Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for IRX3
Z-value: 0.83
Transcription factors associated with IRX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX3
|
ENSG00000177508.11 | IRX3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX3 | hg19_v2_chr16_-_54320675_54320715 | 0.85 | 6.8e-03 | Click! |
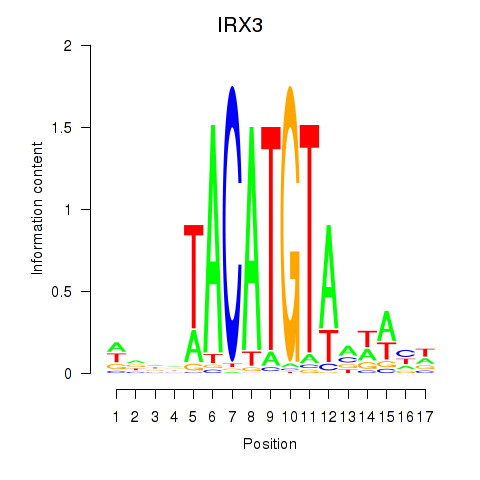
Activity profile of IRX3 motif
Sorted Z-values of IRX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_91574142 | 2.20 |
ENST00000547937.1 |
DCN |
decorin |
| chr12_-_91573132 | 2.01 |
ENST00000550563.1 ENST00000546370.1 |
DCN |
decorin |
| chr12_-_91573316 | 1.92 |
ENST00000393155.1 |
DCN |
decorin |
| chr13_-_38172863 | 1.69 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr12_-_91546926 | 1.68 |
ENST00000550758.1 |
DCN |
decorin |
| chr2_+_189839046 | 1.19 |
ENST00000304636.3 ENST00000317840.5 |
COL3A1 |
collagen, type III, alpha 1 |
| chr3_+_8543393 | 1.13 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr11_-_35547572 | 1.00 |
ENST00000378880.2 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
| chr3_+_157154578 | 0.92 |
ENST00000295927.3 |
PTX3 |
pentraxin 3, long |
| chr1_+_145524891 | 0.81 |
ENST00000369304.3 |
ITGA10 |
integrin, alpha 10 |
| chr5_+_32788945 | 0.71 |
ENST00000326958.1 |
AC026703.1 |
AC026703.1 |
| chr4_-_70626314 | 0.69 |
ENST00000510821.1 |
SULT1B1 |
sulfotransferase family, cytosolic, 1B, member 1 |
| chr19_-_44388116 | 0.68 |
ENST00000587539.1 |
ZNF404 |
zinc finger protein 404 |
| chr5_-_75919253 | 0.67 |
ENST00000296641.4 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr12_+_56114189 | 0.65 |
ENST00000548082.1 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr2_+_233527443 | 0.65 |
ENST00000410095.1 |
EFHD1 |
EF-hand domain family, member D1 |
| chr1_+_97188188 | 0.65 |
ENST00000541987.1 |
PTBP2 |
polypyrimidine tract binding protein 2 |
| chr6_-_117747015 | 0.63 |
ENST00000368508.3 ENST00000368507.3 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
| chr12_+_56114151 | 0.63 |
ENST00000547072.1 ENST00000552930.1 ENST00000257895.5 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr8_-_108510224 | 0.62 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr3_+_8543533 | 0.62 |
ENST00000454244.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr3_-_9994021 | 0.60 |
ENST00000411976.2 ENST00000412055.1 |
PRRT3 |
proline-rich transmembrane protein 3 |
| chr13_+_76362974 | 0.60 |
ENST00000497947.2 |
LMO7 |
LIM domain 7 |
| chr12_-_58329819 | 0.59 |
ENST00000551421.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chr1_-_85870177 | 0.56 |
ENST00000542148.1 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr20_-_18477862 | 0.56 |
ENST00000337227.4 |
RBBP9 |
retinoblastoma binding protein 9 |
| chr1_+_155829286 | 0.55 |
ENST00000368324.4 |
SYT11 |
synaptotagmin XI |
| chr12_-_10605929 | 0.54 |
ENST00000347831.5 ENST00000359151.3 |
KLRC1 |
killer cell lectin-like receptor subfamily C, member 1 |
| chr3_+_12392971 | 0.53 |
ENST00000287820.6 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr9_+_90112767 | 0.52 |
ENST00000408954.3 |
DAPK1 |
death-associated protein kinase 1 |
| chr10_+_60272814 | 0.51 |
ENST00000373886.3 |
BICC1 |
bicaudal C homolog 1 (Drosophila) |
| chr9_+_99690592 | 0.51 |
ENST00000354649.3 |
NUTM2G |
NUT family member 2G |
| chr4_-_70725856 | 0.50 |
ENST00000226444.3 |
SULT1E1 |
sulfotransferase family 1E, estrogen-preferring, member 1 |
| chr2_-_190044480 | 0.50 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr1_+_171060018 | 0.48 |
ENST00000367755.4 ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3 |
flavin containing monooxygenase 3 |
| chr9_-_95244781 | 0.48 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr3_+_8543561 | 0.48 |
ENST00000397386.3 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr4_+_70894130 | 0.47 |
ENST00000526767.1 ENST00000530128.1 ENST00000381057.3 |
HTN3 |
histatin 3 |
| chr12_+_4671352 | 0.47 |
ENST00000542744.1 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr3_+_69928256 | 0.46 |
ENST00000394355.2 |
MITF |
microphthalmia-associated transcription factor |
| chr5_-_75919217 | 0.44 |
ENST00000504899.1 |
F2RL2 |
coagulation factor II (thrombin) receptor-like 2 |
| chr7_-_92777606 | 0.44 |
ENST00000437805.1 ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L |
sterile alpha motif domain containing 9-like |
| chr12_+_1738363 | 0.44 |
ENST00000397196.2 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
| chr12_+_21679220 | 0.43 |
ENST00000256969.2 |
C12orf39 |
chromosome 12 open reading frame 39 |
| chr15_-_56757329 | 0.42 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chr16_+_1578674 | 0.40 |
ENST00000253934.5 |
TMEM204 |
transmembrane protein 204 |
| chr15_+_41057818 | 0.38 |
ENST00000558467.1 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
| chr12_-_91573249 | 0.38 |
ENST00000550099.1 ENST00000546391.1 ENST00000551354.1 |
DCN |
decorin |
| chr9_-_21202204 | 0.38 |
ENST00000239347.3 |
IFNA7 |
interferon, alpha 7 |
| chr12_+_71833550 | 0.37 |
ENST00000266674.5 |
LGR5 |
leucine-rich repeat containing G protein-coupled receptor 5 |
| chr11_-_111782484 | 0.37 |
ENST00000533971.1 |
CRYAB |
crystallin, alpha B |
| chr2_-_127963343 | 0.36 |
ENST00000335247.7 |
CYP27C1 |
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr2_-_158300556 | 0.36 |
ENST00000264192.3 |
CYTIP |
cytohesin 1 interacting protein |
| chr11_+_20044600 | 0.35 |
ENST00000311043.8 |
NAV2 |
neuron navigator 2 |
| chr11_-_111782696 | 0.35 |
ENST00000227251.3 ENST00000526180.1 |
CRYAB |
crystallin, alpha B |
| chr17_-_63822563 | 0.35 |
ENST00000317442.8 |
CEP112 |
centrosomal protein 112kDa |
| chr18_-_70532906 | 0.34 |
ENST00000299430.2 ENST00000397929.1 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr1_-_46598371 | 0.33 |
ENST00000372006.1 ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chrX_-_57147902 | 0.33 |
ENST00000275988.5 ENST00000434397.1 ENST00000333933.3 ENST00000374912.5 |
SPIN2B |
spindlin family, member 2B |
| chrX_-_57147748 | 0.33 |
ENST00000374910.3 |
SPIN2B |
spindlin family, member 2B |
| chr7_+_33169142 | 0.33 |
ENST00000242067.6 ENST00000350941.3 ENST00000396127.2 ENST00000355070.2 ENST00000354265.4 ENST00000425508.2 |
BBS9 |
Bardet-Biedl syndrome 9 |
| chr1_-_46598284 | 0.32 |
ENST00000423209.1 ENST00000262741.5 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr12_+_32655048 | 0.32 |
ENST00000427716.2 ENST00000266482.3 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
| chr12_+_8309630 | 0.30 |
ENST00000396570.3 |
ZNF705A |
zinc finger protein 705A |
| chr1_+_144173162 | 0.30 |
ENST00000356801.6 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
| chr17_-_33760164 | 0.30 |
ENST00000445092.1 ENST00000394562.1 ENST00000447040.2 |
SLFN12 |
schlafen family member 12 |
| chr3_-_49851313 | 0.29 |
ENST00000333486.3 |
UBA7 |
ubiquitin-like modifier activating enzyme 7 |
| chr10_+_90660832 | 0.29 |
ENST00000371924.1 |
STAMBPL1 |
STAM binding protein-like 1 |
| chr17_-_29648761 | 0.28 |
ENST00000247270.3 ENST00000462804.2 |
EVI2A |
ecotropic viral integration site 2A |
| chr20_-_33732952 | 0.28 |
ENST00000541621.1 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr15_+_49170083 | 0.27 |
ENST00000530028.2 |
EID1 |
EP300 interacting inhibitor of differentiation 1 |
| chr10_-_90712520 | 0.27 |
ENST00000224784.6 |
ACTA2 |
actin, alpha 2, smooth muscle, aorta |
| chr6_-_15586238 | 0.26 |
ENST00000462989.2 |
DTNBP1 |
dystrobrevin binding protein 1 |
| chr15_-_63448973 | 0.26 |
ENST00000462430.1 |
RPS27L |
ribosomal protein S27-like |
| chr7_-_16685422 | 0.26 |
ENST00000306999.2 |
ANKMY2 |
ankyrin repeat and MYND domain containing 2 |
| chr3_-_127541194 | 0.26 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr22_+_20193905 | 0.26 |
ENST00000609602.1 |
LINC00896 |
long intergenic non-protein coding RNA 896 |
| chr12_+_133757995 | 0.26 |
ENST00000536435.2 ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268 |
zinc finger protein 268 |
| chr5_+_140571902 | 0.26 |
ENST00000239446.4 |
PCDHB10 |
protocadherin beta 10 |
| chr7_+_141811539 | 0.25 |
ENST00000550469.2 ENST00000477922.3 |
RP11-1220K2.2 |
Putative inactive maltase-glucoamylase-like protein LOC93432 |
| chr3_+_138066539 | 0.25 |
ENST00000289104.4 |
MRAS |
muscle RAS oncogene homolog |
| chr12_-_127174806 | 0.25 |
ENST00000545853.1 ENST00000537478.1 |
RP11-407A16.3 |
RP11-407A16.3 |
| chr19_-_22379753 | 0.25 |
ENST00000397121.2 |
ZNF676 |
zinc finger protein 676 |
| chr17_-_67264947 | 0.24 |
ENST00000586811.1 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr17_-_33760269 | 0.24 |
ENST00000452764.3 |
SLFN12 |
schlafen family member 12 |
| chr11_+_126152954 | 0.24 |
ENST00000392679.1 |
TIRAP |
toll-interleukin 1 receptor (TIR) domain containing adaptor protein |
| chr19_-_44952635 | 0.24 |
ENST00000592308.1 ENST00000588931.1 ENST00000291187.4 |
ZNF229 |
zinc finger protein 229 |
| chr15_-_89089860 | 0.24 |
ENST00000558413.1 ENST00000564406.1 ENST00000268148.8 |
DET1 |
de-etiolated homolog 1 (Arabidopsis) |
| chr4_+_56212505 | 0.23 |
ENST00000505210.1 |
SRD5A3 |
steroid 5 alpha-reductase 3 |
| chr16_+_21689835 | 0.23 |
ENST00000286149.4 ENST00000388958.3 |
OTOA |
otoancorin |
| chr19_-_9092018 | 0.23 |
ENST00000397910.4 |
MUC16 |
mucin 16, cell surface associated |
| chr20_+_54933971 | 0.23 |
ENST00000371384.3 ENST00000437418.1 |
FAM210B |
family with sequence similarity 210, member B |
| chr6_-_111804905 | 0.23 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr2_+_79347577 | 0.23 |
ENST00000233735.1 |
REG1A |
regenerating islet-derived 1 alpha |
| chr14_+_100485712 | 0.23 |
ENST00000544450.2 |
EVL |
Enah/Vasp-like |
| chr3_+_51895621 | 0.23 |
ENST00000333127.3 |
IQCF2 |
IQ motif containing F2 |
| chr3_+_142838091 | 0.23 |
ENST00000309575.3 |
CHST2 |
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
| chr5_+_137203465 | 0.23 |
ENST00000239926.4 |
MYOT |
myotilin |
| chr3_-_151102529 | 0.22 |
ENST00000302632.3 |
P2RY12 |
purinergic receptor P2Y, G-protein coupled, 12 |
| chr15_+_67420441 | 0.22 |
ENST00000558894.1 |
SMAD3 |
SMAD family member 3 |
| chr7_+_20687017 | 0.22 |
ENST00000258738.6 |
ABCB5 |
ATP-binding cassette, sub-family B (MDR/TAP), member 5 |
| chr10_-_28287968 | 0.22 |
ENST00000305242.5 |
ARMC4 |
armadillo repeat containing 4 |
| chr14_+_95047725 | 0.22 |
ENST00000554760.1 ENST00000554866.1 ENST00000329597.7 ENST00000556775.1 |
SERPINA5 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr14_+_95047744 | 0.22 |
ENST00000553511.1 ENST00000554633.1 ENST00000555681.1 ENST00000554276.1 |
SERPINA5 |
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 5 |
| chr12_-_10573149 | 0.22 |
ENST00000381904.2 ENST00000381903.2 ENST00000396439.2 |
KLRC3 |
killer cell lectin-like receptor subfamily C, member 3 |
| chr12_+_55248289 | 0.22 |
ENST00000308796.6 |
MUCL1 |
mucin-like 1 |
| chr8_+_22429205 | 0.22 |
ENST00000520207.1 |
SORBS3 |
sorbin and SH3 domain containing 3 |
| chr5_+_110559603 | 0.21 |
ENST00000512453.1 |
CAMK4 |
calcium/calmodulin-dependent protein kinase IV |
| chr3_-_127441406 | 0.21 |
ENST00000487473.1 ENST00000484451.1 |
MGLL |
monoglyceride lipase |
| chr12_+_75874460 | 0.21 |
ENST00000266659.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr9_-_21305312 | 0.21 |
ENST00000259555.4 |
IFNA5 |
interferon, alpha 5 |
| chr15_+_92006567 | 0.21 |
ENST00000554333.1 |
RP11-661P17.1 |
RP11-661P17.1 |
| chr16_+_22501658 | 0.21 |
ENST00000415833.2 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr3_-_183543301 | 0.21 |
ENST00000318631.3 ENST00000431348.1 |
MAP6D1 |
MAP6 domain containing 1 |
| chr5_+_169010638 | 0.20 |
ENST00000265295.4 ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1 |
spindle apparatus coiled-coil protein 1 |
| chr2_-_88427568 | 0.20 |
ENST00000393750.3 ENST00000295834.3 |
FABP1 |
fatty acid binding protein 1, liver |
| chrX_-_83757399 | 0.20 |
ENST00000373177.2 ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX |
highly divergent homeobox |
| chr1_-_154909329 | 0.20 |
ENST00000368467.3 |
PMVK |
phosphomevalonate kinase |
| chr1_-_112032284 | 0.20 |
ENST00000414219.1 |
ADORA3 |
adenosine A3 receptor |
| chr15_+_25200108 | 0.20 |
ENST00000577949.1 ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF SNRPN |
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr5_+_175490540 | 0.20 |
ENST00000515817.1 |
FAM153B |
family with sequence similarity 153, member B |
| chr4_-_129209944 | 0.20 |
ENST00000520121.1 |
PGRMC2 |
progesterone receptor membrane component 2 |
| chr12_-_92536433 | 0.20 |
ENST00000551563.2 ENST00000546975.1 ENST00000549802.1 |
C12orf79 |
chromosome 12 open reading frame 79 |
| chr1_-_89488510 | 0.19 |
ENST00000564665.1 ENST00000370481.4 |
GBP3 |
guanylate binding protein 3 |
| chr3_-_50378235 | 0.19 |
ENST00000357043.2 |
RASSF1 |
Ras association (RalGDS/AF-6) domain family member 1 |
| chr14_+_22386325 | 0.19 |
ENST00000390439.2 |
TRAV13-2 |
T cell receptor alpha variable 13-2 |
| chr5_-_93447333 | 0.19 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr12_-_13256571 | 0.19 |
ENST00000545401.1 ENST00000432710.2 ENST00000351606.6 |
GSG1 |
germ cell associated 1 |
| chr11_-_134123142 | 0.19 |
ENST00000392595.2 ENST00000341541.3 ENST00000352327.5 ENST00000392594.3 |
THYN1 |
thymocyte nuclear protein 1 |
| chr19_-_36909528 | 0.19 |
ENST00000392161.3 ENST00000392171.1 |
ZFP82 |
ZFP82 zinc finger protein |
| chr10_+_91092241 | 0.18 |
ENST00000371811.4 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
| chr9_+_42704004 | 0.18 |
ENST00000457288.1 |
CBWD7 |
COBW domain containing 7 |
| chr1_-_198509804 | 0.18 |
ENST00000489986.1 ENST00000367382.1 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
| chr4_-_83769996 | 0.18 |
ENST00000511338.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr2_-_207629997 | 0.18 |
ENST00000454776.2 |
MDH1B |
malate dehydrogenase 1B, NAD (soluble) |
| chr2_+_46926048 | 0.18 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr8_+_58890917 | 0.18 |
ENST00000522992.1 |
RP11-1112C15.1 |
RP11-1112C15.1 |
| chr5_+_81601166 | 0.18 |
ENST00000439350.1 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chrX_+_52513455 | 0.18 |
ENST00000446098.1 |
XAGE1C |
X antigen family, member 1C |
| chr10_-_4285923 | 0.18 |
ENST00000418372.1 ENST00000608792.1 |
LINC00702 |
long intergenic non-protein coding RNA 702 |
| chr1_-_227505289 | 0.18 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr20_+_1099233 | 0.18 |
ENST00000246015.4 ENST00000335877.6 ENST00000438768.2 |
PSMF1 |
proteasome (prosome, macropain) inhibitor subunit 1 (PI31) |
| chr3_+_14058794 | 0.18 |
ENST00000424053.1 ENST00000528067.1 ENST00000429201.1 |
TPRXL |
tetra-peptide repeat homeobox-like |
| chr19_-_40596828 | 0.18 |
ENST00000414720.2 ENST00000455521.1 ENST00000340963.5 ENST00000595773.1 |
ZNF780A |
zinc finger protein 780A |
| chr16_+_33204156 | 0.18 |
ENST00000398667.4 |
TP53TG3C |
TP53 target 3C |
| chr17_-_295730 | 0.17 |
ENST00000329099.4 |
FAM101B |
family with sequence similarity 101, member B |
| chr4_-_83812402 | 0.17 |
ENST00000395310.2 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr6_-_111804393 | 0.17 |
ENST00000368802.3 ENST00000368805.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr5_-_177207634 | 0.17 |
ENST00000513554.1 ENST00000440605.3 |
FAM153A |
family with sequence similarity 153, member A |
| chr12_-_96794143 | 0.17 |
ENST00000543119.2 |
CDK17 |
cyclin-dependent kinase 17 |
| chr1_-_3566627 | 0.17 |
ENST00000419924.2 ENST00000270708.7 |
WRAP73 |
WD repeat containing, antisense to TP73 |
| chr5_+_54455946 | 0.17 |
ENST00000503787.1 ENST00000296734.6 ENST00000515370.1 |
GPX8 |
glutathione peroxidase 8 (putative) |
| chr1_+_196788887 | 0.17 |
ENST00000320493.5 ENST00000367424.4 ENST00000367421.3 |
CFHR1 CFHR2 |
complement factor H-related 1 complement factor H-related 2 |
| chr3_-_122283079 | 0.17 |
ENST00000471785.1 ENST00000466126.1 |
PARP9 |
poly (ADP-ribose) polymerase family, member 9 |
| chr11_+_59807748 | 0.17 |
ENST00000278855.2 ENST00000532905.1 |
PLAC1L |
oocyte secreted protein 2 |
| chr2_+_161993465 | 0.16 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chrX_+_36065053 | 0.16 |
ENST00000313548.4 |
CHDC2 |
calponin homology domain containing 2 |
| chr2_-_65593784 | 0.16 |
ENST00000443619.2 |
SPRED2 |
sprouty-related, EVH1 domain containing 2 |
| chr5_+_140810132 | 0.16 |
ENST00000252085.3 |
PCDHGA12 |
protocadherin gamma subfamily A, 12 |
| chr3_-_178969403 | 0.16 |
ENST00000314235.5 ENST00000392685.2 |
KCNMB3 |
potassium large conductance calcium-activated channel, subfamily M beta member 3 |
| chr12_-_91348949 | 0.16 |
ENST00000358859.2 |
CCER1 |
coiled-coil glutamate-rich protein 1 |
| chr11_-_118213360 | 0.16 |
ENST00000529594.1 |
CD3D |
CD3d molecule, delta (CD3-TCR complex) |
| chr19_-_14049184 | 0.16 |
ENST00000339560.5 |
PODNL1 |
podocan-like 1 |
| chr6_+_29429217 | 0.16 |
ENST00000396792.2 |
OR2H1 |
olfactory receptor, family 2, subfamily H, member 1 |
| chr6_-_79944336 | 0.16 |
ENST00000344726.5 ENST00000275036.7 |
HMGN3 |
high mobility group nucleosomal binding domain 3 |
| chr2_+_106679690 | 0.16 |
ENST00000409944.1 |
C2orf40 |
chromosome 2 open reading frame 40 |
| chr6_+_63921399 | 0.16 |
ENST00000356170.3 |
FKBP1C |
FK506 binding protein 1C |
| chr15_+_25200074 | 0.16 |
ENST00000390687.4 ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr14_-_106471723 | 0.16 |
ENST00000390595.2 |
IGHV1-3 |
immunoglobulin heavy variable 1-3 |
| chr13_+_21714653 | 0.16 |
ENST00000382533.4 |
SAP18 |
Sin3A-associated protein, 18kDa |
| chr19_-_54974894 | 0.16 |
ENST00000333834.4 |
LENG9 |
leukocyte receptor cluster (LRC) member 9 |
| chr6_+_28249332 | 0.16 |
ENST00000259883.3 |
PGBD1 |
piggyBac transposable element derived 1 |
| chr14_+_57671888 | 0.16 |
ENST00000391612.1 |
AL391152.1 |
AL391152.1 |
| chr14_-_106586656 | 0.16 |
ENST00000390602.2 |
IGHV3-13 |
immunoglobulin heavy variable 3-13 |
| chr20_+_56964169 | 0.16 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr5_+_115420688 | 0.15 |
ENST00000274458.4 |
COMMD10 |
COMM domain containing 10 |
| chr6_-_35765088 | 0.15 |
ENST00000259938.2 |
CLPS |
colipase, pancreatic |
| chr19_+_38085731 | 0.15 |
ENST00000589117.1 |
ZNF540 |
zinc finger protein 540 |
| chr8_-_7220490 | 0.15 |
ENST00000400078.2 |
ZNF705G |
zinc finger protein 705G |
| chr1_-_148025848 | 0.15 |
ENST00000310701.10 ENST00000369219.1 ENST00000444640.1 ENST00000431121.1 ENST00000436356.1 ENST00000448574.1 ENST00000458135.1 ENST00000392972.3 ENST00000426874.1 |
NBPF14 |
neuroblastoma breakpoint family, member 14 |
| chr14_+_52164820 | 0.15 |
ENST00000554167.1 |
FRMD6 |
FERM domain containing 6 |
| chr3_-_3221358 | 0.15 |
ENST00000424814.1 ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN |
cereblon |
| chr6_+_127587755 | 0.15 |
ENST00000368314.1 ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146 |
ring finger protein 146 |
| chrX_+_150866779 | 0.15 |
ENST00000370353.3 |
PRRG3 |
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chrX_-_19988382 | 0.15 |
ENST00000356980.3 ENST00000379687.3 ENST00000379682.4 |
CXorf23 |
chromosome X open reading frame 23 |
| chr2_+_109204909 | 0.15 |
ENST00000393310.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr1_+_144151520 | 0.15 |
ENST00000369372.4 |
NBPF8 |
neuroblastoma breakpoint family, member 8 |
| chr11_+_114166536 | 0.14 |
ENST00000299964.3 |
NNMT |
nicotinamide N-methyltransferase |
| chr5_+_112073544 | 0.14 |
ENST00000257430.4 ENST00000508376.2 |
APC |
adenomatous polyposis coli |
| chr7_-_80141328 | 0.14 |
ENST00000398291.3 |
GNAT3 |
guanine nucleotide binding protein, alpha transducing 3 |
| chr1_+_145516560 | 0.14 |
ENST00000537888.1 |
PEX11B |
peroxisomal biogenesis factor 11 beta |
| chr17_-_29641084 | 0.14 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr1_+_53480598 | 0.14 |
ENST00000430330.2 ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2 |
sterol carrier protein 2 |
| chr7_+_13141097 | 0.14 |
ENST00000411542.1 |
AC011288.2 |
AC011288.2 |
| chr1_+_178310581 | 0.14 |
ENST00000462775.1 |
RASAL2 |
RAS protein activator like 2 |
| chr19_-_20748541 | 0.14 |
ENST00000427401.4 ENST00000594419.1 |
ZNF737 |
zinc finger protein 737 |
| chr3_-_149095652 | 0.14 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr9_-_135230336 | 0.14 |
ENST00000224140.5 ENST00000372169.2 ENST00000393220.1 |
SETX |
senataxin |
| chr21_+_45553535 | 0.14 |
ENST00000348499.5 ENST00000389690.3 ENST00000449622.1 |
C21orf33 |
chromosome 21 open reading frame 33 |
| chr13_+_100741269 | 0.14 |
ENST00000376286.4 ENST00000376279.3 ENST00000376285.1 |
PCCA |
propionyl CoA carboxylase, alpha polypeptide |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.3 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.3 | 1.7 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.2 | 0.7 | GO:0006711 | estrogen catabolic process(GO:0006711) |
| 0.2 | 0.5 | GO:1990927 | negative regulation of synaptic vesicle recycling(GO:1903422) negative regulation of membrane invagination(GO:1905154) calcium ion regulated lysosome exocytosis(GO:1990927) |
| 0.2 | 0.5 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.2 | 1.0 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.1 | 0.5 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.1 | 0.4 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.1 | 0.6 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.1 | 0.5 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.3 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.1 | 1.0 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.1 | 0.3 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.1 | 0.6 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.4 | GO:0061107 | seminal vesicle development(GO:0061107) |
| 0.1 | 2.2 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.1 | 0.2 | GO:2000340 | interleukin-15 production(GO:0032618) chemokine (C-X-C motif) ligand 1 production(GO:0072566) regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000338) positive regulation of chemokine (C-X-C motif) ligand 1 production(GO:2000340) |
| 0.1 | 0.1 | GO:1902567 | negative regulation of eosinophil activation(GO:1902567) |
| 0.1 | 0.2 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.1 | 0.5 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.3 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.1 | 0.2 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.2 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.1 | 0.1 | GO:0042100 | B cell proliferation(GO:0042100) |
| 0.0 | 1.1 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0002818 | intracellular defense response(GO:0002818) |
| 0.0 | 0.2 | GO:0061767 | negative regulation of lung blood pressure(GO:0061767) |
| 0.0 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.0 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.4 | GO:0021564 | vagus nerve development(GO:0021564) |
| 0.0 | 0.1 | GO:0071030 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.7 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.6 | GO:1900038 | negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.4 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 1.6 | GO:0042572 | retinol metabolic process(GO:0042572) |
| 0.0 | 0.2 | GO:0048749 | compound eye development(GO:0048749) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0006844 | acyl carnitine transport(GO:0006844) acyl carnitine transmembrane transport(GO:1902616) |
| 0.0 | 0.2 | GO:1904124 | microglial cell migration(GO:1904124) regulation of microglial cell migration(GO:1904139) |
| 0.0 | 0.0 | GO:0052203 | modulation by symbiont of host molecular function(GO:0052055) modulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052203) modulation by host of symbiont catalytic activity(GO:0052422) |
| 0.0 | 0.7 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.0 | 0.3 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.4 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.2 | GO:0032000 | positive regulation of fatty acid beta-oxidation(GO:0032000) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0033159 | negative regulation of protein import into nucleus, translocation(GO:0033159) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.1 | GO:0050653 | chondroitin sulfate proteoglycan biosynthetic process, polysaccharide chain biosynthetic process(GO:0050653) |
| 0.0 | 0.1 | GO:1902159 | regulation of cyclic nucleotide-gated ion channel activity(GO:1902159) |
| 0.0 | 0.4 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.6 | GO:0071447 | cellular response to hydroperoxide(GO:0071447) |
| 0.0 | 0.1 | GO:0032380 | regulation of intracellular lipid transport(GO:0032377) regulation of intracellular sterol transport(GO:0032380) regulation of intracellular cholesterol transport(GO:0032383) |
| 0.0 | 0.2 | GO:0021796 | cerebral cortex regionalization(GO:0021796) |
| 0.0 | 0.1 | GO:0019626 | short-chain fatty acid catabolic process(GO:0019626) |
| 0.0 | 0.3 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0001812 | positive regulation of type I hypersensitivity(GO:0001812) |
| 0.0 | 0.5 | GO:0090110 | cargo loading into COPII-coated vesicle(GO:0090110) |
| 0.0 | 0.7 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.0 | GO:0032831 | regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032829) positive regulation of CD4-positive, CD25-positive, alpha-beta regulatory T cell differentiation(GO:0032831) |
| 0.0 | 0.3 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.2 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.1 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.2 | GO:0002371 | dendritic cell cytokine production(GO:0002371) |
| 0.0 | 0.2 | GO:1900028 | negative regulation of ruffle assembly(GO:1900028) |
| 0.0 | 0.2 | GO:0070212 | protein poly-ADP-ribosylation(GO:0070212) |
| 0.0 | 0.1 | GO:2000502 | negative regulation of natural killer cell chemotaxis(GO:2000502) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.1 | GO:0034196 | acylglycerol transport(GO:0034196) triglyceride transport(GO:0034197) |
| 0.0 | 0.1 | GO:0044375 | regulation of peroxisome size(GO:0044375) |
| 0.0 | 0.1 | GO:0000066 | mitochondrial ornithine transport(GO:0000066) |
| 0.0 | 0.8 | GO:0033119 | negative regulation of RNA splicing(GO:0033119) |
| 0.0 | 0.1 | GO:1903385 | regulation of homophilic cell adhesion(GO:1903385) |
| 0.0 | 0.4 | GO:1902857 | positive regulation of nonmotile primary cilium assembly(GO:1902857) |
| 0.0 | 0.4 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.1 | GO:0002447 | eosinophil activation involved in immune response(GO:0002278) eosinophil mediated immunity(GO:0002447) eosinophil activation(GO:0043307) eosinophil degranulation(GO:0043308) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.2 | GO:0045059 | positive thymic T cell selection(GO:0045059) |
| 0.0 | 0.2 | GO:0098543 | detection of bacterium(GO:0016045) detection of other organism(GO:0098543) |
| 0.0 | 0.3 | GO:0001973 | adenosine receptor signaling pathway(GO:0001973) |
| 0.0 | 0.1 | GO:0046121 | deoxyribonucleoside catabolic process(GO:0046121) |
| 0.0 | 0.1 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0009256 | 10-formyltetrahydrofolate metabolic process(GO:0009256) |
| 0.0 | 0.0 | GO:0019442 | tryptophan catabolic process to acetyl-CoA(GO:0019442) |
| 0.0 | 0.2 | GO:0006987 | activation of signaling protein activity involved in unfolded protein response(GO:0006987) |
| 0.0 | 0.1 | GO:0061002 | negative regulation of dendritic spine morphogenesis(GO:0061002) |
| 0.0 | 0.1 | GO:0014826 | cellular magnesium ion homeostasis(GO:0010961) vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.1 | GO:0061358 | negative regulation of Wnt protein secretion(GO:0061358) |
| 0.0 | 0.1 | GO:1902573 | positive regulation of serine-type endopeptidase activity(GO:1900005) positive regulation of serine-type peptidase activity(GO:1902573) |
| 0.0 | 0.3 | GO:0060159 | regulation of dopamine receptor signaling pathway(GO:0060159) |
| 0.0 | 0.2 | GO:0010745 | negative regulation of macrophage derived foam cell differentiation(GO:0010745) |
| 0.0 | 0.0 | GO:0021503 | neural fold bending(GO:0021503) |
| 0.0 | 0.1 | GO:0032053 | ciliary basal body organization(GO:0032053) |
| 0.0 | 0.2 | GO:0045919 | positive regulation of cytolysis(GO:0045919) |
| 0.0 | 0.2 | GO:0034501 | protein localization to kinetochore(GO:0034501) |
| 0.0 | 0.0 | GO:0021784 | postganglionic parasympathetic fiber development(GO:0021784) |
| 0.0 | 0.2 | GO:0003356 | regulation of cilium beat frequency(GO:0003356) |
| 0.0 | 0.1 | GO:0019509 | L-methionine biosynthetic process from methylthioadenosine(GO:0019509) |
| 0.0 | 0.2 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.1 | GO:0060059 | embryonic retina morphogenesis in camera-type eye(GO:0060059) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.1 | GO:0019075 | virus maturation(GO:0019075) |
| 0.0 | 0.1 | GO:0002074 | extraocular skeletal muscle development(GO:0002074) |
| 0.0 | 0.1 | GO:2000676 | positive regulation of type B pancreatic cell apoptotic process(GO:2000676) |
| 0.0 | 0.2 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.1 | GO:0034551 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.2 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
| 0.0 | 0.0 | GO:0033686 | positive regulation of luteinizing hormone secretion(GO:0033686) |
| 0.0 | 0.4 | GO:0042276 | error-prone translesion synthesis(GO:0042276) |
| 0.0 | 0.1 | GO:0034356 | NAD biosynthesis via nicotinamide riboside salvage pathway(GO:0034356) |
| 0.0 | 0.1 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.1 | GO:0022417 | protein maturation by protein folding(GO:0022417) |
| 0.0 | 0.2 | GO:0071415 | cellular response to purine-containing compound(GO:0071415) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.0 | GO:0071033 | nuclear retention of pre-mRNA at the site of transcription(GO:0071033) |
| 0.0 | 0.3 | GO:0000028 | ribosomal small subunit assembly(GO:0000028) |
| 0.0 | 0.5 | GO:2000144 | positive regulation of DNA-templated transcription, initiation(GO:2000144) |
| 0.0 | 0.1 | GO:0006015 | 5-phosphoribose 1-diphosphate biosynthetic process(GO:0006015) 5-phosphoribose 1-diphosphate metabolic process(GO:0046391) |
| 0.0 | 0.1 | GO:1902748 | melanocyte migration(GO:0097324) positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:0044241 | lipid digestion(GO:0044241) |
| 0.0 | 0.4 | GO:0001945 | lymph vessel development(GO:0001945) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 8.2 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.3 | 0.8 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.2 | 0.5 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.4 | GO:0097182 | protein C inhibitor-TMPRSS7 complex(GO:0036024) protein C inhibitor-TMPRSS11E complex(GO:0036025) protein C inhibitor-PLAT complex(GO:0036026) protein C inhibitor-PLAU complex(GO:0036027) protein C inhibitor-thrombin complex(GO:0036028) protein C inhibitor-KLK3 complex(GO:0036029) protein C inhibitor-plasma kallikrein complex(GO:0036030) serine protease inhibitor complex(GO:0097180) protein C inhibitor-coagulation factor V complex(GO:0097181) protein C inhibitor-coagulation factor Xa complex(GO:0097182) protein C inhibitor-coagulation factor XI complex(GO:0097183) |
| 0.1 | 0.5 | GO:0048787 | presynaptic active zone membrane(GO:0048787) |
| 0.1 | 1.2 | GO:0098643 | fibrillar collagen trimer(GO:0005583) banded collagen fibril(GO:0098643) |
| 0.1 | 0.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.2 | GO:0043224 | nuclear SCF ubiquitin ligase complex(GO:0043224) |
| 0.0 | 0.3 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.1 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.0 | 0.1 | GO:0034272 | phosphatidylinositol 3-kinase complex, class III, type I(GO:0034271) phosphatidylinositol 3-kinase complex, class III, type II(GO:0034272) |
| 0.0 | 0.7 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.4 | GO:0031464 | Cul4A-RING E3 ubiquitin ligase complex(GO:0031464) |
| 0.0 | 0.7 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.2 | GO:0071144 | SMAD2-SMAD3 protein complex(GO:0071144) |
| 0.0 | 0.3 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.4 | GO:0034464 | BBSome(GO:0034464) |
| 0.0 | 0.5 | GO:0005687 | U4 snRNP(GO:0005687) |
| 0.0 | 0.2 | GO:0042105 | alpha-beta T cell receptor complex(GO:0042105) |
| 0.0 | 0.6 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.1 | GO:0002081 | outer acrosomal membrane(GO:0002081) |
| 0.0 | 0.3 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.5 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0005863 | striated muscle myosin thick filament(GO:0005863) |
| 0.0 | 0.1 | GO:0097013 | phagocytic vesicle lumen(GO:0097013) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.0 | GO:0071821 | FANCM-MHF complex(GO:0071821) |
| 0.0 | 0.0 | GO:0005903 | brush border(GO:0005903) |
| 0.0 | 0.1 | GO:1990393 | 3M complex(GO:1990393) |
| 0.0 | 1.5 | GO:0031594 | neuromuscular junction(GO:0031594) |
| 0.0 | 0.3 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.1 | GO:0002189 | ribose phosphate diphosphokinase complex(GO:0002189) |
| 0.0 | 0.2 | GO:0000940 | condensed chromosome outer kinetochore(GO:0000940) |
| 0.0 | 0.2 | GO:0033180 | proton-transporting V-type ATPase, V1 domain(GO:0033180) |
| 0.0 | 0.1 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 1.0 | GO:1904724 | tertiary granule lumen(GO:1904724) |
| 0.0 | 0.1 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.1 | GO:0030139 | endocytic vesicle(GO:0030139) |
| 0.0 | 0.2 | GO:0042641 | actomyosin(GO:0042641) |
| 0.0 | 0.2 | GO:0016471 | vacuolar proton-transporting V-type ATPase complex(GO:0016471) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.1 | GO:0015057 | thrombin receptor activity(GO:0015057) |
| 0.1 | 0.8 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.1 | GO:0070052 | collagen V binding(GO:0070052) |
| 0.1 | 0.4 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.1 | 0.5 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.1 | 0.9 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.1 | 8.0 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.1 | 0.3 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.1 | 0.4 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.1 | 0.5 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.1 | 0.6 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 1.2 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.5 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.1 | 0.2 | GO:0035663 | Toll-like receptor 2 binding(GO:0035663) |
| 0.1 | 0.5 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.2 | GO:0047751 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) cholestenone 5-alpha-reductase activity(GO:0047751) |
| 0.1 | 0.7 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.2 | GO:0005173 | stem cell factor receptor binding(GO:0005173) |
| 0.1 | 1.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.1 | 0.2 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.1 | 0.6 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.3 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.1 | GO:0001147 | transcription termination site sequence-specific DNA binding(GO:0001147) transcription termination site DNA binding(GO:0001160) |
| 0.0 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.5 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.2 | GO:0031962 | mineralocorticoid receptor binding(GO:0031962) |
| 0.0 | 0.1 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.3 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.7 | GO:0004062 | aryl sulfotransferase activity(GO:0004062) |
| 0.0 | 0.1 | GO:0030760 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.2 | GO:0005324 | long-chain fatty acid transporter activity(GO:0005324) |
| 0.0 | 0.1 | GO:0017129 | triglyceride binding(GO:0017129) |
| 0.0 | 0.1 | GO:0008955 | peptidoglycan glycosyltransferase activity(GO:0008955) |
| 0.0 | 0.1 | GO:0015227 | acyl carnitine transmembrane transporter activity(GO:0015227) |
| 0.0 | 0.2 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0005220 | inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.1 | GO:0004346 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.5 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.1 | GO:0008746 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.3 | GO:0004571 | mannosyl-oligosaccharide 1,2-alpha-mannosidase activity(GO:0004571) |
| 0.0 | 0.4 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.1 | GO:0004139 | deoxyribose-phosphate aldolase activity(GO:0004139) |
| 0.0 | 0.7 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 0.2 | GO:0015562 | efflux transmembrane transporter activity(GO:0015562) |
| 0.0 | 0.1 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0030109 | HLA-B specific inhibitory MHC class I receptor activity(GO:0030109) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.3 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.6 | GO:0017075 | syntaxin-1 binding(GO:0017075) |
| 0.0 | 0.3 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.0 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.1 | GO:0016681 | ubiquinol-cytochrome-c reductase activity(GO:0008121) oxidoreductase activity, acting on diphenols and related substances as donors, cytochrome as acceptor(GO:0016681) |
| 0.0 | 0.2 | GO:0019788 | NEDD8 transferase activity(GO:0019788) |
| 0.0 | 0.1 | GO:0038047 | beta-endorphin receptor activity(GO:0004979) morphine receptor activity(GO:0038047) |
| 0.0 | 0.8 | GO:0036002 | pre-mRNA binding(GO:0036002) |
| 0.0 | 0.1 | GO:0000064 | L-ornithine transmembrane transporter activity(GO:0000064) |
| 0.0 | 0.1 | GO:0009374 | biotin carboxylase activity(GO:0004075) biotin binding(GO:0009374) |
| 0.0 | 0.1 | GO:0019863 | IgE binding(GO:0019863) |
| 0.0 | 0.1 | GO:0016402 | pristanoyl-CoA oxidase activity(GO:0016402) |
| 0.0 | 0.1 | GO:0016428 | tRNA (cytosine-5-)-methyltransferase activity(GO:0016428) |
| 0.0 | 0.1 | GO:0031727 | CCR2 chemokine receptor binding(GO:0031727) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.2 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0097677 | STAT family protein binding(GO:0097677) |
| 0.0 | 0.4 | GO:0003887 | DNA-directed DNA polymerase activity(GO:0003887) |
| 0.0 | 0.5 | GO:0031369 | translation initiation factor binding(GO:0031369) |
| 0.0 | 0.0 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.0 | GO:0048030 | disaccharide binding(GO:0048030) |
| 0.0 | 0.0 | GO:0004833 | tryptophan 2,3-dioxygenase activity(GO:0004833) |
| 0.0 | 0.2 | GO:0009931 | calcium-dependent protein serine/threonine kinase activity(GO:0009931) |
| 0.0 | 0.8 | GO:0030971 | receptor tyrosine kinase binding(GO:0030971) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0015403 | thiamine uptake transmembrane transporter activity(GO:0015403) uptake transmembrane transporter activity(GO:0015563) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0004534 | 5'-3' exoribonuclease activity(GO:0004534) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 8.9 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.1 | 1.1 | PID THROMBIN PAR4 PATHWAY | PAR4-mediated thrombin signaling events |
| 0.0 | 1.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.2 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 1.7 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.3 | ST JAK STAT PATHWAY | Jak-STAT Pathway |
| 0.0 | 0.8 | PID INTEGRIN CS PATHWAY | Integrin family cell surface interactions |
| 0.0 | 0.5 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.7 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.5 | PID NETRIN PATHWAY | Netrin-mediated signaling events |
| 0.0 | 0.6 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 8.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 1.2 | REACTOME CYTOSOLIC SULFONATION OF SMALL MOLECULES | Genes involved in Cytosolic sulfonation of small molecules |
| 0.0 | 0.2 | REACTOME ADP SIGNALLING THROUGH P2RY12 | Genes involved in ADP signalling through P2Y purinoceptor 12 |
| 0.0 | 0.5 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 1.7 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 1.1 | REACTOME THROMBIN SIGNALLING THROUGH PROTEINASE ACTIVATED RECEPTORS PARS | Genes involved in Thrombin signalling through proteinase activated receptors (PARs) |
| 0.0 | 0.7 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION | Genes involved in TRAF6 mediated IRF7 activation |
| 0.0 | 0.6 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 0.4 | REACTOME TETRAHYDROBIOPTERIN BH4 SYNTHESIS RECYCLING SALVAGE AND REGULATION | Genes involved in Tetrahydrobiopterin (BH4) synthesis, recycling, salvage and regulation |
| 0.0 | 0.3 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.2 | REACTOME ANDROGEN BIOSYNTHESIS | Genes involved in Androgen biosynthesis |
| 0.0 | 0.5 | REACTOME ANTIGEN PRESENTATION FOLDING ASSEMBLY AND PEPTIDE LOADING OF CLASS I MHC | Genes involved in Antigen Presentation: Folding, assembly and peptide loading of class I MHC |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.3 | REACTOME NUCLEOTIDE LIKE PURINERGIC RECEPTORS | Genes involved in Nucleotide-like (purinergic) receptors |
| 0.0 | 0.4 | REACTOME SIGNALING BY NODAL | Genes involved in Signaling by NODAL |
| 0.0 | 0.2 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.4 | REACTOME METABOLISM OF STEROID HORMONES AND VITAMINS A AND D | Genes involved in Metabolism of steroid hormones and vitamins A and D |
| 0.0 | 0.1 | REACTOME SYNTHESIS OF PIPS AT THE LATE ENDOSOME MEMBRANE | Genes involved in Synthesis of PIPs at the late endosome membrane |