Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for HSF4
Z-value: 0.65
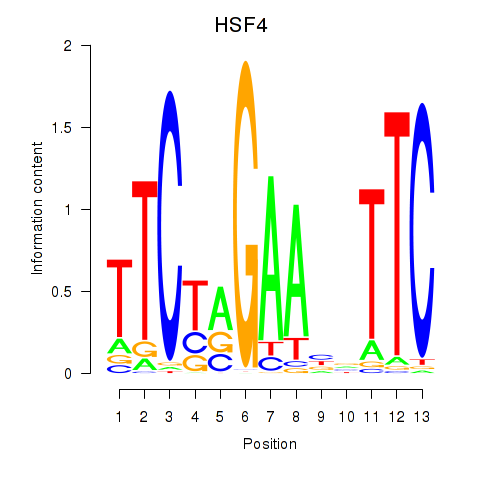
Transcription factors associated with HSF4
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSF4
|
ENSG00000102878.11 | HSF4 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSF4 | hg19_v2_chr16_+_67198683_67198715 | -0.38 | 3.5e-01 | Click! |
Activity profile of HSF4 motif
Sorted Z-values of HSF4 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HSF4
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr7_-_56119156 | 0.67 |
ENST00000421312.1 ENST00000416592.1 |
PSPH |
phosphoserine phosphatase |
| chr7_-_56118981 | 0.66 |
ENST00000419984.2 ENST00000413218.1 ENST00000424596.1 |
PSPH |
phosphoserine phosphatase |
| chr7_-_56119238 | 0.40 |
ENST00000275605.3 ENST00000395471.3 |
PSPH |
phosphoserine phosphatase |
| chr19_-_49567124 | 0.34 |
ENST00000301411.3 |
NTF4 |
neurotrophin 4 |
| chr2_-_238499303 | 0.29 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr4_+_41258786 | 0.29 |
ENST00000503431.1 ENST00000284440.4 ENST00000508768.1 ENST00000512788.1 |
UCHL1 |
ubiquitin carboxyl-terminal esterase L1 (ubiquitin thiolesterase) |
| chr19_-_6720686 | 0.26 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chr8_+_143916217 | 0.25 |
ENST00000220940.1 |
GML |
glycosylphosphatidylinositol anchored molecule like |
| chr19_+_6135646 | 0.25 |
ENST00000588304.1 ENST00000588485.1 ENST00000588722.1 ENST00000591403.1 ENST00000586696.1 ENST00000589401.1 ENST00000252669.5 |
ACSBG2 |
acyl-CoA synthetase bubblegum family member 2 |
| chr3_-_195603566 | 0.24 |
ENST00000424563.1 ENST00000411741.1 |
TNK2 |
tyrosine kinase, non-receptor, 2 |
| chr9_-_138591341 | 0.23 |
ENST00000298466.5 ENST00000425225.1 |
SOHLH1 |
spermatogenesis and oogenesis specific basic helix-loop-helix 1 |
| chr17_+_33474826 | 0.22 |
ENST00000268876.5 ENST00000433649.1 ENST00000378449.1 |
UNC45B |
unc-45 homolog B (C. elegans) |
| chr17_+_33474860 | 0.21 |
ENST00000394570.2 |
UNC45B |
unc-45 homolog B (C. elegans) |
| chr5_+_147258266 | 0.21 |
ENST00000296694.4 |
SCGB3A2 |
secretoglobin, family 3A, member 2 |
| chr11_+_107799118 | 0.20 |
ENST00000320578.2 |
RAB39A |
RAB39A, member RAS oncogene family |
| chr8_-_81083731 | 0.20 |
ENST00000379096.5 |
TPD52 |
tumor protein D52 |
| chr2_-_238499131 | 0.20 |
ENST00000538644.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr12_+_2904102 | 0.18 |
ENST00000001008.4 |
FKBP4 |
FK506 binding protein 4, 59kDa |
| chr7_-_139763521 | 0.17 |
ENST00000263549.3 |
PARP12 |
poly (ADP-ribose) polymerase family, member 12 |
| chrX_+_108779870 | 0.17 |
ENST00000372107.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr14_+_96722539 | 0.17 |
ENST00000553356.1 |
BDKRB1 |
bradykinin receptor B1 |
| chr2_-_69180012 | 0.17 |
ENST00000481498.1 |
GKN2 |
gastrokine 2 |
| chr17_-_9694614 | 0.16 |
ENST00000330255.5 ENST00000571134.1 |
DHRS7C |
dehydrogenase/reductase (SDR family) member 7C |
| chr19_-_49565254 | 0.16 |
ENST00000593537.1 |
NTF4 |
neurotrophin 4 |
| chr3_+_136676707 | 0.15 |
ENST00000329582.4 |
IL20RB |
interleukin 20 receptor beta |
| chr3_+_136676851 | 0.14 |
ENST00000309741.5 |
IL20RB |
interleukin 20 receptor beta |
| chr3_-_126327398 | 0.14 |
ENST00000383572.2 |
TXNRD3NB |
thioredoxin reductase 3 neighbor |
| chr19_-_14628645 | 0.14 |
ENST00000598235.1 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr13_-_20805109 | 0.14 |
ENST00000241124.6 |
GJB6 |
gap junction protein, beta 6, 30kDa |
| chr16_-_8955570 | 0.13 |
ENST00000567554.1 |
CARHSP1 |
calcium regulated heat stable protein 1, 24kDa |
| chr16_-_8955601 | 0.13 |
ENST00000569398.1 ENST00000568968.1 |
CARHSP1 |
calcium regulated heat stable protein 1, 24kDa |
| chr1_+_212782012 | 0.12 |
ENST00000341491.4 ENST00000366985.1 |
ATF3 |
activating transcription factor 3 |
| chr11_+_65769550 | 0.12 |
ENST00000312175.2 ENST00000445560.2 ENST00000530204.1 |
BANF1 |
barrier to autointegration factor 1 |
| chr19_-_14629224 | 0.12 |
ENST00000254322.2 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr11_+_86013253 | 0.12 |
ENST00000533986.1 ENST00000278483.3 |
C11orf73 |
chromosome 11 open reading frame 73 |
| chr9_+_74764278 | 0.12 |
ENST00000238018.4 ENST00000376989.3 |
GDA |
guanine deaminase |
| chr16_+_76587314 | 0.12 |
ENST00000563764.1 |
RP11-58C22.1 |
Uncharacterized protein |
| chr11_-_111782484 | 0.12 |
ENST00000533971.1 |
CRYAB |
crystallin, alpha B |
| chr19_-_55672037 | 0.11 |
ENST00000588076.1 |
DNAAF3 |
dynein, axonemal, assembly factor 3 |
| chr9_+_34989638 | 0.11 |
ENST00000453597.3 ENST00000335998.3 ENST00000312316.5 |
DNAJB5 |
DnaJ (Hsp40) homolog, subfamily B, member 5 |
| chr18_+_11751493 | 0.11 |
ENST00000269162.5 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr22_+_29168652 | 0.11 |
ENST00000249064.4 ENST00000444523.1 ENST00000448492.2 ENST00000421503.2 |
CCDC117 |
coiled-coil domain containing 117 |
| chr11_+_65769946 | 0.10 |
ENST00000533166.1 |
BANF1 |
barrier to autointegration factor 1 |
| chr15_-_89755034 | 0.10 |
ENST00000563254.1 |
RLBP1 |
retinaldehyde binding protein 1 |
| chr4_+_5527117 | 0.10 |
ENST00000505296.1 |
C4orf6 |
chromosome 4 open reading frame 6 |
| chr4_+_5526883 | 0.10 |
ENST00000195455.2 |
C4orf6 |
chromosome 4 open reading frame 6 |
| chr11_+_65770227 | 0.10 |
ENST00000527348.1 |
BANF1 |
barrier to autointegration factor 1 |
| chr12_+_70132632 | 0.10 |
ENST00000378815.6 ENST00000483530.2 ENST00000325555.9 |
RAB3IP |
RAB3A interacting protein |
| chr5_-_133706695 | 0.10 |
ENST00000521755.1 ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3 |
cyclin-dependent kinase-like 3 |
| chr15_+_79165222 | 0.10 |
ENST00000559930.1 |
MORF4L1 |
mortality factor 4 like 1 |
| chr1_+_154947148 | 0.10 |
ENST00000368436.1 ENST00000308987.5 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chr6_+_36165133 | 0.10 |
ENST00000446974.1 ENST00000454960.1 |
BRPF3 |
bromodomain and PHD finger containing, 3 |
| chr4_-_152147579 | 0.10 |
ENST00000304527.4 ENST00000455740.1 ENST00000424281.1 ENST00000409598.4 |
SH3D19 |
SH3 domain containing 19 |
| chr2_-_69180083 | 0.09 |
ENST00000328895.4 |
GKN2 |
gastrokine 2 |
| chr6_+_54172653 | 0.09 |
ENST00000370869.3 |
TINAG |
tubulointerstitial nephritis antigen |
| chr20_+_10015678 | 0.09 |
ENST00000378392.1 ENST00000378380.3 |
ANKEF1 |
ankyrin repeat and EF-hand domain containing 1 |
| chr11_+_20385327 | 0.09 |
ENST00000451739.2 ENST00000532505.1 |
HTATIP2 |
HIV-1 Tat interactive protein 2, 30kDa |
| chr15_-_86338100 | 0.09 |
ENST00000536947.1 |
KLHL25 |
kelch-like family member 25 |
| chr15_-_86338134 | 0.09 |
ENST00000337975.5 |
KLHL25 |
kelch-like family member 25 |
| chr11_-_111794446 | 0.09 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr11_+_20385231 | 0.09 |
ENST00000530266.1 ENST00000421577.2 ENST00000443524.2 ENST00000419348.2 |
HTATIP2 |
HIV-1 Tat interactive protein 2, 30kDa |
| chr14_-_68141535 | 0.09 |
ENST00000554659.1 |
VTI1B |
vesicle transport through interaction with t-SNAREs 1B |
| chr15_+_44092784 | 0.09 |
ENST00000458412.1 |
HYPK |
huntingtin interacting protein K |
| chr6_+_4890226 | 0.09 |
ENST00000343762.5 |
CDYL |
chromodomain protein, Y-like |
| chr19_-_344786 | 0.09 |
ENST00000264819.4 |
MIER2 |
mesoderm induction early response 1, family member 2 |
| chr17_-_54911250 | 0.09 |
ENST00000575658.1 ENST00000397861.2 |
C17orf67 |
chromosome 17 open reading frame 67 |
| chr6_-_76782371 | 0.09 |
ENST00000369950.3 ENST00000369963.3 |
IMPG1 |
interphotoreceptor matrix proteoglycan 1 |
| chr11_-_111782696 | 0.09 |
ENST00000227251.3 ENST00000526180.1 |
CRYAB |
crystallin, alpha B |
| chr1_+_154947126 | 0.09 |
ENST00000368439.1 |
CKS1B |
CDC28 protein kinase regulatory subunit 1B |
| chr22_-_46644182 | 0.09 |
ENST00000404583.1 ENST00000404744.1 |
CDPF1 |
cysteine-rich, DPF motif domain containing 1 |
| chr19_+_50354462 | 0.09 |
ENST00000601675.1 |
PTOV1 |
prostate tumor overexpressed 1 |
| chrX_+_108780062 | 0.08 |
ENST00000372106.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr4_+_76932326 | 0.08 |
ENST00000513353.1 ENST00000341029.5 |
ART3 |
ADP-ribosyltransferase 3 |
| chr17_-_39928106 | 0.08 |
ENST00000540235.1 |
JUP |
junction plakoglobin |
| chr11_-_82997013 | 0.08 |
ENST00000529073.1 ENST00000529611.1 |
CCDC90B |
coiled-coil domain containing 90B |
| chr19_+_50354393 | 0.08 |
ENST00000391842.1 |
PTOV1 |
prostate tumor overexpressed 1 |
| chr19_+_50354430 | 0.08 |
ENST00000599732.1 |
PTOV1 |
prostate tumor overexpressed 1 |
| chr7_+_75931861 | 0.08 |
ENST00000248553.6 |
HSPB1 |
heat shock 27kDa protein 1 |
| chr11_-_108408895 | 0.08 |
ENST00000443411.1 ENST00000533052.1 |
EXPH5 |
exophilin 5 |
| chr2_+_198365095 | 0.08 |
ENST00000409468.1 |
HSPE1 |
heat shock 10kDa protein 1 |
| chr7_+_157129660 | 0.08 |
ENST00000429029.2 ENST00000262177.4 ENST00000417758.1 ENST00000452797.2 ENST00000443280.1 |
DNAJB6 |
DnaJ (Hsp40) homolog, subfamily B, member 6 |
| chr11_-_82997371 | 0.08 |
ENST00000525503.1 |
CCDC90B |
coiled-coil domain containing 90B |
| chr20_-_60573188 | 0.08 |
ENST00000474089.1 |
TAF4 |
TAF4 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 135kDa |
| chr1_+_67773044 | 0.08 |
ENST00000262345.1 ENST00000371000.1 |
IL12RB2 |
interleukin 12 receptor, beta 2 |
| chr19_+_2269485 | 0.08 |
ENST00000582888.4 ENST00000602676.2 ENST00000322297.4 ENST00000583542.4 |
OAZ1 |
ornithine decarboxylase antizyme 1 |
| chr18_-_6929829 | 0.08 |
ENST00000583840.1 ENST00000581571.1 ENST00000578497.1 ENST00000579012.1 |
LINC00668 |
long intergenic non-protein coding RNA 668 |
| chr4_-_17513702 | 0.07 |
ENST00000428702.2 ENST00000508623.1 ENST00000513615.1 |
QDPR |
quinoid dihydropteridine reductase |
| chr19_+_751122 | 0.07 |
ENST00000215582.6 |
MISP |
mitotic spindle positioning |
| chr22_+_32871224 | 0.07 |
ENST00000452138.1 ENST00000382058.3 ENST00000397426.1 |
FBXO7 |
F-box protein 7 |
| chr20_+_18794370 | 0.07 |
ENST00000377428.2 |
SCP2D1 |
SCP2 sterol-binding domain containing 1 |
| chr1_+_28586006 | 0.07 |
ENST00000253063.3 |
SESN2 |
sestrin 2 |
| chr11_+_20385666 | 0.07 |
ENST00000532081.1 ENST00000531058.1 |
HTATIP2 |
HIV-1 Tat interactive protein 2, 30kDa |
| chr11_-_82997420 | 0.07 |
ENST00000455220.2 ENST00000529689.1 |
CCDC90B |
coiled-coil domain containing 90B |
| chr20_-_43093968 | 0.07 |
ENST00000306731.4 ENST00000372910.3 |
C20orf62 |
chromosome 20 open reading frame 62 |
| chr14_-_57277163 | 0.07 |
ENST00000555006.1 |
OTX2 |
orthodenticle homeobox 2 |
| chr19_-_42721819 | 0.07 |
ENST00000336034.4 ENST00000598200.1 ENST00000598727.1 ENST00000596251.1 |
DEDD2 |
death effector domain containing 2 |
| chr4_-_159644507 | 0.07 |
ENST00000307720.3 |
PPID |
peptidylprolyl isomerase D |
| chr9_+_123970052 | 0.07 |
ENST00000373823.3 |
GSN |
gelsolin |
| chrX_+_15767971 | 0.07 |
ENST00000479740.1 ENST00000454127.2 |
CA5B |
carbonic anhydrase VB, mitochondrial |
| chr11_+_63953587 | 0.07 |
ENST00000305218.4 ENST00000538945.1 |
STIP1 |
stress-induced-phosphoprotein 1 |
| chr21_+_17792672 | 0.07 |
ENST00000602620.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr11_-_111781610 | 0.07 |
ENST00000525823.1 |
CRYAB |
crystallin, alpha B |
| chr8_-_89339705 | 0.07 |
ENST00000286614.6 |
MMP16 |
matrix metallopeptidase 16 (membrane-inserted) |
| chr12_+_70132458 | 0.07 |
ENST00000247833.7 |
RAB3IP |
RAB3A interacting protein |
| chr17_-_76573465 | 0.07 |
ENST00000585328.1 ENST00000389840.5 |
DNAH17 |
dynein, axonemal, heavy chain 17 |
| chr19_-_2308127 | 0.06 |
ENST00000404279.1 |
LINGO3 |
leucine rich repeat and Ig domain containing 3 |
| chr1_-_26633067 | 0.06 |
ENST00000421827.2 ENST00000374215.1 ENST00000374223.1 ENST00000357089.4 ENST00000535108.1 ENST00000314675.7 ENST00000436301.2 ENST00000423664.1 ENST00000374221.3 |
UBXN11 |
UBX domain protein 11 |
| chr1_-_45987526 | 0.06 |
ENST00000372079.1 ENST00000262746.1 ENST00000447184.1 ENST00000319248.8 |
PRDX1 |
peroxiredoxin 1 |
| chr6_-_106773291 | 0.06 |
ENST00000343245.3 |
ATG5 |
autophagy related 5 |
| chr7_+_130020932 | 0.06 |
ENST00000484324.1 |
CPA1 |
carboxypeptidase A1 (pancreatic) |
| chr22_-_41252962 | 0.06 |
ENST00000216218.3 |
ST13 |
suppression of tumorigenicity 13 (colon carcinoma) (Hsp70 interacting protein) |
| chr21_+_45937497 | 0.06 |
ENST00000354333.5 |
C21orf90 |
chromosome 21 open reading frame 90 |
| chr4_-_25865159 | 0.06 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr7_+_142378566 | 0.06 |
ENST00000390398.3 |
TRBV25-1 |
T cell receptor beta variable 25-1 |
| chr1_-_24469602 | 0.06 |
ENST00000270800.1 |
IL22RA1 |
interleukin 22 receptor, alpha 1 |
| chr11_+_63953691 | 0.06 |
ENST00000543847.1 |
STIP1 |
stress-induced-phosphoprotein 1 |
| chr11_-_111781554 | 0.06 |
ENST00000526167.1 ENST00000528961.1 |
CRYAB |
crystallin, alpha B |
| chr16_+_68119764 | 0.06 |
ENST00000570212.1 ENST00000562926.1 |
NFATC3 |
nuclear factor of activated T-cells, cytoplasmic, calcineurin-dependent 3 |
| chr21_-_40720974 | 0.06 |
ENST00000380748.1 |
HMGN1 |
high mobility group nucleosome binding domain 1 |
| chr11_-_111781454 | 0.06 |
ENST00000533280.1 |
CRYAB |
crystallin, alpha B |
| chr1_+_27248203 | 0.06 |
ENST00000321265.5 |
NUDC |
nudC nuclear distribution protein |
| chr3_+_158787041 | 0.06 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chrX_-_20236970 | 0.06 |
ENST00000379548.4 |
RPS6KA3 |
ribosomal protein S6 kinase, 90kDa, polypeptide 3 |
| chrX_+_150345054 | 0.06 |
ENST00000218316.3 |
GPR50 |
G protein-coupled receptor 50 |
| chr11_-_10590118 | 0.05 |
ENST00000529598.1 |
LYVE1 |
lymphatic vessel endothelial hyaluronan receptor 1 |
| chr18_+_57567180 | 0.05 |
ENST00000316660.6 ENST00000269518.9 |
PMAIP1 |
phorbol-12-myristate-13-acetate-induced protein 1 |
| chr2_+_198365122 | 0.05 |
ENST00000604458.1 |
HSPE1-MOB4 |
HSPE1-MOB4 readthrough |
| chr2_-_68479614 | 0.05 |
ENST00000234310.3 |
PPP3R1 |
protein phosphatase 3, regulatory subunit B, alpha |
| chr5_+_118690466 | 0.05 |
ENST00000503646.1 |
TNFAIP8 |
tumor necrosis factor, alpha-induced protein 8 |
| chr11_-_134123142 | 0.05 |
ENST00000392595.2 ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1 |
thymocyte nuclear protein 1 |
| chr16_+_29911666 | 0.05 |
ENST00000563177.1 ENST00000483405.1 |
ASPHD1 |
aspartate beta-hydroxylase domain containing 1 |
| chr20_-_43589109 | 0.05 |
ENST00000372813.3 |
TOMM34 |
translocase of outer mitochondrial membrane 34 |
| chrX_+_150866779 | 0.05 |
ENST00000370353.3 |
PRRG3 |
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr11_+_117063295 | 0.05 |
ENST00000525478.1 ENST00000532062.1 |
SIDT2 |
SID1 transmembrane family, member 2 |
| chr8_+_142402089 | 0.04 |
ENST00000521578.1 ENST00000520105.1 ENST00000523147.1 |
PTP4A3 |
protein tyrosine phosphatase type IVA, member 3 |
| chr21_+_31768348 | 0.04 |
ENST00000355459.2 |
KRTAP13-1 |
keratin associated protein 13-1 |
| chr1_-_149459549 | 0.04 |
ENST00000369175.3 |
FAM72C |
family with sequence similarity 72, member C |
| chr6_-_41909466 | 0.04 |
ENST00000414200.2 |
CCND3 |
cyclin D3 |
| chr4_-_185269050 | 0.04 |
ENST00000511465.1 |
RP11-290F5.2 |
RP11-290F5.2 |
| chr5_+_133707252 | 0.04 |
ENST00000506787.1 ENST00000507277.1 |
UBE2B |
ubiquitin-conjugating enzyme E2B |
| chr3_+_186501336 | 0.04 |
ENST00000323963.5 ENST00000440191.2 ENST00000356531.5 |
EIF4A2 |
eukaryotic translation initiation factor 4A2 |
| chr6_-_41909561 | 0.04 |
ENST00000372991.4 |
CCND3 |
cyclin D3 |
| chr14_-_23388338 | 0.04 |
ENST00000555209.1 ENST00000554256.1 ENST00000557403.1 ENST00000557549.1 ENST00000555676.1 ENST00000557571.1 ENST00000557464.1 ENST00000554618.1 ENST00000556862.1 ENST00000555722.1 ENST00000346528.5 ENST00000542016.2 ENST00000399922.2 ENST00000557227.1 ENST00000359890.3 |
RBM23 |
RNA binding motif protein 23 |
| chr1_-_205904950 | 0.04 |
ENST00000340781.4 |
SLC26A9 |
solute carrier family 26 (anion exchanger), member 9 |
| chr7_+_26240776 | 0.04 |
ENST00000337620.4 |
CBX3 |
chromobox homolog 3 |
| chr11_+_75273101 | 0.04 |
ENST00000533603.1 ENST00000358171.3 ENST00000526242.1 |
SERPINH1 |
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr1_-_173638976 | 0.04 |
ENST00000333279.2 |
ANKRD45 |
ankyrin repeat domain 45 |
| chr6_-_41909191 | 0.04 |
ENST00000512426.1 ENST00000372987.4 |
CCND3 |
cyclin D3 |
| chr1_-_6260896 | 0.04 |
ENST00000497965.1 |
RPL22 |
ribosomal protein L22 |
| chr1_+_196857144 | 0.04 |
ENST00000367416.2 ENST00000251424.4 ENST00000367418.2 |
CFHR4 |
complement factor H-related 4 |
| chr17_-_8286484 | 0.04 |
ENST00000582556.1 ENST00000584164.1 ENST00000293842.5 ENST00000584343.1 ENST00000578812.1 ENST00000583011.1 |
RPL26 |
ribosomal protein L26 |
| chr12_-_57505121 | 0.03 |
ENST00000538913.2 ENST00000537215.2 ENST00000454075.3 ENST00000554825.1 ENST00000553275.1 ENST00000300134.3 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr19_-_52133588 | 0.03 |
ENST00000570106.2 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
| chr4_+_74275057 | 0.03 |
ENST00000511370.1 |
ALB |
albumin |
| chr11_-_89956461 | 0.03 |
ENST00000320585.6 |
CHORDC1 |
cysteine and histidine-rich domain (CHORD) containing 1 |
| chr7_+_130020180 | 0.03 |
ENST00000481342.1 ENST00000011292.3 ENST00000604896.1 |
CPA1 |
carboxypeptidase A1 (pancreatic) |
| chr12_-_123011476 | 0.03 |
ENST00000528279.1 ENST00000344591.4 ENST00000526560.2 |
RSRC2 |
arginine/serine-rich coiled-coil 2 |
| chr11_-_89956227 | 0.03 |
ENST00000457199.2 ENST00000530765.1 |
CHORDC1 |
cysteine and histidine-rich domain (CHORD) containing 1 |
| chrX_-_39923656 | 0.03 |
ENST00000413905.1 |
BCOR |
BCL6 corepressor |
| chr15_+_85359911 | 0.03 |
ENST00000258888.5 |
ALPK3 |
alpha-kinase 3 |
| chr20_+_5451845 | 0.03 |
ENST00000430097.2 |
RP5-828H9.1 |
RP5-828H9.1 |
| chr11_+_75273246 | 0.03 |
ENST00000526397.1 ENST00000529643.1 ENST00000525492.1 ENST00000530284.1 ENST00000532356.1 ENST00000524558.1 |
SERPINH1 |
serpin peptidase inhibitor, clade H (heat shock protein 47), member 1, (collagen binding protein 1) |
| chr19_+_859425 | 0.03 |
ENST00000327726.6 |
CFD |
complement factor D (adipsin) |
| chr5_+_133706865 | 0.03 |
ENST00000265339.2 |
UBE2B |
ubiquitin-conjugating enzyme E2B |
| chr1_-_228613026 | 0.03 |
ENST00000366696.1 |
HIST3H3 |
histone cluster 3, H3 |
| chr21_-_40720995 | 0.03 |
ENST00000380749.5 |
HMGN1 |
high mobility group nucleosome binding domain 1 |
| chr1_+_196743912 | 0.03 |
ENST00000367425.4 |
CFHR3 |
complement factor H-related 3 |
| chr15_+_89921280 | 0.03 |
ENST00000560596.1 ENST00000558692.1 ENST00000538734.2 ENST00000559235.1 |
LINC00925 |
long intergenic non-protein coding RNA 925 |
| chr21_+_35445811 | 0.03 |
ENST00000399312.2 |
MRPS6 |
mitochondrial ribosomal protein S6 |
| chr8_+_62200509 | 0.03 |
ENST00000519846.1 ENST00000518592.1 ENST00000325897.4 |
CLVS1 |
clavesin 1 |
| chrX_+_123095155 | 0.03 |
ENST00000371160.1 ENST00000435103.1 |
STAG2 |
stromal antigen 2 |
| chr15_+_79165151 | 0.03 |
ENST00000331268.5 |
MORF4L1 |
mortality factor 4 like 1 |
| chr10_+_131265443 | 0.03 |
ENST00000306010.7 |
MGMT |
O-6-methylguanine-DNA methyltransferase |
| chr12_-_9885888 | 0.03 |
ENST00000327839.3 |
CLECL1 |
C-type lectin-like 1 |
| chr10_-_115423792 | 0.03 |
ENST00000369360.3 ENST00000360478.3 ENST00000359988.3 ENST00000369358.4 |
NRAP |
nebulin-related anchoring protein |
| chr2_-_100759037 | 0.03 |
ENST00000317233.4 ENST00000423966.1 ENST00000416492.1 |
AFF3 |
AF4/FMR2 family, member 3 |
| chr5_+_159895275 | 0.03 |
ENST00000517927.1 |
MIR146A |
microRNA 146a |
| chr17_+_1936687 | 0.02 |
ENST00000570477.1 |
DPH1 |
diphthamide biosynthesis 1 |
| chr12_-_123011536 | 0.02 |
ENST00000331738.7 ENST00000354654.2 |
RSRC2 |
arginine/serine-rich coiled-coil 2 |
| chr15_-_58357932 | 0.02 |
ENST00000347587.3 |
ALDH1A2 |
aldehyde dehydrogenase 1 family, member A2 |
| chr18_+_11751466 | 0.02 |
ENST00000535121.1 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr2_-_69664586 | 0.02 |
ENST00000303698.3 ENST00000394305.1 ENST00000410022.2 |
NFU1 |
NFU1 iron-sulfur cluster scaffold homolog (S. cerevisiae) |
| chr1_-_53704157 | 0.02 |
ENST00000371466.4 ENST00000371470.3 |
MAGOH |
mago-nashi homolog, proliferation-associated (Drosophila) |
| chr10_+_24528108 | 0.02 |
ENST00000438429.1 |
KIAA1217 |
KIAA1217 |
| chr22_+_41253080 | 0.02 |
ENST00000541156.1 ENST00000414396.1 ENST00000357137.4 |
XPNPEP3 |
X-prolyl aminopeptidase (aminopeptidase P) 3, putative |
| chrX_-_100548045 | 0.02 |
ENST00000372907.3 ENST00000372905.2 |
TAF7L |
TAF7-like RNA polymerase II, TATA box binding protein (TBP)-associated factor, 50kDa |
| chr11_-_14913765 | 0.02 |
ENST00000334636.5 |
CYP2R1 |
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr18_+_13611763 | 0.02 |
ENST00000585931.1 |
LDLRAD4 |
low density lipoprotein receptor class A domain containing 4 |
| chr15_+_79165112 | 0.02 |
ENST00000426013.2 |
MORF4L1 |
mortality factor 4 like 1 |
| chr17_-_74722672 | 0.02 |
ENST00000397625.4 ENST00000445478.2 |
JMJD6 |
jumonji domain containing 6 |
| chr20_+_57430162 | 0.02 |
ENST00000450130.1 ENST00000349036.3 ENST00000423897.1 |
GNAS |
GNAS complex locus |
| chr16_-_23568651 | 0.02 |
ENST00000563232.1 ENST00000563459.1 ENST00000449606.1 |
EARS2 |
glutamyl-tRNA synthetase 2, mitochondrial |
| chr12_-_76817036 | 0.02 |
ENST00000546946.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr1_+_154966058 | 0.02 |
ENST00000392487.1 |
LENEP |
lens epithelial protein |
| chr1_-_99470368 | 0.02 |
ENST00000263177.4 |
LPPR5 |
Lipid phosphate phosphatase-related protein type 5 |
| chr17_+_74723031 | 0.02 |
ENST00000586200.1 |
METTL23 |
methyltransferase like 23 |
| chr20_+_55108302 | 0.02 |
ENST00000371325.1 |
FAM209B |
family with sequence similarity 209, member B |
| chr17_+_74722912 | 0.02 |
ENST00000589977.1 ENST00000591571.1 ENST00000592849.1 ENST00000586738.1 ENST00000588783.1 ENST00000588563.1 ENST00000586752.1 ENST00000588302.1 ENST00000590964.1 ENST00000341249.6 ENST00000588822.1 |
METTL23 |
methyltransferase like 23 |
| chr14_-_23540747 | 0.02 |
ENST00000555566.1 ENST00000338631.6 ENST00000557515.1 ENST00000397341.3 |
ACIN1 |
apoptotic chromatin condensation inducer 1 |
| chr22_+_48972118 | 0.02 |
ENST00000358295.5 |
FAM19A5 |
family with sequence similarity 19 (chemokine (C-C motif)-like), member A5 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.1 | 0.5 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.1 | 0.5 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.1 | 0.3 | GO:0007412 | axon target recognition(GO:0007412) |
| 0.1 | 0.3 | GO:0002894 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 0.3 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 0.1 | GO:0061394 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) |
| 0.0 | 0.5 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.1 | GO:0051066 | dihydrobiopterin metabolic process(GO:0051066) |
| 0.0 | 0.1 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.3 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.1 | GO:2000298 | regulation of Rho-dependent protein serine/threonine kinase activity(GO:2000298) |
| 0.0 | 0.1 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0071492 | cellular response to UV-A(GO:0071492) |
| 0.0 | 0.1 | GO:0044855 | plasma membrane raft distribution(GO:0044855) plasma membrane raft localization(GO:0044856) plasma membrane raft polarization(GO:0044858) regulation of plasma membrane raft polarization(GO:1903906) |
| 0.0 | 0.2 | GO:0090385 | phagosome-lysosome fusion(GO:0090385) |
| 0.0 | 0.1 | GO:0060715 | syncytiotrophoblast cell differentiation involved in labyrinthine layer development(GO:0060715) |
| 0.0 | 0.0 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.1 | GO:2000619 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.0 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.2 | GO:0070436 | Grb2-EGFR complex(GO:0070436) |
| 0.0 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.2 | GO:0071682 | endocytic vesicle lumen(GO:0071682) |
| 0.0 | 0.3 | GO:0000177 | cytoplasmic exosome (RNase complex)(GO:0000177) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.7 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.1 | 0.5 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.3 | GO:0031694 | alpha-2A adrenergic receptor binding(GO:0031694) |
| 0.1 | 0.2 | GO:0032767 | copper-dependent protein binding(GO:0032767) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.2 | GO:0004947 | bradykinin receptor activity(GO:0004947) |
| 0.0 | 0.2 | GO:0061575 | cyclin-dependent protein serine/threonine kinase activator activity(GO:0061575) |
| 0.0 | 0.1 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0005502 | 11-cis retinal binding(GO:0005502) |
| 0.0 | 0.1 | GO:0070404 | NADH binding(GO:0070404) |
| 0.0 | 0.4 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.3 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.0 | GO:0004727 | prenylated protein tyrosine phosphatase activity(GO:0004727) |
| 0.0 | 0.1 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.7 | REACTOME AMINO ACID SYNTHESIS AND INTERCONVERSION TRANSAMINATION | Genes involved in Amino acid synthesis and interconversion (transamination) |
| 0.0 | 0.3 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.3 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |