOME GLUCURONIDATION | Genes involved in Glucuronidation |
| 0.0 | 0.2 | REACTOME G2 M DNA DAMAGE CHECKPOINT | Genes involved in G2/M DNA damage checkpoint |