Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for IRF6_IRF4_IRF5
Z-value: 0.64
Transcription factors associated with IRF6_IRF4_IRF5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
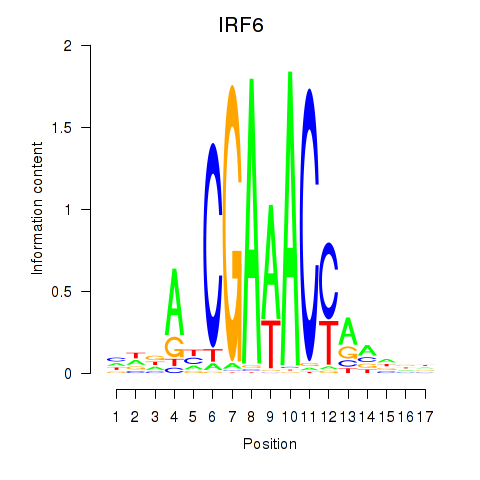
IRF6
|
ENSG00000117595.6 | IRF6 |
|
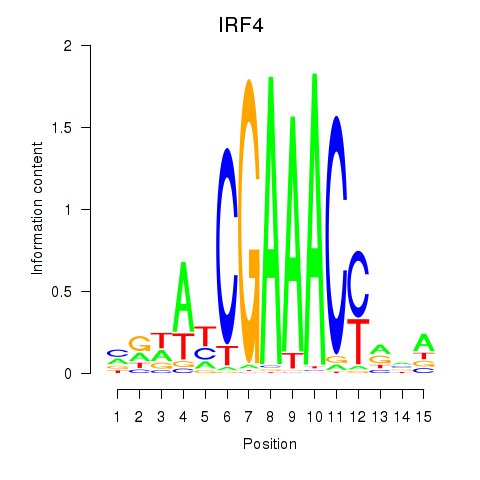
IRF4
|
ENSG00000137265.10 | IRF4 |
|
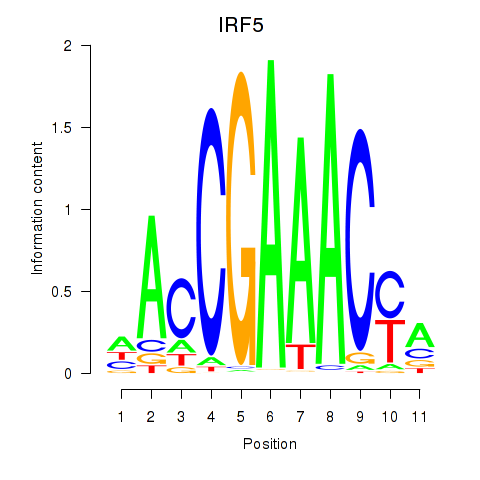
IRF5
|
ENSG00000128604.14 | IRF5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRF6 | hg19_v2_chr1_-_209979465_209979494 | 0.97 | 9.1e-05 | Click! |
| IRF5 | hg19_v2_chr7_+_128577972_128578047 | 0.93 | 8.5e-04 | Click! |
| IRF4 | hg19_v2_chr6_+_391739_391759 | -0.29 | 4.8e-01 | Click! |
Activity profile of IRF6_IRF4_IRF5 motif
Sorted Z-values of IRF6_IRF4_IRF5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRF6_IRF4_IRF5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr11_-_118134997 | 2.35 |
ENST00000278937.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr11_-_118135160 | 1.89 |
ENST00000438295.2 |
MPZL2 |
myelin protein zero-like 2 |
| chr8_+_8559406 | 0.85 |
ENST00000519106.1 |
CLDN23 |
claudin 23 |
| chr12_+_113344755 | 0.80 |
ENST00000550883.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr10_-_10836919 | 0.74 |
ENST00000602763.1 ENST00000415590.2 ENST00000434919.2 |
SFTA1P |
surfactant associated 1, pseudogene |
| chr15_+_41136586 | 0.73 |
ENST00000431806.1 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr11_+_32851487 | 0.70 |
ENST00000257836.3 |
PRRG4 |
proline rich Gla (G-carboxyglutamic acid) 4 (transmembrane) |
| chr12_+_113344811 | 0.68 |
ENST00000551241.1 ENST00000553185.1 ENST00000550689.1 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr17_-_39677971 | 0.66 |
ENST00000393976.2 |
KRT15 |
keratin 15 |
| chr12_+_113376157 | 0.62 |
ENST00000228928.7 |
OAS3 |
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr10_-_10836865 | 0.59 |
ENST00000446372.2 |
SFTA1P |
surfactant associated 1, pseudogene |
| chr12_+_113344582 | 0.59 |
ENST00000202917.5 ENST00000445409.2 ENST00000452357.2 |
OAS1 |
2'-5'-oligoadenylate synthetase 1, 40/46kDa |
| chr1_+_86889769 | 0.50 |
ENST00000370565.4 |
CLCA2 |
chloride channel accessory 2 |
| chr2_-_70780572 | 0.48 |
ENST00000450929.1 |
TGFA |
transforming growth factor, alpha |
| chr9_-_32526184 | 0.46 |
ENST00000545044.1 ENST00000379868.1 |
DDX58 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr11_+_130029457 | 0.46 |
ENST00000278742.5 |
ST14 |
suppression of tumorigenicity 14 (colon carcinoma) |
| chr5_+_68788594 | 0.44 |
ENST00000396442.2 ENST00000380766.2 |
OCLN |
occludin |
| chr17_-_7164410 | 0.43 |
ENST00000574070.1 |
CLDN7 |
claudin 7 |
| chr10_+_134210672 | 0.40 |
ENST00000305233.5 ENST00000368609.4 |
PWWP2B |
PWWP domain containing 2B |
| chr1_-_59043166 | 0.39 |
ENST00000371225.2 |
TACSTD2 |
tumor-associated calcium signal transducer 2 |
| chr9_-_32526299 | 0.38 |
ENST00000379882.1 ENST00000379883.2 |
DDX58 |
DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 |
| chr21_+_42733870 | 0.38 |
ENST00000330714.3 ENST00000436410.1 ENST00000435611.1 |
MX2 |
myxovirus (influenza virus) resistance 2 (mouse) |
| chr7_-_20256965 | 0.37 |
ENST00000400331.5 ENST00000332878.4 |
MACC1 |
metastasis associated in colon cancer 1 |
| chr2_+_163175394 | 0.36 |
ENST00000446271.1 ENST00000429691.2 |
GCA |
grancalcin, EF-hand calcium binding protein |
| chr4_-_89080003 | 0.36 |
ENST00000237612.3 |
ABCG2 |
ATP-binding cassette, sub-family G (WHITE), member 2 |
| chr1_-_160990886 | 0.36 |
ENST00000537746.1 |
F11R |
F11 receptor |
| chr1_-_159915386 | 0.36 |
ENST00000361509.3 ENST00000368094.1 |
IGSF9 |
immunoglobulin superfamily, member 9 |
| chr15_-_45670924 | 0.34 |
ENST00000396659.3 |
GATM |
glycine amidinotransferase (L-arginine:glycine amidinotransferase) |
| chr2_+_120770686 | 0.33 |
ENST00000331393.4 ENST00000443124.1 |
EPB41L5 |
erythrocyte membrane protein band 4.1 like 5 |
| chr21_+_42792442 | 0.33 |
ENST00000398600.2 |
MX1 |
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr2_+_68961934 | 0.31 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr2_+_68961905 | 0.31 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr4_+_89378261 | 0.30 |
ENST00000264350.3 |
HERC5 |
HECT and RLD domain containing E3 ubiquitin protein ligase 5 |
| chr4_+_144257915 | 0.29 |
ENST00000262995.4 |
GAB1 |
GRB2-associated binding protein 1 |
| chr4_-_100575781 | 0.28 |
ENST00000511828.1 |
RP11-766F14.2 |
Protein LOC285556 |
| chr16_+_30751953 | 0.27 |
ENST00000483578.1 |
RP11-2C24.4 |
RP11-2C24.4 |
| chr11_-_104817919 | 0.26 |
ENST00000533252.1 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr6_+_43738444 | 0.25 |
ENST00000324450.6 ENST00000417285.2 ENST00000413642.3 ENST00000372055.4 ENST00000482630.2 ENST00000425836.2 ENST00000372064.4 ENST00000372077.4 ENST00000519767.1 |
VEGFA |
vascular endothelial growth factor A |
| chr11_+_128563652 | 0.25 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr7_+_30323923 | 0.23 |
ENST00000323037.4 |
ZNRF2 |
zinc and ring finger 2 |
| chr11_+_65554493 | 0.23 |
ENST00000335987.3 |
OVOL1 |
ovo-like zinc finger 1 |
| chr6_+_106546808 | 0.22 |
ENST00000369089.3 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr10_+_71561649 | 0.22 |
ENST00000398978.3 ENST00000354547.3 ENST00000357811.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr3_-_182880541 | 0.22 |
ENST00000470251.1 ENST00000265598.3 |
LAMP3 |
lysosomal-associated membrane protein 3 |
| chr12_+_46123448 | 0.22 |
ENST00000334344.6 |
ARID2 |
AT rich interactive domain 2 (ARID, RFX-like) |
| chr14_-_102552659 | 0.22 |
ENST00000441629.2 |
HSP90AA1 |
heat shock protein 90kDa alpha (cytosolic), class A member 1 |
| chr21_-_34914394 | 0.22 |
ENST00000361093.5 ENST00000381815.4 |
GART |
phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| chr21_+_42798094 | 0.21 |
ENST00000398598.3 ENST00000455164.2 ENST00000424365.1 |
MX1 |
myxovirus (influenza virus) resistance 1, interferon-inducible protein p78 (mouse) |
| chr10_+_71561630 | 0.21 |
ENST00000398974.3 ENST00000398971.3 ENST00000398968.3 ENST00000398966.3 ENST00000398964.3 ENST00000398969.3 ENST00000356340.3 ENST00000398972.3 ENST00000398973.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr2_-_163175133 | 0.19 |
ENST00000421365.2 ENST00000263642.2 |
IFIH1 |
interferon induced with helicase C domain 1 |
| chr1_+_179262905 | 0.19 |
ENST00000539888.1 ENST00000540564.1 ENST00000535686.1 ENST00000367619.3 |
SOAT1 |
sterol O-acyltransferase 1 |
| chr13_-_46961580 | 0.18 |
ENST00000378787.3 ENST00000378797.2 ENST00000429979.1 ENST00000378781.3 |
KIAA0226L |
KIAA0226-like |
| chr13_-_76056250 | 0.17 |
ENST00000377636.3 ENST00000431480.2 ENST00000377625.2 ENST00000425511.1 |
TBC1D4 |
TBC1 domain family, member 4 |
| chr2_+_68962014 | 0.17 |
ENST00000467265.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr5_-_35230434 | 0.17 |
ENST00000504500.1 |
PRLR |
prolactin receptor |
| chr6_+_26087509 | 0.15 |
ENST00000397022.3 ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE |
hemochromatosis |
| chr2_+_113299990 | 0.15 |
ENST00000537335.1 ENST00000417433.2 |
POLR1B |
polymerase (RNA) I polypeptide B, 128kDa |
| chr20_+_20033158 | 0.15 |
ENST00000340348.6 ENST00000377309.2 ENST00000389656.3 ENST00000377306.1 ENST00000245957.5 ENST00000377303.2 ENST00000475466.1 |
C20orf26 |
chromosome 20 open reading frame 26 |
| chr4_+_144258021 | 0.14 |
ENST00000262994.4 |
GAB1 |
GRB2-associated binding protein 1 |
| chr2_-_220252603 | 0.14 |
ENST00000322176.7 ENST00000273075.4 |
DNPEP |
aspartyl aminopeptidase |
| chrX_-_110655391 | 0.14 |
ENST00000356915.2 ENST00000356220.3 |
DCX |
doublecortin |
| chrX_-_30877837 | 0.14 |
ENST00000378930.3 |
TAB3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr2_-_55920952 | 0.13 |
ENST00000447944.2 |
PNPT1 |
polyribonucleotide nucleotidyltransferase 1 |
| chr12_-_31477072 | 0.13 |
ENST00000454658.2 |
FAM60A |
family with sequence similarity 60, member A |
| chr12_+_113376249 | 0.13 |
ENST00000551007.1 ENST00000548514.1 |
OAS3 |
2'-5'-oligoadenylate synthetase 3, 100kDa |
| chr16_-_74734672 | 0.13 |
ENST00000306247.7 ENST00000575686.1 |
MLKL |
mixed lineage kinase domain-like |
| chr1_-_153514241 | 0.13 |
ENST00000368718.1 ENST00000359215.1 |
S100A5 |
S100 calcium binding protein A5 |
| chr2_-_231989808 | 0.13 |
ENST00000258400.3 |
HTR2B |
5-hydroxytryptamine (serotonin) receptor 2B, G protein-coupled |
| chr2_+_7017796 | 0.13 |
ENST00000382040.3 |
RSAD2 |
radical S-adenosyl methionine domain containing 2 |
| chr6_-_137365402 | 0.12 |
ENST00000541547.1 |
IL20RA |
interleukin 20 receptor, alpha |
| chr15_-_26108355 | 0.12 |
ENST00000356865.6 |
ATP10A |
ATPase, class V, type 10A |
| chr6_+_89855765 | 0.12 |
ENST00000275072.4 |
PM20D2 |
peptidase M20 domain containing 2 |
| chr1_-_109940550 | 0.12 |
ENST00000256637.6 |
SORT1 |
sortilin 1 |
| chr6_+_26087646 | 0.12 |
ENST00000309234.6 |
HFE |
hemochromatosis |
| chr4_-_11431188 | 0.12 |
ENST00000510712.1 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr19_+_41222998 | 0.11 |
ENST00000263370.2 |
ITPKC |
inositol-trisphosphate 3-kinase C |
| chr14_+_39736582 | 0.11 |
ENST00000556148.1 ENST00000348007.3 |
CTAGE5 |
CTAGE family, member 5 |
| chr7_-_77045617 | 0.11 |
ENST00000257626.7 |
GSAP |
gamma-secretase activating protein |
| chr5_-_176057518 | 0.11 |
ENST00000393693.2 |
SNCB |
synuclein, beta |
| chr3_-_179169181 | 0.11 |
ENST00000497513.1 |
GNB4 |
guanine nucleotide binding protein (G protein), beta polypeptide 4 |
| chr3_+_118892362 | 0.11 |
ENST00000497685.1 ENST00000264234.3 |
UPK1B |
uroplakin 1B |
| chr8_-_22926526 | 0.11 |
ENST00000347739.3 ENST00000542226.1 |
TNFRSF10B |
tumor necrosis factor receptor superfamily, member 10b |
| chr12_-_50616382 | 0.10 |
ENST00000552783.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr10_+_71561704 | 0.10 |
ENST00000520267.1 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr2_-_75938115 | 0.10 |
ENST00000321027.3 |
GCFC2 |
GC-rich sequence DNA-binding factor 2 |
| chr15_-_30114231 | 0.10 |
ENST00000356107.6 ENST00000545208.2 |
TJP1 |
tight junction protein 1 |
| chr11_+_100862811 | 0.10 |
ENST00000303130.2 |
TMEM133 |
transmembrane protein 133 |
| chr2_-_170219079 | 0.09 |
ENST00000263816.3 |
LRP2 |
low density lipoprotein receptor-related protein 2 |
| chr20_-_25038804 | 0.09 |
ENST00000323482.4 |
ACSS1 |
acyl-CoA synthetase short-chain family member 1 |
| chr11_+_63655987 | 0.09 |
ENST00000509502.2 ENST00000512060.1 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr8_-_22926623 | 0.09 |
ENST00000276431.4 |
TNFRSF10B |
tumor necrosis factor receptor superfamily, member 10b |
| chr5_-_35938674 | 0.09 |
ENST00000397366.1 ENST00000513623.1 ENST00000514524.1 ENST00000397367.2 |
CAPSL |
calcyphosine-like |
| chr3_+_127634069 | 0.09 |
ENST00000405109.1 |
KBTBD12 |
kelch repeat and BTB (POZ) domain containing 12 |
| chr19_+_7600584 | 0.09 |
ENST00000600737.1 |
PNPLA6 |
patatin-like phospholipase domain containing 6 |
| chr6_-_27799305 | 0.09 |
ENST00000357549.2 |
HIST1H4K |
histone cluster 1, H4k |
| chr15_+_74287009 | 0.09 |
ENST00000395135.3 |
PML |
promyelocytic leukemia |
| chr9_-_139640458 | 0.09 |
ENST00000476567.1 |
LCN6 |
lipocalin 6 |
| chr4_+_147096837 | 0.09 |
ENST00000296581.5 ENST00000502781.1 |
LSM6 |
LSM6 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr18_+_42260861 | 0.09 |
ENST00000282030.5 |
SETBP1 |
SET binding protein 1 |
| chr2_-_111435610 | 0.09 |
ENST00000447014.1 ENST00000420328.1 ENST00000535254.1 ENST00000409311.1 ENST00000302759.6 |
BUB1 |
BUB1 mitotic checkpoint serine/threonine kinase |
| chr5_-_95297534 | 0.08 |
ENST00000513343.1 ENST00000431061.2 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chrX_-_54824673 | 0.08 |
ENST00000218436.6 |
ITIH6 |
inter-alpha-trypsin inhibitor heavy chain family, member 6 |
| chr4_+_57371509 | 0.08 |
ENST00000360096.2 |
ARL9 |
ADP-ribosylation factor-like 9 |
| chr15_+_90544532 | 0.08 |
ENST00000268154.4 |
ZNF710 |
zinc finger protein 710 |
| chr4_-_88141755 | 0.08 |
ENST00000273963.5 |
KLHL8 |
kelch-like family member 8 |
| chr16_+_12058961 | 0.08 |
ENST00000053243.1 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr15_+_28624878 | 0.08 |
ENST00000450328.2 |
GOLGA8F |
golgin A8 family, member F |
| chr9_-_33264676 | 0.08 |
ENST00000472232.3 ENST00000379704.2 |
BAG1 |
BCL2-associated athanogene |
| chr7_+_141478242 | 0.08 |
ENST00000247881.2 |
TAS2R4 |
taste receptor, type 2, member 4 |
| chr1_-_54355430 | 0.08 |
ENST00000371399.1 ENST00000072644.1 ENST00000412288.1 |
YIPF1 |
Yip1 domain family, member 1 |
| chr9_+_95820966 | 0.08 |
ENST00000375472.3 ENST00000465709.1 |
SUSD3 |
sushi domain containing 3 |
| chr10_+_15139394 | 0.08 |
ENST00000441850.1 |
RPP38 |
ribonuclease P/MRP 38kDa subunit |
| chr11_-_85522172 | 0.08 |
ENST00000316356.4 ENST00000389960.4 ENST00000527523.1 |
SYTL2 |
synaptotagmin-like 2 |
| chr3_+_45429998 | 0.08 |
ENST00000265537.3 ENST00000415258.1 ENST00000431023.1 ENST00000414984.1 |
LARS2 |
leucyl-tRNA synthetase 2, mitochondrial |
| chr22_+_36044411 | 0.08 |
ENST00000409652.4 |
APOL6 |
apolipoprotein L, 6 |
| chr12_-_71148413 | 0.08 |
ENST00000440835.2 ENST00000549308.1 ENST00000550661.1 |
PTPRR |
protein tyrosine phosphatase, receptor type, R |
| chr6_-_167797887 | 0.07 |
ENST00000476779.2 ENST00000460930.2 ENST00000397829.4 ENST00000366827.2 |
TCP10 |
t-complex 10 |
| chr9_-_103115185 | 0.07 |
ENST00000374902.4 |
TEX10 |
testis expressed 10 |
| chr4_-_75719896 | 0.07 |
ENST00000395743.3 |
BTC |
betacellulin |
| chr12_+_46123682 | 0.07 |
ENST00000422737.1 |
ARID2 |
AT rich interactive domain 2 (ARID, RFX-like) |
| chr7_+_37960163 | 0.07 |
ENST00000199448.4 ENST00000559325.1 ENST00000423717.1 |
EPDR1 |
ependymin related 1 |
| chr10_+_123923205 | 0.07 |
ENST00000369004.3 ENST00000260733.3 |
TACC2 |
transforming, acidic coiled-coil containing protein 2 |
| chr14_+_94577074 | 0.07 |
ENST00000444961.1 ENST00000448882.1 ENST00000557098.1 ENST00000554800.1 ENST00000556544.1 ENST00000298902.5 ENST00000555819.1 ENST00000557634.1 ENST00000555744.1 |
IFI27 |
interferon, alpha-inducible protein 27 |
| chr1_-_44497024 | 0.07 |
ENST00000372306.3 ENST00000372310.3 ENST00000475075.2 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr4_+_144303093 | 0.07 |
ENST00000505913.1 |
GAB1 |
GRB2-associated binding protein 1 |
| chr18_-_33647487 | 0.07 |
ENST00000590898.1 ENST00000357384.4 ENST00000319040.6 ENST00000588737.1 ENST00000399022.4 |
RPRD1A |
regulation of nuclear pre-mRNA domain containing 1A |
| chr19_-_41222775 | 0.07 |
ENST00000324464.3 ENST00000450541.1 ENST00000594720.1 |
ADCK4 |
aarF domain containing kinase 4 |
| chr16_+_12059050 | 0.07 |
ENST00000396495.3 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr17_-_46799872 | 0.07 |
ENST00000290294.3 |
PRAC1 |
prostate cancer susceptibility candidate 1 |
| chr18_-_812517 | 0.07 |
ENST00000584307.1 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chrM_+_9207 | 0.07 |
ENST00000362079.2 |
MT-CO3 |
mitochondrially encoded cytochrome c oxidase III |
| chr17_+_7387677 | 0.07 |
ENST00000322644.6 |
POLR2A |
polymerase (RNA) II (DNA directed) polypeptide A, 220kDa |
| chr7_-_33080506 | 0.06 |
ENST00000381626.2 ENST00000409467.1 ENST00000449201.1 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
| chr18_-_812231 | 0.06 |
ENST00000314574.4 |
YES1 |
v-yes-1 Yamaguchi sarcoma viral oncogene homolog 1 |
| chr1_-_44497118 | 0.06 |
ENST00000537678.1 ENST00000466926.1 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr17_-_79885576 | 0.06 |
ENST00000574686.1 ENST00000357736.4 |
MAFG |
v-maf avian musculoaponeurotic fibrosarcoma oncogene homolog G |
| chr17_+_79373540 | 0.06 |
ENST00000307745.7 |
RP11-1055B8.7 |
BAH and coiled-coil domain-containing protein 1 |
| chr6_-_100016678 | 0.06 |
ENST00000523799.1 ENST00000520429.1 |
CCNC |
cyclin C |
| chr15_+_43425672 | 0.06 |
ENST00000260403.2 |
TMEM62 |
transmembrane protein 62 |
| chr6_-_82462425 | 0.06 |
ENST00000369754.3 ENST00000320172.6 ENST00000369756.3 |
FAM46A |
family with sequence similarity 46, member A |
| chr8_+_40018977 | 0.06 |
ENST00000520487.1 |
RP11-470M17.2 |
RP11-470M17.2 |
| chr17_+_18380051 | 0.06 |
ENST00000581545.1 ENST00000582333.1 ENST00000328114.6 ENST00000412421.2 ENST00000583322.1 ENST00000584941.1 |
LGALS9C |
lectin, galactoside-binding, soluble, 9C |
| chr3_+_159706537 | 0.06 |
ENST00000305579.2 ENST00000480787.1 ENST00000466512.1 |
IL12A |
interleukin 12A (natural killer cell stimulatory factor 1, cytotoxic lymphocyte maturation factor 1, p35) |
| chr2_+_85811525 | 0.06 |
ENST00000306384.4 |
VAMP5 |
vesicle-associated membrane protein 5 |
| chr9_-_133814455 | 0.06 |
ENST00000448616.1 |
FIBCD1 |
fibrinogen C domain containing 1 |
| chr9_-_77643307 | 0.06 |
ENST00000376834.3 ENST00000376830.3 |
C9orf41 |
chromosome 9 open reading frame 41 |
| chr4_-_48782259 | 0.06 |
ENST00000507711.1 ENST00000358350.4 ENST00000537810.1 ENST00000264319.7 |
FRYL |
FRY-like |
| chr10_+_71562180 | 0.06 |
ENST00000517713.1 ENST00000522165.1 ENST00000520133.1 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr15_+_74287118 | 0.06 |
ENST00000563500.1 |
PML |
promyelocytic leukemia |
| chr2_-_160473114 | 0.06 |
ENST00000392783.2 |
BAZ2B |
bromodomain adjacent to zinc finger domain, 2B |
| chr6_+_139349817 | 0.06 |
ENST00000367660.3 |
ABRACL |
ABRA C-terminal like |
| chr16_-_28518153 | 0.06 |
ENST00000356897.1 |
IL27 |
interleukin 27 |
| chr11_-_47736896 | 0.05 |
ENST00000525123.1 ENST00000528244.1 ENST00000532595.1 ENST00000529154.1 ENST00000530969.1 |
AGBL2 |
ATP/GTP binding protein-like 2 |
| chr2_-_106810742 | 0.05 |
ENST00000409501.3 ENST00000428048.2 ENST00000441952.1 ENST00000457835.1 ENST00000540130.1 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
| chrX_-_45710920 | 0.05 |
ENST00000456532.1 |
RP5-1158E12.3 |
RP5-1158E12.3 |
| chr2_-_106810783 | 0.05 |
ENST00000283148.7 |
UXS1 |
UDP-glucuronate decarboxylase 1 |
| chr5_-_61699698 | 0.05 |
ENST00000506390.1 ENST00000199320.4 |
DIMT1 |
DIM1 dimethyladenosine transferase 1 homolog (S. cerevisiae) |
| chr15_-_30113676 | 0.05 |
ENST00000400011.2 |
TJP1 |
tight junction protein 1 |
| chr5_-_95297678 | 0.05 |
ENST00000237853.4 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr7_-_150779995 | 0.05 |
ENST00000462940.1 ENST00000492838.1 ENST00000392818.3 ENST00000488752.1 ENST00000476627.1 |
TMUB1 |
transmembrane and ubiquitin-like domain containing 1 |
| chr6_-_94129244 | 0.05 |
ENST00000369303.4 ENST00000369297.1 |
EPHA7 |
EPH receptor A7 |
| chr11_+_72983246 | 0.05 |
ENST00000393590.2 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr18_+_3466248 | 0.05 |
ENST00000581029.1 ENST00000581442.1 ENST00000579007.1 |
RP11-838N2.4 |
RP11-838N2.4 |
| chrX_+_65382433 | 0.05 |
ENST00000374727.3 |
HEPH |
hephaestin |
| chr2_+_32390925 | 0.05 |
ENST00000440718.1 ENST00000379343.2 ENST00000282587.5 ENST00000435660.1 ENST00000538303.1 ENST00000357055.3 ENST00000406369.1 |
SLC30A6 |
solute carrier family 30 (zinc transporter), member 6 |
| chr2_-_112642267 | 0.05 |
ENST00000341068.3 |
ANAPC1 |
anaphase promoting complex subunit 1 |
| chr2_-_74753332 | 0.05 |
ENST00000451518.1 ENST00000404568.3 |
DQX1 |
DEAQ box RNA-dependent ATPase 1 |
| chr12_-_99548645 | 0.05 |
ENST00000549025.2 |
ANKS1B |
ankyrin repeat and sterile alpha motif domain containing 1B |
| chr9_-_90589402 | 0.05 |
ENST00000375871.4 ENST00000605159.1 ENST00000336654.5 |
CDK20 |
cyclin-dependent kinase 20 |
| chr1_+_179050512 | 0.05 |
ENST00000367627.3 |
TOR3A |
torsin family 3, member A |
| chr15_+_83478370 | 0.05 |
ENST00000286760.4 |
WHAMM |
WAS protein homolog associated with actin, golgi membranes and microtubules |
| chr12_-_104350898 | 0.05 |
ENST00000552940.1 ENST00000547975.1 ENST00000549478.1 ENST00000546540.1 ENST00000546819.1 ENST00000378090.4 ENST00000547945.1 |
C12orf73 |
chromosome 12 open reading frame 73 |
| chr2_-_74753305 | 0.05 |
ENST00000393951.2 |
DQX1 |
DEAQ box RNA-dependent ATPase 1 |
| chr16_-_88851618 | 0.05 |
ENST00000301015.9 |
PIEZO1 |
piezo-type mechanosensitive ion channel component 1 |
| chr11_-_19082216 | 0.04 |
ENST00000329773.2 |
MRGPRX2 |
MAS-related GPR, member X2 |
| chr4_-_88141615 | 0.04 |
ENST00000545252.1 ENST00000425278.2 ENST00000498875.2 |
KLHL8 |
kelch-like family member 8 |
| chr1_+_179051160 | 0.04 |
ENST00000367625.4 ENST00000352445.6 |
TOR3A |
torsin family 3, member A |
| chr10_+_15139330 | 0.04 |
ENST00000378202.5 ENST00000378197.4 |
RPP38 |
ribonuclease P/MRP 38kDa subunit |
| chr13_-_100624012 | 0.04 |
ENST00000267294.4 |
ZIC5 |
Zic family member 5 |
| chr2_-_7005785 | 0.04 |
ENST00000256722.5 ENST00000404168.1 ENST00000458098.1 |
CMPK2 |
cytidine monophosphate (UMP-CMP) kinase 2, mitochondrial |
| chr6_-_88411911 | 0.04 |
ENST00000257787.5 |
AKIRIN2 |
akirin 2 |
| chr3_-_46037299 | 0.04 |
ENST00000296137.2 |
FYCO1 |
FYVE and coiled-coil domain containing 1 |
| chr2_+_124782857 | 0.04 |
ENST00000431078.1 |
CNTNAP5 |
contactin associated protein-like 5 |
| chr12_+_62654119 | 0.04 |
ENST00000353364.3 ENST00000549523.1 ENST00000280377.5 |
USP15 |
ubiquitin specific peptidase 15 |
| chr17_+_4643337 | 0.04 |
ENST00000592813.1 |
ZMYND15 |
zinc finger, MYND-type containing 15 |
| chr9_+_33265011 | 0.04 |
ENST00000419016.2 |
CHMP5 |
charged multivesicular body protein 5 |
| chr1_-_168464875 | 0.04 |
ENST00000422253.1 |
RP5-968D22.3 |
RP5-968D22.3 |
| chr13_+_98794810 | 0.04 |
ENST00000595437.1 |
FARP1 |
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived) |
| chr19_+_42300548 | 0.04 |
ENST00000344550.4 |
CEACAM3 |
carcinoembryonic antigen-related cell adhesion molecule 3 |
| chr3_-_151176497 | 0.04 |
ENST00000282466.3 |
IGSF10 |
immunoglobulin superfamily, member 10 |
| chr1_-_53018654 | 0.04 |
ENST00000257177.4 ENST00000355809.4 ENST00000528642.1 ENST00000470626.1 ENST00000371544.3 |
ZCCHC11 |
zinc finger, CCHC domain containing 11 |
| chr2_-_75937994 | 0.04 |
ENST00000409857.3 ENST00000470503.1 ENST00000541687.1 ENST00000442309.1 |
GCFC2 |
GC-rich sequence DNA-binding factor 2 |
| chr10_+_121410882 | 0.04 |
ENST00000369085.3 |
BAG3 |
BCL2-associated athanogene 3 |
| chr12_+_70133152 | 0.04 |
ENST00000550536.1 ENST00000362025.5 |
RAB3IP |
RAB3A interacting protein |
| chr1_+_110993795 | 0.04 |
ENST00000271331.3 |
PROK1 |
prokineticin 1 |
| chr19_+_18942761 | 0.04 |
ENST00000599848.1 |
UPF1 |
UPF1 regulator of nonsense transcripts homolog (yeast) |
| chr15_-_65117807 | 0.04 |
ENST00000559239.1 ENST00000268043.4 ENST00000333425.6 |
PIF1 |
PIF1 5'-to-3' DNA helicase |
| chr2_-_197664366 | 0.03 |
ENST00000409364.3 ENST00000263956.3 |
GTF3C3 |
general transcription factor IIIC, polypeptide 3, 102kDa |
| chr19_+_36705504 | 0.03 |
ENST00000456324.1 |
ZNF146 |
zinc finger protein 146 |
| chr12_+_4758264 | 0.03 |
ENST00000266544.5 |
NDUFA9 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 9, 39kDa |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0034343 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.1 | 0.1 | GO:1902003 | regulation of beta-amyloid formation(GO:1902003) |
| 0.1 | 0.5 | GO:0060672 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.1 | 0.3 | GO:0002590 | regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002589) negative regulation of antigen processing and presentation of peptide antigen via MHC class I(GO:0002590) regulation of T cell antigen processing and presentation(GO:0002625) positive regulation of iron ion transport(GO:0034758) positive regulation of iron ion transmembrane transport(GO:0034761) regulation of iron ion import(GO:1900390) regulation of ferrous iron import into cell(GO:1903989) positive regulation of ferrous iron import into cell(GO:1903991) regulation of ferrous iron binding(GO:1904432) positive regulation of ferrous iron binding(GO:1904434) regulation of transferrin receptor binding(GO:1904435) positive regulation of transferrin receptor binding(GO:1904437) regulation of ferrous iron import across plasma membrane(GO:1904438) positive regulation of ferrous iron import across plasma membrane(GO:1904440) response to iron ion starvation(GO:1990641) |
| 0.1 | 0.3 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.1 | 0.9 | GO:0003373 | dynamin polymerization involved in membrane fission(GO:0003373) dynamin polymerization involved in mitochondrial fission(GO:0003374) |
| 0.1 | 0.2 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.1 | 0.8 | GO:0060700 | regulation of ribonuclease activity(GO:0060700) |
| 0.1 | 0.2 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.1 | GO:1901350 | cell-cell signaling involved in cell-cell junction organization(GO:1901350) |
| 0.0 | 0.7 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.4 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.0 | 0.2 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.2 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 1.2 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.1 | GO:0002857 | positive regulation of natural killer cell mediated immune response to tumor cell(GO:0002857) positive regulation of natural killer cell mediated cytotoxicity directed against tumor cell target(GO:0002860) |
| 0.0 | 0.4 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.2 | GO:0031339 | negative regulation of vesicle fusion(GO:0031339) |
| 0.0 | 0.5 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.2 | GO:0038161 | prolactin signaling pathway(GO:0038161) |
| 0.0 | 0.3 | GO:1903265 | positive regulation of tumor necrosis factor-mediated signaling pathway(GO:1903265) |
| 0.0 | 0.4 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.4 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.1 | GO:0010513 | positive regulation of phosphatidylinositol biosynthetic process(GO:0010513) |
| 0.0 | 0.1 | GO:0034165 | regulation of toll-like receptor 9 signaling pathway(GO:0034163) positive regulation of toll-like receptor 9 signaling pathway(GO:0034165) |
| 0.0 | 0.1 | GO:0019541 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.0 | 4.4 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0060982 | coronary artery morphogenesis(GO:0060982) |
| 0.0 | 0.1 | GO:0001172 | transcription, RNA-templated(GO:0001172) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.2 | GO:0006189 | 'de novo' IMP biosynthetic process(GO:0006189) |
| 0.0 | 0.1 | GO:0000294 | nuclear-transcribed mRNA catabolic process, endonucleolytic cleavage-dependent decay(GO:0000294) |
| 0.0 | 0.0 | GO:0032899 | regulation of neurotrophin production(GO:0032899) |
| 0.0 | 0.1 | GO:0030578 | PML body organization(GO:0030578) |
| 0.0 | 0.0 | GO:0009138 | pyrimidine nucleoside diphosphate metabolic process(GO:0009138) |
| 0.0 | 0.1 | GO:0007000 | nucleolus organization(GO:0007000) |
| 0.0 | 0.1 | GO:0051005 | nerve growth factor signaling pathway(GO:0038180) negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.0 | 0.1 | GO:0070940 | dephosphorylation of RNA polymerase II C-terminal domain(GO:0070940) |
| 0.0 | 0.4 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.0 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0035610 | protein side chain deglutamylation(GO:0035610) |
| 0.0 | 0.3 | GO:0035855 | megakaryocyte development(GO:0035855) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:0030936 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.1 | 0.2 | GO:0097232 | lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.3 | GO:1990357 | terminal web(GO:1990357) |
| 0.0 | 0.4 | GO:0036056 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.0 | 0.3 | GO:0097169 | AIM2 inflammasome complex(GO:0097169) |
| 0.0 | 0.1 | GO:0071008 | U2-type post-mRNA release spliceosomal complex(GO:0071008) |
| 0.0 | 0.9 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.0 | 0.4 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.9 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.1 | GO:0044530 | supraspliceosomal complex(GO:0044530) |
| 0.0 | 0.1 | GO:0000942 | condensed nuclear chromosome outer kinetochore(GO:0000942) |
| 0.0 | 0.1 | GO:0005655 | nucleolar ribonuclease P complex(GO:0005655) |
| 0.0 | 0.1 | GO:0042788 | polysomal ribosome(GO:0042788) |
| 0.0 | 1.7 | GO:0005923 | bicellular tight junction(GO:0005923) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.2 | GO:0016882 | cyclo-ligase activity(GO:0016882) |
| 0.0 | 0.3 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.4 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0004925 | prolactin receptor activity(GO:0004925) |
| 0.0 | 0.2 | GO:0045569 | TRAIL binding(GO:0045569) |
| 0.0 | 0.2 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.0 | 0.1 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.1 | GO:0034046 | poly(G) binding(GO:0034046) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.3 | GO:0039706 | co-receptor binding(GO:0039706) |
| 0.0 | 0.2 | GO:0030911 | TPR domain binding(GO:0030911) |
| 0.0 | 0.1 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.1 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.0 | 0.5 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.0 | GO:0019912 | cyclin-dependent protein kinase activating kinase activity(GO:0019912) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.3 | GO:0050700 | CARD domain binding(GO:0050700) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.1 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 0.0 | GO:0017116 | single-stranded DNA-dependent ATP-dependent DNA helicase activity(GO:0017116) DNA/RNA helicase activity(GO:0033677) |
| 0.0 | 0.1 | GO:0043176 | amine binding(GO:0043176) serotonin binding(GO:0051378) |
| 0.0 | 0.3 | GO:0016769 | transferase activity, transferring nitrogenous groups(GO:0016769) |
| 0.0 | 0.0 | GO:0009041 | uridylate kinase activity(GO:0009041) |
| 0.0 | 0.0 | GO:0033754 | indoleamine 2,3-dioxygenase activity(GO:0033754) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.2 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.3 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 2.9 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.6 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.0 | 0.4 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.7 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.7 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 0.1 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.3 | REACTOME VEGF LIGAND RECEPTOR INTERACTIONS | Genes involved in VEGF ligand-receptor interactions |
| 0.0 | 0.2 | REACTOME PURINE RIBONUCLEOSIDE MONOPHOSPHATE BIOSYNTHESIS | Genes involved in Purine ribonucleoside monophosphate biosynthesis |
| 0.0 | 0.1 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |