Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
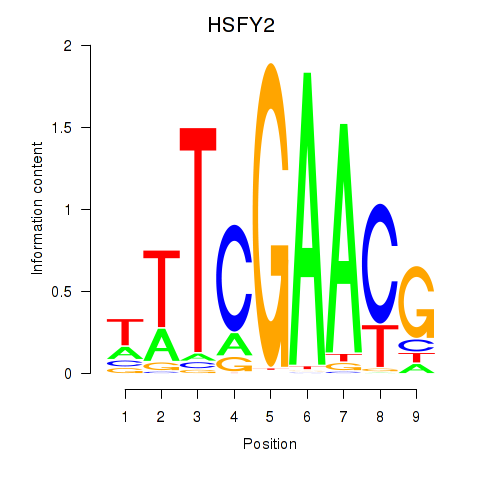
Results for HSFY2
Z-value: 0.84
Transcription factors associated with HSFY2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
HSFY2
|
ENSG00000169953.11 | HSFY2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| HSFY2 | hg19_v2_chrY_-_20935572_20935621 | -0.05 | 9.0e-01 | Click! |
Activity profile of HSFY2 motif
Sorted Z-values of HSFY2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of HSFY2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_+_102142296 | 0.81 |
ENST00000376162.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr1_-_57045228 | 0.71 |
ENST00000371250.3 |
PPAP2B |
phosphatidic acid phosphatase type 2B |
| chr1_+_183605200 | 0.58 |
ENST00000304685.4 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
| chr13_+_76378357 | 0.51 |
ENST00000489941.2 ENST00000525373.1 |
LMO7 |
LIM domain 7 |
| chr2_+_201170703 | 0.49 |
ENST00000358677.5 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr13_+_76378305 | 0.48 |
ENST00000526371.1 ENST00000526528.1 |
LMO7 |
LIM domain 7 |
| chr17_+_68165657 | 0.48 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr12_-_91572278 | 0.45 |
ENST00000425043.1 ENST00000420120.2 ENST00000441303.2 ENST00000456569.2 |
DCN |
decorin |
| chr3_-_20227619 | 0.44 |
ENST00000425061.1 ENST00000443724.1 ENST00000421451.1 ENST00000452020.1 ENST00000417364.1 ENST00000306698.2 ENST00000419233.2 ENST00000263753.4 ENST00000383774.1 ENST00000437051.1 ENST00000412868.1 ENST00000429446.3 ENST00000442720.1 |
SGOL1 |
shugoshin-like 1 (S. pombe) |
| chr8_-_72987810 | 0.43 |
ENST00000262209.4 |
TRPA1 |
transient receptor potential cation channel, subfamily A, member 1 |
| chr1_+_114522049 | 0.42 |
ENST00000369551.1 ENST00000320334.4 |
OLFML3 |
olfactomedin-like 3 |
| chr14_+_104394770 | 0.38 |
ENST00000409874.4 ENST00000339063.5 |
TDRD9 |
tudor domain containing 9 |
| chr8_+_120428546 | 0.36 |
ENST00000259526.3 |
NOV |
nephroblastoma overexpressed |
| chr10_-_49860525 | 0.35 |
ENST00000435790.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr5_-_19988339 | 0.33 |
ENST00000382275.1 |
CDH18 |
cadherin 18, type 2 |
| chr1_+_155829286 | 0.33 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr3_+_101546827 | 0.31 |
ENST00000461724.1 ENST00000483180.1 ENST00000394054.2 |
NFKBIZ |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, zeta |
| chr12_+_75874460 | 0.31 |
ENST00000266659.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr4_-_13629269 | 0.30 |
ENST00000040738.5 |
BOD1L1 |
biorientation of chromosomes in cell division 1-like 1 |
| chr10_-_129691195 | 0.30 |
ENST00000368671.3 |
CLRN3 |
clarin 3 |
| chr7_-_138363824 | 0.28 |
ENST00000419765.3 |
SVOPL |
SVOP-like |
| chr2_+_109335929 | 0.28 |
ENST00000283195.6 |
RANBP2 |
RAN binding protein 2 |
| chr16_+_15596123 | 0.28 |
ENST00000452191.2 |
C16orf45 |
chromosome 16 open reading frame 45 |
| chr4_-_113558079 | 0.28 |
ENST00000445203.2 |
C4orf21 |
chromosome 4 open reading frame 21 |
| chr16_+_31539197 | 0.27 |
ENST00000564707.1 |
AHSP |
alpha hemoglobin stabilizing protein |
| chr18_+_3247779 | 0.27 |
ENST00000578611.1 ENST00000583449.1 |
MYL12A |
myosin, light chain 12A, regulatory, non-sarcomeric |
| chr12_+_75874984 | 0.27 |
ENST00000550491.1 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr2_-_169769787 | 0.27 |
ENST00000451987.1 |
SPC25 |
SPC25, NDC80 kinetochore complex component |
| chr12_+_75874580 | 0.27 |
ENST00000456650.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr13_-_33112823 | 0.26 |
ENST00000504114.1 |
N4BP2L2 |
NEDD4 binding protein 2-like 2 |
| chr1_-_92371839 | 0.25 |
ENST00000370399.2 |
TGFBR3 |
transforming growth factor, beta receptor III |
| chr4_-_113558014 | 0.25 |
ENST00000503172.1 ENST00000505019.1 ENST00000309071.5 |
C4orf21 |
chromosome 4 open reading frame 21 |
| chr1_+_93646238 | 0.25 |
ENST00000448243.1 ENST00000370276.1 |
CCDC18 |
coiled-coil domain containing 18 |
| chr3_-_50374869 | 0.24 |
ENST00000327761.3 |
RASSF1 |
Ras association (RalGDS/AF-6) domain family member 1 |
| chr6_+_26458152 | 0.24 |
ENST00000312541.5 |
BTN2A1 |
butyrophilin, subfamily 2, member A1 |
| chr12_-_88535747 | 0.24 |
ENST00000309041.7 |
CEP290 |
centrosomal protein 290kDa |
| chr11_+_102980126 | 0.24 |
ENST00000375735.2 |
DYNC2H1 |
dynein, cytoplasmic 2, heavy chain 1 |
| chr14_+_85996507 | 0.24 |
ENST00000554746.1 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr11_+_12132117 | 0.24 |
ENST00000256194.4 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr17_-_8113542 | 0.23 |
ENST00000578549.1 ENST00000535053.1 ENST00000582368.1 |
AURKB |
aurora kinase B |
| chr20_+_30327063 | 0.23 |
ENST00000300403.6 ENST00000340513.4 |
TPX2 |
TPX2, microtubule-associated |
| chr6_+_73331918 | 0.22 |
ENST00000402622.2 ENST00000355635.3 ENST00000403813.2 ENST00000414165.2 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr15_-_85197501 | 0.22 |
ENST00000434634.2 |
WDR73 |
WD repeat domain 73 |
| chr22_-_32341336 | 0.22 |
ENST00000248984.3 |
C22orf24 |
chromosome 22 open reading frame 24 |
| chr1_+_12851545 | 0.21 |
ENST00000332296.7 |
PRAMEF1 |
PRAME family member 1 |
| chr1_-_197115818 | 0.21 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr3_-_50375657 | 0.21 |
ENST00000395126.3 |
RASSF1 |
Ras association (RalGDS/AF-6) domain family member 1 |
| chr16_+_31494323 | 0.21 |
ENST00000569576.1 ENST00000330498.3 |
SLC5A2 |
solute carrier family 5 (sodium/glucose cotransporter), member 2 |
| chr8_+_27629459 | 0.21 |
ENST00000523566.1 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr1_-_149900122 | 0.21 |
ENST00000271628.8 |
SF3B4 |
splicing factor 3b, subunit 4, 49kDa |
| chr11_-_63376013 | 0.20 |
ENST00000540943.1 |
PLA2G16 |
phospholipase A2, group XVI |
| chr7_+_29603394 | 0.20 |
ENST00000319694.2 |
PRR15 |
proline rich 15 |
| chr11_+_124824000 | 0.20 |
ENST00000529051.1 ENST00000344762.5 |
CCDC15 |
coiled-coil domain containing 15 |
| chr16_+_31483374 | 0.19 |
ENST00000394863.3 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
| chr6_+_26458171 | 0.19 |
ENST00000493173.1 ENST00000541522.1 ENST00000429381.1 ENST00000469185.1 |
BTN2A1 |
butyrophilin, subfamily 2, member A1 |
| chr1_+_214776516 | 0.19 |
ENST00000366955.3 |
CENPF |
centromere protein F, 350/400kDa |
| chr12_+_66217911 | 0.18 |
ENST00000403681.2 |
HMGA2 |
high mobility group AT-hook 2 |
| chr15_+_40886199 | 0.18 |
ENST00000346991.5 ENST00000528975.1 ENST00000527044.1 |
CASC5 |
cancer susceptibility candidate 5 |
| chr15_+_41186609 | 0.18 |
ENST00000220509.5 |
VPS18 |
vacuolar protein sorting 18 homolog (S. cerevisiae) |
| chr14_+_85996471 | 0.18 |
ENST00000330753.4 |
FLRT2 |
fibronectin leucine rich transmembrane protein 2 |
| chr12_-_132628847 | 0.18 |
ENST00000397333.3 |
DDX51 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 51 |
| chr7_-_93520191 | 0.18 |
ENST00000545378.1 |
TFPI2 |
tissue factor pathway inhibitor 2 |
| chr3_-_149093499 | 0.18 |
ENST00000472441.1 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr8_-_42065187 | 0.18 |
ENST00000270189.6 ENST00000352041.3 ENST00000220809.4 |
PLAT |
plasminogen activator, tissue |
| chr6_+_31637944 | 0.18 |
ENST00000375864.4 |
LY6G5B |
lymphocyte antigen 6 complex, locus G5B |
| chr14_-_85996332 | 0.17 |
ENST00000380722.1 |
RP11-497E19.1 |
RP11-497E19.1 |
| chr1_-_89458415 | 0.17 |
ENST00000321792.5 ENST00000370491.3 |
RBMXL1 CCBL2 |
RNA binding motif protein, X-linked-like 1 cysteine conjugate-beta lyase 2 |
| chr10_+_13203543 | 0.17 |
ENST00000378714.3 ENST00000479669.1 ENST00000484800.2 |
MCM10 |
minichromosome maintenance complex component 10 |
| chr4_-_104119528 | 0.17 |
ENST00000380026.3 ENST00000503705.1 ENST00000265148.3 |
CENPE |
centromere protein E, 312kDa |
| chr20_-_44718538 | 0.17 |
ENST00000290231.6 ENST00000372291.3 |
NCOA5 |
nuclear receptor coactivator 5 |
| chr2_+_65216462 | 0.17 |
ENST00000234256.3 |
SLC1A4 |
solute carrier family 1 (glutamate/neutral amino acid transporter), member 4 |
| chr1_+_12916941 | 0.17 |
ENST00000240189.2 |
PRAMEF2 |
PRAME family member 2 |
| chr5_+_42756903 | 0.17 |
ENST00000361970.5 ENST00000388827.4 |
CCDC152 |
coiled-coil domain containing 152 |
| chr19_+_58341656 | 0.16 |
ENST00000442832.4 ENST00000594901.1 |
ZNF587B |
zinc finger protein 587B |
| chr7_+_138943265 | 0.16 |
ENST00000483726.1 |
UBN2 |
ubinuclein 2 |
| chr6_+_36839616 | 0.16 |
ENST00000359359.2 ENST00000510325.2 |
C6orf89 |
chromosome 6 open reading frame 89 |
| chrX_-_73072534 | 0.16 |
ENST00000429829.1 |
XIST |
X inactive specific transcript (non-protein coding) |
| chr6_+_73331776 | 0.16 |
ENST00000370398.1 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr4_+_150999418 | 0.16 |
ENST00000296550.7 |
DCLK2 |
doublecortin-like kinase 2 |
| chr16_+_20817839 | 0.16 |
ENST00000348433.6 ENST00000568501.1 ENST00000566276.1 |
AC004381.6 |
Putative RNA exonuclease NEF-sp |
| chr6_+_73331520 | 0.16 |
ENST00000342056.2 ENST00000355194.4 |
KCNQ5 |
potassium voltage-gated channel, KQT-like subfamily, member 5 |
| chr10_+_17272608 | 0.15 |
ENST00000421459.2 |
VIM |
vimentin |
| chr22_-_17073700 | 0.15 |
ENST00000359963.3 |
CCT8L2 |
chaperonin containing TCP1, subunit 8 (theta)-like 2 |
| chr19_-_16770915 | 0.15 |
ENST00000593459.1 ENST00000358726.6 ENST00000597711.1 ENST00000487416.2 |
CTC-429P9.4 SMIM7 |
Small integral membrane protein 7; Uncharacterized protein small integral membrane protein 7 |
| chr6_+_107077471 | 0.15 |
ENST00000369044.1 |
QRSL1 |
glutaminyl-tRNA synthase (glutamine-hydrolyzing)-like 1 |
| chr8_+_22224811 | 0.15 |
ENST00000381237.1 |
SLC39A14 |
solute carrier family 39 (zinc transporter), member 14 |
| chr19_+_53073526 | 0.15 |
ENST00000596514.1 ENST00000391785.3 ENST00000301093.2 |
ZNF701 |
zinc finger protein 701 |
| chr19_-_58514129 | 0.15 |
ENST00000552184.1 ENST00000546715.1 ENST00000536132.1 ENST00000547828.1 ENST00000547121.1 ENST00000551380.1 |
ZNF606 |
zinc finger protein 606 |
| chr7_-_93520259 | 0.15 |
ENST00000222543.5 |
TFPI2 |
tissue factor pathway inhibitor 2 |
| chr20_-_271304 | 0.14 |
ENST00000400269.3 ENST00000360321.2 |
C20orf96 |
chromosome 20 open reading frame 96 |
| chr9_-_6015607 | 0.14 |
ENST00000259569.5 |
RANBP6 |
RAN binding protein 6 |
| chr9_-_99180597 | 0.14 |
ENST00000375256.4 |
ZNF367 |
zinc finger protein 367 |
| chr15_+_90118723 | 0.14 |
ENST00000560985.1 |
TICRR |
TOPBP1-interacting checkpoint and replication regulator |
| chr1_-_198990166 | 0.14 |
ENST00000427439.1 |
RP11-16L9.3 |
RP11-16L9.3 |
| chr3_+_186383741 | 0.14 |
ENST00000232003.4 |
HRG |
histidine-rich glycoprotein |
| chr1_-_13002348 | 0.14 |
ENST00000355096.2 |
PRAMEF6 |
PRAME family member 6 |
| chr11_+_17281900 | 0.14 |
ENST00000530527.1 |
NUCB2 |
nucleobindin 2 |
| chr6_-_44281043 | 0.14 |
ENST00000244571.4 |
AARS2 |
alanyl-tRNA synthetase 2, mitochondrial |
| chr1_-_93645818 | 0.14 |
ENST00000370280.1 ENST00000479918.1 |
TMED5 |
transmembrane emp24 protein transport domain containing 5 |
| chrX_+_57618269 | 0.14 |
ENST00000374888.1 |
ZXDB |
zinc finger, X-linked, duplicated B |
| chr12_-_118406777 | 0.13 |
ENST00000339824.5 |
KSR2 |
kinase suppressor of ras 2 |
| chr11_+_102980251 | 0.13 |
ENST00000334267.7 ENST00000398093.3 |
DYNC2H1 |
dynein, cytoplasmic 2, heavy chain 1 |
| chr10_+_35415851 | 0.13 |
ENST00000374726.3 |
CREM |
cAMP responsive element modulator |
| chr3_+_142838091 | 0.13 |
ENST00000309575.3 |
CHST2 |
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
| chr19_+_45147098 | 0.13 |
ENST00000425690.3 ENST00000344956.4 ENST00000403059.4 |
PVR |
poliovirus receptor |
| chr12_+_109785708 | 0.13 |
ENST00000310903.5 |
MYO1H |
myosin IH |
| chr8_+_17780483 | 0.13 |
ENST00000517730.1 ENST00000518537.1 ENST00000523055.1 ENST00000519253.1 |
PCM1 |
pericentriolar material 1 |
| chr8_+_27631903 | 0.13 |
ENST00000305188.8 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr8_-_42065075 | 0.13 |
ENST00000429089.2 ENST00000519510.1 ENST00000429710.2 ENST00000524009.1 |
PLAT |
plasminogen activator, tissue |
| chr6_-_169654139 | 0.13 |
ENST00000366787.3 |
THBS2 |
thrombospondin 2 |
| chr11_+_111126707 | 0.13 |
ENST00000280325.4 |
C11orf53 |
chromosome 11 open reading frame 53 |
| chr20_+_42086525 | 0.13 |
ENST00000244020.3 |
SRSF6 |
serine/arginine-rich splicing factor 6 |
| chr19_-_58220517 | 0.13 |
ENST00000512439.2 ENST00000426889.1 |
ZNF154 |
zinc finger protein 154 |
| chr1_+_144173162 | 0.13 |
ENST00000356801.6 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
| chr15_-_50411412 | 0.13 |
ENST00000284509.6 |
ATP8B4 |
ATPase, class I, type 8B, member 4 |
| chr18_+_39535152 | 0.12 |
ENST00000262039.4 ENST00000398870.3 ENST00000586545.1 |
PIK3C3 |
phosphatidylinositol 3-kinase, catalytic subunit type 3 |
| chr8_+_80523962 | 0.12 |
ENST00000518491.1 |
STMN2 |
stathmin-like 2 |
| chr17_-_3301704 | 0.12 |
ENST00000322608.2 |
OR1E1 |
olfactory receptor, family 1, subfamily E, member 1 |
| chr10_+_35416223 | 0.12 |
ENST00000489321.1 ENST00000427847.2 ENST00000345491.3 ENST00000395895.2 ENST00000374728.3 ENST00000487132.1 |
CREM |
cAMP responsive element modulator |
| chrX_+_100353153 | 0.12 |
ENST00000423383.1 ENST00000218507.5 ENST00000403304.2 ENST00000435570.1 |
CENPI |
centromere protein I |
| chr19_+_50529212 | 0.12 |
ENST00000270617.3 ENST00000445728.3 ENST00000601364.1 |
ZNF473 |
zinc finger protein 473 |
| chr11_+_93394805 | 0.12 |
ENST00000325212.6 ENST00000411936.1 ENST00000344196.4 |
KIAA1731 |
KIAA1731 |
| chr19_+_58193337 | 0.12 |
ENST00000601064.1 ENST00000282296.5 ENST00000356715.4 |
ZNF551 |
zinc finger protein 551 |
| chr15_-_57025759 | 0.12 |
ENST00000267807.7 |
ZNF280D |
zinc finger protein 280D |
| chr8_+_17780346 | 0.12 |
ENST00000325083.8 |
PCM1 |
pericentriolar material 1 |
| chr3_+_155838337 | 0.12 |
ENST00000490337.1 ENST00000389636.5 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr7_-_121784285 | 0.12 |
ENST00000417368.2 |
AASS |
aminoadipate-semialdehyde synthase |
| chr1_+_213123915 | 0.12 |
ENST00000366968.4 ENST00000490792.1 |
VASH2 |
vasohibin 2 |
| chr20_+_18488137 | 0.12 |
ENST00000450074.1 ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B |
Sec23 homolog B (S. cerevisiae) |
| chr7_+_120590803 | 0.12 |
ENST00000315870.5 ENST00000339121.5 ENST00000445699.1 |
ING3 |
inhibitor of growth family, member 3 |
| chr11_-_71318487 | 0.12 |
ENST00000343767.3 |
AP000867.1 |
AP000867.1 |
| chr11_-_77850629 | 0.12 |
ENST00000376156.3 ENST00000525870.1 ENST00000530454.1 ENST00000525755.1 ENST00000527099.1 ENST00000525761.1 ENST00000299626.5 |
ALG8 |
ALG8, alpha-1,3-glucosyltransferase |
| chrX_+_21857717 | 0.12 |
ENST00000379484.5 |
MBTPS2 |
membrane-bound transcription factor peptidase, site 2 |
| chr3_+_8543393 | 0.12 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chrX_+_36065053 | 0.12 |
ENST00000313548.4 |
CHDC2 |
calponin homology domain containing 2 |
| chr3_-_149293990 | 0.12 |
ENST00000472417.1 |
WWTR1 |
WW domain containing transcription regulator 1 |
| chr16_+_21312170 | 0.12 |
ENST00000338573.5 ENST00000561968.1 |
CRYM-AS1 |
CRYM antisense RNA 1 |
| chr22_-_36013368 | 0.12 |
ENST00000442617.1 ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB |
myoglobin |
| chr2_+_169757750 | 0.12 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr20_-_8000426 | 0.12 |
ENST00000527925.1 ENST00000246024.2 |
TMX4 |
thioredoxin-related transmembrane protein 4 |
| chr5_-_37249397 | 0.12 |
ENST00000425232.2 ENST00000274258.7 |
C5orf42 |
chromosome 5 open reading frame 42 |
| chr13_+_114462193 | 0.11 |
ENST00000375353.3 |
TMEM255B |
transmembrane protein 255B |
| chr8_-_63998590 | 0.11 |
ENST00000260116.4 |
TTPA |
tocopherol (alpha) transfer protein |
| chr2_-_97405775 | 0.11 |
ENST00000264963.4 ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L |
lectin, mannose-binding 2-like |
| chr9_-_115983568 | 0.11 |
ENST00000446284.1 ENST00000414250.1 |
FKBP15 |
FK506 binding protein 15, 133kDa |
| chr10_+_70587279 | 0.11 |
ENST00000298596.6 ENST00000399169.4 ENST00000399165.4 ENST00000399162.2 |
STOX1 |
storkhead box 1 |
| chr7_-_82792215 | 0.11 |
ENST00000333891.9 ENST00000423517.2 |
PCLO |
piccolo presynaptic cytomatrix protein |
| chr20_+_39969519 | 0.11 |
ENST00000373257.3 |
LPIN3 |
lipin 3 |
| chr20_+_33292068 | 0.11 |
ENST00000374810.3 ENST00000374809.2 ENST00000451665.1 |
TP53INP2 |
tumor protein p53 inducible nuclear protein 2 |
| chr10_+_45495898 | 0.11 |
ENST00000298299.3 |
ZNF22 |
zinc finger protein 22 |
| chr6_-_11382478 | 0.11 |
ENST00000397378.3 ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
| chr1_-_11159887 | 0.11 |
ENST00000544779.1 ENST00000304457.7 ENST00000376936.4 |
EXOSC10 |
exosome component 10 |
| chrX_+_49028265 | 0.11 |
ENST00000376322.3 ENST00000376327.5 |
PLP2 |
proteolipid protein 2 (colonic epithelium-enriched) |
| chr2_+_109237717 | 0.11 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr12_-_7125770 | 0.11 |
ENST00000261407.4 |
LPCAT3 |
lysophosphatidylcholine acyltransferase 3 |
| chr13_+_37581115 | 0.11 |
ENST00000481013.1 |
EXOSC8 |
exosome component 8 |
| chr9_-_114937465 | 0.11 |
ENST00000355396.3 |
SUSD1 |
sushi domain containing 1 |
| chr5_+_134240588 | 0.11 |
ENST00000254908.6 |
PCBD2 |
pterin-4 alpha-carbinolamine dehydratase/dimerization cofactor of hepatocyte nuclear factor 1 alpha (TCF1) 2 |
| chr19_-_1876156 | 0.11 |
ENST00000565797.1 |
CTB-31O20.2 |
CTB-31O20.2 |
| chr17_-_61523535 | 0.11 |
ENST00000584031.1 ENST00000392976.1 |
CYB561 |
cytochrome b561 |
| chr10_-_90611566 | 0.11 |
ENST00000371930.4 |
ANKRD22 |
ankyrin repeat domain 22 |
| chr7_+_99070464 | 0.11 |
ENST00000331410.5 ENST00000483089.1 ENST00000448667.1 ENST00000493485.1 |
ZNF789 |
zinc finger protein 789 |
| chr12_+_12202774 | 0.10 |
ENST00000589718.1 |
BCL2L14 |
BCL2-like 14 (apoptosis facilitator) |
| chr11_+_65479702 | 0.10 |
ENST00000530446.1 ENST00000534104.1 ENST00000530605.1 ENST00000528198.1 ENST00000531880.1 ENST00000534650.1 |
KAT5 |
K(lysine) acetyltransferase 5 |
| chr12_+_12202785 | 0.10 |
ENST00000586576.1 ENST00000464885.2 |
BCL2L14 |
BCL2-like 14 (apoptosis facilitator) |
| chr15_+_69365265 | 0.10 |
ENST00000415504.1 |
LINC00277 |
long intergenic non-protein coding RNA 277 |
| chr1_-_53163992 | 0.10 |
ENST00000371538.3 |
SELRC1 |
cytochrome c oxidase assembly factor 7 |
| chrX_+_46940254 | 0.10 |
ENST00000336169.3 |
RGN |
regucalcin |
| chr5_+_108083517 | 0.10 |
ENST00000281092.4 ENST00000536402.1 |
FER |
fer (fps/fes related) tyrosine kinase |
| chr17_-_38574169 | 0.10 |
ENST00000423485.1 |
TOP2A |
topoisomerase (DNA) II alpha 170kDa |
| chr16_-_30006922 | 0.10 |
ENST00000564026.1 |
HIRIP3 |
HIRA interacting protein 3 |
| chr9_-_115983641 | 0.10 |
ENST00000238256.3 |
FKBP15 |
FK506 binding protein 15, 133kDa |
| chr7_-_35293740 | 0.10 |
ENST00000408931.3 |
TBX20 |
T-box 20 |
| chr6_-_169582835 | 0.10 |
ENST00000564830.1 |
RP11-417E7.2 |
RP11-417E7.2 |
| chr9_+_96793076 | 0.10 |
ENST00000375360.3 |
PTPDC1 |
protein tyrosine phosphatase domain containing 1 |
| chr10_-_25010795 | 0.10 |
ENST00000416305.1 ENST00000376410.2 |
ARHGAP21 |
Rho GTPase activating protein 21 |
| chr9_-_114937676 | 0.10 |
ENST00000374270.3 |
SUSD1 |
sushi domain containing 1 |
| chr1_-_26232522 | 0.10 |
ENST00000399728.1 |
STMN1 |
stathmin 1 |
| chr7_-_35077653 | 0.10 |
ENST00000310974.4 |
DPY19L1 |
dpy-19-like 1 (C. elegans) |
| chr22_+_30792846 | 0.10 |
ENST00000312932.9 ENST00000428195.1 |
SEC14L2 |
SEC14-like 2 (S. cerevisiae) |
| chr8_+_23386305 | 0.10 |
ENST00000519973.1 |
SLC25A37 |
solute carrier family 25 (mitochondrial iron transporter), member 37 |
| chr2_-_70475701 | 0.10 |
ENST00000282574.4 |
TIA1 |
TIA1 cytotoxic granule-associated RNA binding protein |
| chr11_+_103907308 | 0.10 |
ENST00000302259.3 |
DDI1 |
DNA-damage inducible 1 homolog 1 (S. cerevisiae) |
| chr1_+_236958554 | 0.10 |
ENST00000366577.5 ENST00000418145.2 |
MTR |
5-methyltetrahydrofolate-homocysteine methyltransferase |
| chr6_+_108882069 | 0.10 |
ENST00000406360.1 |
FOXO3 |
forkhead box O3 |
| chr12_+_22778009 | 0.10 |
ENST00000266517.4 ENST00000335148.3 |
ETNK1 |
ethanolamine kinase 1 |
| chr6_+_108977520 | 0.09 |
ENST00000540898.1 |
FOXO3 |
forkhead box O3 |
| chr11_+_65627865 | 0.09 |
ENST00000308110.4 |
MUS81 |
MUS81 structure-specific endonuclease subunit |
| chr14_-_50154921 | 0.09 |
ENST00000553805.2 ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2 |
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr16_-_18462221 | 0.09 |
ENST00000528301.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr4_+_113558272 | 0.09 |
ENST00000509061.1 ENST00000508577.1 ENST00000513553.1 |
LARP7 |
La ribonucleoprotein domain family, member 7 |
| chr19_+_50528971 | 0.09 |
ENST00000598809.1 ENST00000595661.1 ENST00000391821.2 |
ZNF473 |
zinc finger protein 473 |
| chr2_-_97509729 | 0.09 |
ENST00000418232.1 |
ANKRD23 |
ankyrin repeat domain 23 |
| chr6_-_43543702 | 0.09 |
ENST00000265351.7 |
XPO5 |
exportin 5 |
| chr1_+_15573757 | 0.09 |
ENST00000358897.4 ENST00000417793.1 ENST00000375999.3 ENST00000433640.2 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr12_-_42877408 | 0.09 |
ENST00000552240.1 |
PRICKLE1 |
prickle homolog 1 (Drosophila) |
| chr6_+_163148161 | 0.09 |
ENST00000337019.3 ENST00000366889.2 |
PACRG |
PARK2 co-regulated |
| chr17_+_28705921 | 0.09 |
ENST00000225719.4 |
CPD |
carboxypeptidase D |
| chr6_-_26033796 | 0.09 |
ENST00000259791.2 |
HIST1H2AB |
histone cluster 1, H2ab |
| chr12_+_107168418 | 0.09 |
ENST00000392839.2 ENST00000548914.1 ENST00000355478.2 ENST00000552619.1 ENST00000549643.1 |
RIC8B |
RIC8 guanine nucleotide exchange factor B |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.1 | GO:0090210 | regulation of establishment of blood-brain barrier(GO:0090210) negative regulation of establishment of blood-brain barrier(GO:0090212) |
| 0.1 | 0.3 | GO:1903422 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.1 | 0.4 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.1 | 0.4 | GO:0050955 | thermoception(GO:0050955) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.2 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.1 | 0.3 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.1 | 0.2 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.1 | 0.4 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.1 | 0.3 | GO:0003131 | mesodermal-endodermal cell signaling(GO:0003131) programmed DNA elimination(GO:0031049) chromosome breakage(GO:0031052) histone H2A-S139 phosphorylation(GO:0035978) positive regulation of cellular response to X-ray(GO:2000685) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.3 | GO:0007181 | transforming growth factor beta receptor complex assembly(GO:0007181) response to luteinizing hormone(GO:0034699) |
| 0.1 | 0.3 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.3 | GO:1903976 | negative regulation of glial cell migration(GO:1903976) |
| 0.1 | 0.2 | GO:0070681 | glutaminyl-tRNAGln biosynthesis via transamidation(GO:0070681) |
| 0.1 | 0.1 | GO:1904294 | positive regulation of ERAD pathway(GO:1904294) |
| 0.0 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.0 | 0.1 | GO:0060370 | susceptibility to T cell mediated cytotoxicity(GO:0060370) |
| 0.0 | 0.6 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.3 | GO:1902035 | positive regulation of hematopoietic stem cell proliferation(GO:1902035) |
| 0.0 | 0.1 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:0019853 | L-ascorbic acid biosynthetic process(GO:0019853) regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.1 | GO:0036216 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:0003193 | pulmonary valve formation(GO:0003193) visceral motor neuron differentiation(GO:0021524) foramen ovale closure(GO:0035922) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.3 | GO:0022027 | interkinetic nuclear migration(GO:0022027) |
| 0.0 | 0.1 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0033512 | L-lysine catabolic process to acetyl-CoA via saccharopine(GO:0033512) |
| 0.0 | 0.2 | GO:0007079 | mitotic chromosome movement towards spindle pole(GO:0007079) |
| 0.0 | 0.1 | GO:0019557 | histidine catabolic process to glutamate and formamide(GO:0019556) histidine catabolic process to glutamate and formate(GO:0019557) formamide metabolic process(GO:0043606) |
| 0.0 | 0.1 | GO:2000532 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) L-cystine transport(GO:0015811) |
| 0.0 | 0.1 | GO:1901656 | cellular response to mycotoxin(GO:0036146) glycoside transport(GO:1901656) cellular response to bile acid(GO:1903413) |
| 0.0 | 0.2 | GO:0006398 | mRNA 3'-end processing by stem-loop binding and cleavage(GO:0006398) |
| 0.0 | 0.1 | GO:0034476 | U1 snRNA 3'-end processing(GO:0034473) U5 snRNA 3'-end processing(GO:0034476) |
| 0.0 | 0.1 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.2 | GO:0030174 | regulation of DNA-dependent DNA replication initiation(GO:0030174) |
| 0.0 | 0.4 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.1 | GO:2000690 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.1 | GO:0000915 | assembly of actomyosin apparatus involved in cytokinesis(GO:0000912) actomyosin contractile ring assembly(GO:0000915) actomyosin contractile ring organization(GO:0044837) |
| 0.0 | 0.2 | GO:1903998 | response to isolation stress(GO:0035900) regulation of eating behavior(GO:1903998) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0070934 | CRD-mediated mRNA stabilization(GO:0070934) |
| 0.0 | 0.1 | GO:0060501 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) |
| 0.0 | 0.2 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0039534 | negative regulation of MDA-5 signaling pathway(GO:0039534) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.2 | GO:0048793 | pronephros development(GO:0048793) |
| 0.0 | 0.2 | GO:0021859 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.2 | GO:1903608 | protein localization to cytoplasmic stress granule(GO:1903608) |
| 0.0 | 0.1 | GO:0072434 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) |
| 0.0 | 0.2 | GO:1901727 | positive regulation of histone deacetylase activity(GO:1901727) |
| 0.0 | 0.1 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.1 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.1 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.1 | GO:1904491 | protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.3 | GO:0020027 | hemoglobin metabolic process(GO:0020027) |
| 0.0 | 0.1 | GO:0036116 | medium-chain fatty-acyl-CoA catabolic process(GO:0036114) long-chain fatty-acyl-CoA catabolic process(GO:0036116) palmitic acid metabolic process(GO:1900533) palmitic acid biosynthetic process(GO:1900535) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:1903288 | positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.1 | GO:0090245 | axis elongation involved in somitogenesis(GO:0090245) |
| 0.0 | 0.0 | GO:0021816 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.0 | 0.2 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0035616 | histone H2B conserved C-terminal lysine deubiquitination(GO:0035616) |
| 0.0 | 0.0 | GO:0044205 | 'de novo' UMP biosynthetic process(GO:0044205) |
| 0.0 | 0.2 | GO:0060020 | Bergmann glial cell differentiation(GO:0060020) |
| 0.0 | 0.1 | GO:0051534 | negative regulation of NFAT protein import into nucleus(GO:0051534) |
| 0.0 | 0.1 | GO:0043504 | mitochondrial DNA repair(GO:0043504) |
| 0.0 | 0.1 | GO:0071475 | cellular hyperosmotic salinity response(GO:0071475) |
| 0.0 | 0.1 | GO:0018103 | protein C-linked glycosylation(GO:0018103) peptidyl-tryptophan modification(GO:0018211) protein C-linked glycosylation via tryptophan(GO:0018317) protein C-linked glycosylation via 2'-alpha-mannosyl-L-tryptophan(GO:0018406) |
| 0.0 | 0.0 | GO:0003175 | tricuspid valve development(GO:0003175) |
| 0.0 | 0.4 | GO:0043046 | DNA methylation involved in gamete generation(GO:0043046) |
| 0.0 | 0.3 | GO:0031297 | replication fork processing(GO:0031297) |
| 0.0 | 0.4 | GO:0071498 | cellular response to fluid shear stress(GO:0071498) |
| 0.0 | 0.1 | GO:0051944 | positive regulation of neurotransmitter uptake(GO:0051582) positive regulation of dopamine uptake involved in synaptic transmission(GO:0051586) positive regulation of catecholamine uptake involved in synaptic transmission(GO:0051944) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.0 | GO:0035964 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.1 | GO:0006610 | ribosomal protein import into nucleus(GO:0006610) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0031848 | protection from non-homologous end joining at telomere(GO:0031848) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 0.3 | GO:0034673 | inhibin-betaglycan-ActRII complex(GO:0034673) |
| 0.1 | 0.4 | GO:0071547 | piP-body(GO:0071547) |
| 0.1 | 0.2 | GO:0030956 | glutamyl-tRNA(Gln) amidotransferase complex(GO:0030956) |
| 0.0 | 0.3 | GO:0044614 | nuclear pore cytoplasmic filaments(GO:0044614) |
| 0.0 | 0.3 | GO:0031262 | Ndc80 complex(GO:0031262) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.1 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.2 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0097165 | nuclear stress granule(GO:0097165) |
| 0.0 | 0.4 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.1 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.2 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.3 | GO:0035985 | senescence-associated heterochromatin focus(GO:0035985) |
| 0.0 | 0.3 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.3 | GO:0005833 | hemoglobin complex(GO:0005833) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.1 | GO:0070939 | Dsl1p complex(GO:0070939) |
| 0.0 | 0.2 | GO:0043203 | axon hillock(GO:0043203) |
| 0.0 | 0.1 | GO:1990635 | proximal dendrite(GO:1990635) |
| 0.0 | 0.1 | GO:0031465 | Cul4B-RING E3 ubiquitin ligase complex(GO:0031465) |
| 0.0 | 0.1 | GO:0097149 | centralspindlin complex(GO:0097149) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.2 | GO:0031298 | replication fork protection complex(GO:0031298) |
| 0.0 | 0.5 | GO:0034451 | centriolar satellite(GO:0034451) |
| 0.0 | 0.2 | GO:0000176 | nuclear exosome (RNase complex)(GO:0000176) |
| 0.0 | 0.1 | GO:0032311 | angiogenin-PRI complex(GO:0032311) |
| 0.0 | 0.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.1 | GO:0016222 | procollagen-proline 4-dioxygenase complex(GO:0016222) |
| 0.0 | 0.1 | GO:0070937 | CRD-mediated mRNA stability complex(GO:0070937) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.1 | GO:0042765 | GPI-anchor transamidase complex(GO:0042765) |
| 0.0 | 0.0 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.1 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.1 | GO:0043564 | Ku70:Ku80 complex(GO:0043564) |
| 0.0 | 0.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.1 | 0.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 0.3 | GO:0035501 | MH1 domain binding(GO:0035501) |
| 0.1 | 0.2 | GO:0008431 | vitamin E binding(GO:0008431) |
| 0.1 | 0.3 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.1 | 0.2 | GO:0050567 | glutaminyl-tRNA synthase (glutamine-hydrolyzing) activity(GO:0050567) |
| 0.0 | 0.2 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.1 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.0 | 0.3 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.0 | 0.2 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.1 | GO:0008124 | 4-alpha-hydroxytetrahydrobiopterin dehydratase activity(GO:0008124) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0061505 | DNA topoisomerase type II (ATP-hydrolyzing) activity(GO:0003918) DNA topoisomerase II activity(GO:0061505) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.1 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.2 | GO:0015180 | L-alanine transmembrane transporter activity(GO:0015180) L-cystine transmembrane transporter activity(GO:0015184) alanine transmembrane transporter activity(GO:0022858) |
| 0.0 | 0.1 | GO:0008332 | low voltage-gated calcium channel activity(GO:0008332) |
| 0.0 | 0.5 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.4 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.2 | GO:0035174 | histone serine kinase activity(GO:0035174) |
| 0.0 | 0.2 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.3 | GO:0005114 | type II transforming growth factor beta receptor binding(GO:0005114) |
| 0.0 | 0.2 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.4 | GO:0008140 | cAMP response element binding protein binding(GO:0008140) |
| 0.0 | 0.2 | GO:0070290 | N-acylphosphatidylethanolamine-specific phospholipase D activity(GO:0070290) |
| 0.0 | 0.1 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0016509 | long-chain-3-hydroxyacyl-CoA dehydrogenase activity(GO:0016509) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.1 | GO:0015093 | ferrous iron transmembrane transporter activity(GO:0015093) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.4 | GO:0008569 | ATP-dependent microtubule motor activity, minus-end-directed(GO:0008569) |
| 0.0 | 0.1 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.1 | GO:0034602 | oxoglutarate dehydrogenase (NAD+) activity(GO:0034602) |
| 0.0 | 0.2 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0035312 | 5'-3' exodeoxyribonuclease activity(GO:0035312) |
| 0.0 | 0.4 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.1 | GO:0090554 | phosphatidylcholine-translocating ATPase activity(GO:0090554) |
| 0.0 | 0.3 | GO:0019789 | SUMO transferase activity(GO:0019789) |
| 0.0 | 0.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.2 | GO:0042809 | vitamin D receptor binding(GO:0042809) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0070095 | fructose-6-phosphate binding(GO:0070095) |
| 0.0 | 0.1 | GO:0016812 | hydrolase activity, acting on carbon-nitrogen (but not peptide) bonds, in cyclic amides(GO:0016812) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 1.0 | PID AURORA B PATHWAY | Aurora B signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 1.4 | REACTOME MITOTIC PROMETAPHASE | Genes involved in Mitotic Prometaphase |
| 0.0 | 0.2 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 0.4 | REACTOME SPHINGOLIPID DE NOVO BIOSYNTHESIS | Genes involved in Sphingolipid de novo biosynthesis |
| 0.0 | 0.4 | REACTOME INHIBITION OF VOLTAGE GATED CA2 CHANNELS VIA GBETA GAMMA SUBUNITS | Genes involved in Inhibition of voltage gated Ca2+ channels via Gbeta/gamma subunits |