Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for IRX5
Z-value: 0.24
Transcription factors associated with IRX5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
IRX5
|
ENSG00000176842.10 | IRX5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| IRX5 | hg19_v2_chr16_+_54964740_54964789 | 0.52 | 1.8e-01 | Click! |
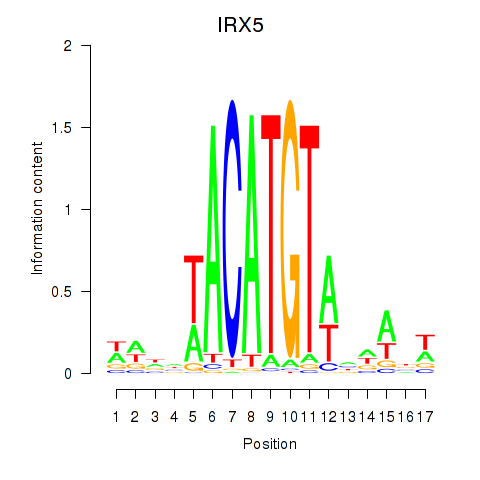
Activity profile of IRX5 motif
Sorted Z-values of IRX5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of IRX5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_108510224 | 0.47 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr11_-_35547572 | 0.34 |
ENST00000378880.2 |
PAMR1 |
peptidase domain containing associated with muscle regeneration 1 |
| chr13_-_38172863 | 0.22 |
ENST00000541481.1 ENST00000379743.4 ENST00000379742.4 ENST00000379749.4 ENST00000541179.1 ENST00000379747.4 |
POSTN |
periostin, osteoblast specific factor |
| chr3_+_12392971 | 0.21 |
ENST00000287820.6 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr12_+_21679220 | 0.20 |
ENST00000256969.2 |
C12orf39 |
chromosome 12 open reading frame 39 |
| chr12_-_58329819 | 0.18 |
ENST00000551421.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chr1_+_145524891 | 0.18 |
ENST00000369304.3 |
ITGA10 |
integrin, alpha 10 |
| chr3_-_9994021 | 0.18 |
ENST00000411976.2 ENST00000412055.1 |
PRRT3 |
proline-rich transmembrane protein 3 |
| chr12_+_56114189 | 0.17 |
ENST00000548082.1 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr6_-_117747015 | 0.17 |
ENST00000368508.3 ENST00000368507.3 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
| chr12_+_56114151 | 0.16 |
ENST00000547072.1 ENST00000552930.1 ENST00000257895.5 |
RDH5 |
retinol dehydrogenase 5 (11-cis/9-cis) |
| chr13_+_76362974 | 0.15 |
ENST00000497947.2 |
LMO7 |
LIM domain 7 |
| chr10_+_90660832 | 0.14 |
ENST00000371924.1 |
STAMBPL1 |
STAM binding protein-like 1 |
| chr1_-_85870177 | 0.14 |
ENST00000542148.1 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr16_+_1578674 | 0.11 |
ENST00000253934.5 |
TMEM204 |
transmembrane protein 204 |
| chr12_+_55248289 | 0.11 |
ENST00000308796.6 |
MUCL1 |
mucin-like 1 |
| chr12_+_71833550 | 0.11 |
ENST00000266674.5 |
LGR5 |
leucine-rich repeat containing G protein-coupled receptor 5 |
| chr7_-_92777606 | 0.11 |
ENST00000437805.1 ENST00000446959.1 ENST00000439952.1 ENST00000414791.1 ENST00000446033.1 ENST00000411955.1 ENST00000318238.4 |
SAMD9L |
sterile alpha motif domain containing 9-like |
| chr15_+_41057818 | 0.11 |
ENST00000558467.1 |
GCHFR |
GTP cyclohydrolase I feedback regulator |
| chr12_+_32655048 | 0.10 |
ENST00000427716.2 ENST00000266482.3 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
| chr1_+_97188188 | 0.10 |
ENST00000541987.1 |
PTBP2 |
polypyrimidine tract binding protein 2 |
| chr16_+_21689835 | 0.09 |
ENST00000286149.4 ENST00000388958.3 |
OTOA |
otoancorin |
| chr15_-_63448973 | 0.09 |
ENST00000462430.1 |
RPS27L |
ribosomal protein S27-like |
| chr9_-_21202204 | 0.08 |
ENST00000239347.3 |
IFNA7 |
interferon, alpha 7 |
| chr12_-_127174806 | 0.08 |
ENST00000545853.1 ENST00000537478.1 |
RP11-407A16.3 |
RP11-407A16.3 |
| chr5_+_169010638 | 0.07 |
ENST00000265295.4 ENST00000506574.1 ENST00000515224.1 ENST00000508247.1 ENST00000513941.1 |
SPDL1 |
spindle apparatus coiled-coil protein 1 |
| chr2_-_127963343 | 0.07 |
ENST00000335247.7 |
CYP27C1 |
cytochrome P450, family 27, subfamily C, polypeptide 1 |
| chr1_-_110284384 | 0.07 |
ENST00000540225.1 |
GSTM3 |
glutathione S-transferase mu 3 (brain) |
| chr19_-_44388116 | 0.07 |
ENST00000587539.1 |
ZNF404 |
zinc finger protein 404 |
| chr20_+_54933971 | 0.07 |
ENST00000371384.3 ENST00000437418.1 |
FAM210B |
family with sequence similarity 210, member B |
| chr5_-_93447333 | 0.07 |
ENST00000395965.3 ENST00000505869.1 ENST00000509163.1 |
FAM172A |
family with sequence similarity 172, member A |
| chr19_-_22379753 | 0.07 |
ENST00000397121.2 |
ZNF676 |
zinc finger protein 676 |
| chr12_-_58329888 | 0.07 |
ENST00000546580.1 |
RP11-620J15.3 |
RP11-620J15.3 |
| chrX_-_52258669 | 0.06 |
ENST00000441417.1 |
XAGE1A |
X antigen family, member 1A |
| chr2_+_210517895 | 0.06 |
ENST00000447185.1 |
MAP2 |
microtubule-associated protein 2 |
| chr19_-_36909528 | 0.06 |
ENST00000392161.3 ENST00000392171.1 |
ZFP82 |
ZFP82 zinc finger protein |
| chr11_+_114166536 | 0.06 |
ENST00000299964.3 |
NNMT |
nicotinamide N-methyltransferase |
| chrX_+_52513455 | 0.06 |
ENST00000446098.1 |
XAGE1C |
X antigen family, member 1C |
| chr7_-_138348969 | 0.06 |
ENST00000436657.1 |
SVOPL |
SVOP-like |
| chr17_-_67264947 | 0.06 |
ENST00000586811.1 |
ABCA5 |
ATP-binding cassette, sub-family A (ABC1), member 5 |
| chr19_-_44952635 | 0.06 |
ENST00000592308.1 ENST00000588931.1 ENST00000291187.4 |
ZNF229 |
zinc finger protein 229 |
| chr10_-_115904361 | 0.06 |
ENST00000428953.1 ENST00000543782.1 |
C10orf118 |
chromosome 10 open reading frame 118 |
| chr1_-_227505289 | 0.05 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chrX_-_19988382 | 0.05 |
ENST00000356980.3 ENST00000379687.3 ENST00000379682.4 |
CXorf23 |
chromosome X open reading frame 23 |
| chr6_+_140175987 | 0.05 |
ENST00000414038.1 ENST00000431609.1 |
RP5-899B16.1 |
RP5-899B16.1 |
| chr12_+_8309630 | 0.05 |
ENST00000396570.3 |
ZNF705A |
zinc finger protein 705A |
| chr8_+_84824920 | 0.05 |
ENST00000523678.1 |
RP11-120I21.2 |
RP11-120I21.2 |
| chr4_+_141264597 | 0.05 |
ENST00000338517.4 ENST00000394203.3 ENST00000506322.1 |
SCOC |
short coiled-coil protein |
| chr1_+_196788887 | 0.05 |
ENST00000320493.5 ENST00000367424.4 ENST00000367421.3 |
CFHR1 CFHR2 |
complement factor H-related 1 complement factor H-related 2 |
| chr15_-_81616446 | 0.05 |
ENST00000302824.6 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
| chrX_-_83757399 | 0.05 |
ENST00000373177.2 ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX |
highly divergent homeobox |
| chr3_-_127541194 | 0.05 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr4_+_56212270 | 0.05 |
ENST00000264228.4 |
SRD5A3 |
steroid 5 alpha-reductase 3 |
| chr11_+_59807748 | 0.04 |
ENST00000278855.2 ENST00000532905.1 |
PLAC1L |
oocyte secreted protein 2 |
| chr7_+_13141097 | 0.04 |
ENST00000411542.1 |
AC011288.2 |
AC011288.2 |
| chr8_-_116504448 | 0.04 |
ENST00000518018.1 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr5_+_54320078 | 0.04 |
ENST00000231009.2 |
GZMK |
granzyme K (granzyme 3; tryptase II) |
| chr10_-_31146615 | 0.04 |
ENST00000444692.2 |
ZNF438 |
zinc finger protein 438 |
| chr15_+_49170083 | 0.04 |
ENST00000530028.2 |
EID1 |
EP300 interacting inhibitor of differentiation 1 |
| chr3_-_81792780 | 0.04 |
ENST00000489715.1 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
| chr1_-_213020991 | 0.04 |
ENST00000332912.3 |
C1orf227 |
chromosome 1 open reading frame 227 |
| chr19_-_40596828 | 0.04 |
ENST00000414720.2 ENST00000455521.1 ENST00000340963.5 ENST00000595773.1 |
ZNF780A |
zinc finger protein 780A |
| chr4_+_113739244 | 0.04 |
ENST00000503271.1 ENST00000503423.1 ENST00000506722.1 |
ANK2 |
ankyrin 2, neuronal |
| chr17_+_75447326 | 0.04 |
ENST00000591088.1 |
SEPT9 |
septin 9 |
| chr15_+_25200108 | 0.04 |
ENST00000577949.1 ENST00000338094.6 ENST00000338327.4 ENST00000579070.1 ENST00000577565.1 |
SNURF SNRPN |
SNRPN upstream reading frame protein small nuclear ribonucleoprotein polypeptide N |
| chr6_-_111804393 | 0.04 |
ENST00000368802.3 ENST00000368805.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chrX_+_52240504 | 0.04 |
ENST00000399805.2 |
XAGE1B |
X antigen family, member 1B |
| chr5_-_177207634 | 0.04 |
ENST00000513554.1 ENST00000440605.3 |
FAM153A |
family with sequence similarity 153, member A |
| chr1_+_158969752 | 0.04 |
ENST00000566111.1 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr1_+_53480598 | 0.04 |
ENST00000430330.2 ENST00000408941.3 ENST00000478274.2 ENST00000484100.1 ENST00000435345.2 ENST00000488965.1 |
SCP2 |
sterol carrier protein 2 |
| chr1_-_3566627 | 0.04 |
ENST00000419924.2 ENST00000270708.7 |
WRAP73 |
WD repeat containing, antisense to TP73 |
| chr7_-_80141328 | 0.04 |
ENST00000398291.3 |
GNAT3 |
guanine nucleotide binding protein, alpha transducing 3 |
| chr2_-_88427568 | 0.04 |
ENST00000393750.3 ENST00000295834.3 |
FABP1 |
fatty acid binding protein 1, liver |
| chr6_+_29429217 | 0.04 |
ENST00000396792.2 |
OR2H1 |
olfactory receptor, family 2, subfamily H, member 1 |
| chr15_+_25200074 | 0.04 |
ENST00000390687.4 ENST00000584968.1 ENST00000346403.6 ENST00000554227.2 |
SNRPN |
small nuclear ribonucleoprotein polypeptide N |
| chr9_+_42704004 | 0.04 |
ENST00000457288.1 |
CBWD7 |
COBW domain containing 7 |
| chr8_-_12668962 | 0.04 |
ENST00000534827.1 |
RP11-252C15.1 |
RP11-252C15.1 |
| chr6_-_111804905 | 0.04 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr3_+_158787041 | 0.03 |
ENST00000471575.1 ENST00000476809.1 ENST00000485419.1 |
IQCJ-SCHIP1 |
IQCJ-SCHIP1 readthrough |
| chr2_-_105372203 | 0.03 |
ENST00000424321.1 |
AC068057.2 |
long intergenic non-protein coding RNA 1114 |
| chrX_-_15333736 | 0.03 |
ENST00000380470.3 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr13_-_41768654 | 0.03 |
ENST00000379483.3 |
KBTBD7 |
kelch repeat and BTB (POZ) domain containing 7 |
| chr13_-_20080080 | 0.03 |
ENST00000400103.2 |
TPTE2 |
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr3_+_138066539 | 0.03 |
ENST00000289104.4 |
MRAS |
muscle RAS oncogene homolog |
| chr5_+_54455946 | 0.03 |
ENST00000503787.1 ENST00000296734.6 ENST00000515370.1 |
GPX8 |
glutathione peroxidase 8 (putative) |
| chr7_-_99573677 | 0.03 |
ENST00000292401.4 |
AZGP1 |
alpha-2-glycoprotein 1, zinc-binding |
| chr12_-_100486668 | 0.03 |
ENST00000550544.1 ENST00000551980.1 ENST00000548045.1 ENST00000545232.2 ENST00000551973.1 |
UHRF1BP1L |
UHRF1 binding protein 1-like |
| chr5_+_81601166 | 0.03 |
ENST00000439350.1 |
ATP6AP1L |
ATPase, H+ transporting, lysosomal accessory protein 1-like |
| chr19_-_50266580 | 0.03 |
ENST00000246801.3 |
TSKS |
testis-specific serine kinase substrate |
| chr15_-_56757329 | 0.03 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chrX_+_108779004 | 0.03 |
ENST00000218004.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chrX_+_36065053 | 0.03 |
ENST00000313548.4 |
CHDC2 |
calponin homology domain containing 2 |
| chr18_+_6729725 | 0.03 |
ENST00000400091.2 ENST00000583410.1 ENST00000584387.1 |
ARHGAP28 |
Rho GTPase activating protein 28 |
| chr5_+_175490540 | 0.03 |
ENST00000515817.1 |
FAM153B |
family with sequence similarity 153, member B |
| chr12_+_75874460 | 0.03 |
ENST00000266659.3 |
GLIPR1 |
GLI pathogenesis-related 1 |
| chr6_+_26204825 | 0.03 |
ENST00000360441.4 |
HIST1H4E |
histone cluster 1, H4e |
| chr7_-_99573640 | 0.03 |
ENST00000411734.1 |
AZGP1 |
alpha-2-glycoprotein 1, zinc-binding |
| chr11_+_114310237 | 0.03 |
ENST00000539119.1 |
REXO2 |
RNA exonuclease 2 |
| chr3_-_3221358 | 0.03 |
ENST00000424814.1 ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN |
cereblon |
| chr1_+_202317815 | 0.03 |
ENST00000608999.1 ENST00000336894.4 ENST00000480184.1 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr3_-_50378235 | 0.03 |
ENST00000357043.2 |
RASSF1 |
Ras association (RalGDS/AF-6) domain family member 1 |
| chr9_-_4666421 | 0.03 |
ENST00000381895.5 |
SPATA6L |
spermatogenesis associated 6-like |
| chr19_-_54974894 | 0.03 |
ENST00000333834.4 |
LENG9 |
leukocyte receptor cluster (LRC) member 9 |
| chr12_-_76462713 | 0.03 |
ENST00000552056.1 |
NAP1L1 |
nucleosome assembly protein 1-like 1 |
| chr3_+_180586536 | 0.03 |
ENST00000465551.1 |
FXR1 |
fragile X mental retardation, autosomal homolog 1 |
| chr15_+_92006567 | 0.03 |
ENST00000554333.1 |
RP11-661P17.1 |
RP11-661P17.1 |
| chr1_-_89488510 | 0.03 |
ENST00000564665.1 ENST00000370481.4 |
GBP3 |
guanylate binding protein 3 |
| chr8_+_22225041 | 0.03 |
ENST00000289952.5 ENST00000524285.1 |
SLC39A14 |
solute carrier family 39 (zinc transporter), member 14 |
| chr22_-_24096562 | 0.03 |
ENST00000398465.3 |
VPREB3 |
pre-B lymphocyte 3 |
| chr9_+_105757590 | 0.03 |
ENST00000374798.3 ENST00000487798.1 |
CYLC2 |
cylicin, basic protein of sperm head cytoskeleton 2 |
| chr3_+_155838337 | 0.02 |
ENST00000490337.1 ENST00000389636.5 |
KCNAB1 |
potassium voltage-gated channel, shaker-related subfamily, beta member 1 |
| chr3_-_149470229 | 0.02 |
ENST00000473414.1 |
COMMD2 |
COMM domain containing 2 |
| chr18_+_22006580 | 0.02 |
ENST00000284202.4 |
IMPACT |
impact RWD domain protein |
| chr1_+_10509971 | 0.02 |
ENST00000320498.4 |
CORT |
cortistatin |
| chr11_+_60223225 | 0.02 |
ENST00000524807.1 ENST00000345732.4 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr9_+_96846740 | 0.02 |
ENST00000288976.3 |
PTPDC1 |
protein tyrosine phosphatase domain containing 1 |
| chr9_-_125667618 | 0.02 |
ENST00000423239.2 |
RC3H2 |
ring finger and CCCH-type domains 2 |
| chr13_-_20110902 | 0.02 |
ENST00000390680.2 ENST00000382977.4 ENST00000382975.4 ENST00000457266.2 |
TPTE2 |
transmembrane phosphoinositide 3-phosphatase and tensin homolog 2 |
| chr1_-_241683001 | 0.02 |
ENST00000366560.3 |
FH |
fumarate hydratase |
| chr19_-_48059113 | 0.02 |
ENST00000391901.3 ENST00000314121.4 ENST00000448976.1 |
ZNF541 |
zinc finger protein 541 |
| chr17_-_29641084 | 0.02 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr3_-_46068969 | 0.02 |
ENST00000542109.1 ENST00000395946.2 |
XCR1 |
chemokine (C motif) receptor 1 |
| chr6_+_22569784 | 0.02 |
ENST00000510882.2 |
HDGFL1 |
hepatoma derived growth factor-like 1 |
| chr14_+_22554680 | 0.02 |
ENST00000390451.2 |
TRAV23DV6 |
T cell receptor alpha variable 23/delta variable 6 |
| chr17_+_35732955 | 0.02 |
ENST00000300618.4 |
C17orf78 |
chromosome 17 open reading frame 78 |
| chr3_-_52713729 | 0.02 |
ENST00000296302.7 ENST00000356770.4 ENST00000337303.4 ENST00000409057.1 ENST00000410007.1 ENST00000409114.3 ENST00000409767.1 ENST00000423351.1 |
PBRM1 |
polybromo 1 |
| chr1_-_3566590 | 0.02 |
ENST00000424367.1 ENST00000378322.3 |
WRAP73 |
WD repeat containing, antisense to TP73 |
| chr17_-_47723943 | 0.02 |
ENST00000510476.1 ENST00000503676.1 |
SPOP |
speckle-type POZ protein |
| chr11_+_114310102 | 0.02 |
ENST00000265881.5 |
REXO2 |
RNA exonuclease 2 |
| chr12_+_53848549 | 0.02 |
ENST00000439930.3 ENST00000548933.1 ENST00000562264.1 |
PCBP2 |
poly(rC) binding protein 2 |
| chr5_-_13944652 | 0.02 |
ENST00000265104.4 |
DNAH5 |
dynein, axonemal, heavy chain 5 |
| chr1_-_7913089 | 0.02 |
ENST00000361696.5 |
UTS2 |
urotensin 2 |
| chr3_+_160473996 | 0.02 |
ENST00000498165.1 |
PPM1L |
protein phosphatase, Mg2+/Mn2+ dependent, 1L |
| chr2_-_40657397 | 0.02 |
ENST00000408028.2 ENST00000332839.4 ENST00000406391.2 ENST00000542024.1 ENST00000542756.1 ENST00000405901.3 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr1_+_63989004 | 0.02 |
ENST00000371088.4 |
EFCAB7 |
EF-hand calcium binding domain 7 |
| chr10_-_91403625 | 0.02 |
ENST00000322191.6 ENST00000342512.3 ENST00000371774.2 |
PANK1 |
pantothenate kinase 1 |
| chr3_+_186383741 | 0.02 |
ENST00000232003.4 |
HRG |
histidine-rich glycoprotein |
| chr4_+_169842707 | 0.02 |
ENST00000503290.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr18_+_33709834 | 0.02 |
ENST00000358232.6 ENST00000351393.6 ENST00000442325.2 ENST00000423854.2 ENST00000350494.6 ENST00000542824.1 |
ELP2 |
elongator acetyltransferase complex subunit 2 |
| chr4_+_130017268 | 0.02 |
ENST00000425929.1 ENST00000508673.1 ENST00000508622.1 |
C4orf33 |
chromosome 4 open reading frame 33 |
| chrX_+_15767971 | 0.02 |
ENST00000479740.1 ENST00000454127.2 |
CA5B |
carbonic anhydrase VB, mitochondrial |
| chr7_+_80267973 | 0.02 |
ENST00000394788.3 ENST00000447544.2 |
CD36 |
CD36 molecule (thrombospondin receptor) |
| chr5_+_140566 | 0.02 |
ENST00000502646.1 |
PLEKHG4B |
pleckstrin homology domain containing, family G (with RhoGef domain) member 4B |
| chr19_+_35849362 | 0.01 |
ENST00000327809.4 |
FFAR3 |
free fatty acid receptor 3 |
| chr5_+_128300810 | 0.01 |
ENST00000262462.4 |
SLC27A6 |
solute carrier family 27 (fatty acid transporter), member 6 |
| chr12_+_58335360 | 0.01 |
ENST00000300145.3 |
XRCC6BP1 |
XRCC6 binding protein 1 |
| chr2_-_242041607 | 0.01 |
ENST00000434791.1 ENST00000401626.2 ENST00000439144.1 ENST00000406593.1 ENST00000495694.1 ENST00000407095.3 ENST00000391980.2 |
MTERFD2 |
MTERF domain containing 2 |
| chr9_-_4666337 | 0.01 |
ENST00000381890.5 |
SPATA6L |
spermatogenesis associated 6-like |
| chr10_-_14050522 | 0.01 |
ENST00000342409.2 |
FRMD4A |
FERM domain containing 4A |
| chr8_-_67976509 | 0.01 |
ENST00000518747.1 |
COPS5 |
COP9 signalosome subunit 5 |
| chr12_-_10282742 | 0.01 |
ENST00000298523.5 ENST00000396484.2 ENST00000310002.4 |
CLEC7A |
C-type lectin domain family 7, member A |
| chr2_+_169757750 | 0.01 |
ENST00000375363.3 ENST00000429379.2 ENST00000421979.1 |
G6PC2 |
glucose-6-phosphatase, catalytic, 2 |
| chr20_+_44330651 | 0.01 |
ENST00000305479.2 |
WFDC13 |
WAP four-disulfide core domain 13 |
| chrX_+_12809463 | 0.01 |
ENST00000380663.3 ENST00000380668.5 ENST00000398491.2 ENST00000489404.1 |
PRPS2 |
phosphoribosyl pyrophosphate synthetase 2 |
| chr1_+_155579979 | 0.01 |
ENST00000452804.2 ENST00000538143.1 ENST00000245564.2 ENST00000368341.4 |
MSTO1 |
misato 1, mitochondrial distribution and morphology regulator |
| chr18_-_44702668 | 0.01 |
ENST00000256433.3 |
IER3IP1 |
immediate early response 3 interacting protein 1 |
| chr20_-_13971255 | 0.01 |
ENST00000284951.5 ENST00000378072.5 |
SEL1L2 |
sel-1 suppressor of lin-12-like 2 (C. elegans) |
| chr2_+_201173667 | 0.01 |
ENST00000409755.3 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr21_+_38792602 | 0.01 |
ENST00000398960.2 ENST00000398956.2 |
DYRK1A |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1A |
| chr3_-_139199565 | 0.01 |
ENST00000511956.1 |
RBP2 |
retinol binding protein 2, cellular |
| chr12_-_10282681 | 0.01 |
ENST00000533022.1 |
CLEC7A |
C-type lectin domain family 7, member A |
| chr17_+_25958174 | 0.01 |
ENST00000313648.6 ENST00000577392.1 ENST00000584661.1 ENST00000413914.2 |
LGALS9 |
lectin, galactoside-binding, soluble, 9 |
| chr3_-_108476231 | 0.01 |
ENST00000295755.6 |
RETNLB |
resistin like beta |
| chr8_+_67976593 | 0.01 |
ENST00000262210.5 ENST00000412460.1 |
CSPP1 |
centrosome and spindle pole associated protein 1 |
| chrY_-_6742068 | 0.01 |
ENST00000215479.5 |
AMELY |
amelogenin, Y-linked |
| chr7_+_116502527 | 0.01 |
ENST00000361183.3 |
CAPZA2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr2_-_32490859 | 0.01 |
ENST00000404025.2 |
NLRC4 |
NLR family, CARD domain containing 4 |
| chr19_-_23456996 | 0.01 |
ENST00000594653.1 |
RP11-15H20.7 |
RP11-15H20.7 |
| chr18_-_69246167 | 0.01 |
ENST00000566582.1 |
RP11-510D19.1 |
RP11-510D19.1 |
| chr8_-_16035454 | 0.01 |
ENST00000355282.2 |
MSR1 |
macrophage scavenger receptor 1 |
| chr3_+_108855558 | 0.01 |
ENST00000467240.1 ENST00000477643.1 ENST00000479039.1 ENST00000593799.1 |
RP11-59E19.1 |
RP11-59E19.1 |
| chr1_+_15668240 | 0.01 |
ENST00000444385.1 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr15_-_51630772 | 0.01 |
ENST00000557858.1 ENST00000558328.1 ENST00000396404.4 ENST00000561075.1 ENST00000405011.2 ENST00000559980.1 ENST00000453807.2 ENST00000396402.1 |
CYP19A1 |
cytochrome P450, family 19, subfamily A, polypeptide 1 |
| chr1_-_115124257 | 0.01 |
ENST00000369541.3 |
BCAS2 |
breast carcinoma amplified sequence 2 |
| chr2_-_32490801 | 0.01 |
ENST00000360906.5 ENST00000342905.6 |
NLRC4 |
NLR family, CARD domain containing 4 |
| chr3_+_53528659 | 0.01 |
ENST00000350061.5 |
CACNA1D |
calcium channel, voltage-dependent, L type, alpha 1D subunit |
| chrX_-_138790348 | 0.00 |
ENST00000414978.1 ENST00000519895.1 |
MCF2 |
MCF.2 cell line derived transforming sequence |
| chr18_-_69246186 | 0.00 |
ENST00000568095.1 |
RP11-510D19.1 |
RP11-510D19.1 |
| chr17_-_3301704 | 0.00 |
ENST00000322608.2 |
OR1E1 |
olfactory receptor, family 1, subfamily E, member 1 |
| chr1_+_196912902 | 0.00 |
ENST00000476712.2 ENST00000367415.5 |
CFHR2 |
complement factor H-related 2 |
| chr12_-_10282836 | 0.00 |
ENST00000304084.8 ENST00000353231.5 ENST00000525605.1 |
CLEC7A |
C-type lectin domain family 7, member A |
| chr1_-_145826450 | 0.00 |
ENST00000462900.2 |
GPR89A |
G protein-coupled receptor 89A |
| chr1_-_89641680 | 0.00 |
ENST00000294671.2 |
GBP7 |
guanylate binding protein 7 |
| chr1_-_244006528 | 0.00 |
ENST00000336199.5 ENST00000263826.5 |
AKT3 |
v-akt murine thymoma viral oncogene homolog 3 |
| chr12_-_120315074 | 0.00 |
ENST00000261833.7 ENST00000392521.2 |
CIT |
citron (rho-interacting, serine/threonine kinase 21) |
| chr9_+_71819927 | 0.00 |
ENST00000535702.1 |
TJP2 |
tight junction protein 2 |
| chr1_+_240408560 | 0.00 |
ENST00000441342.1 ENST00000545751.1 |
FMN2 |
formin 2 |
| chr20_+_43990576 | 0.00 |
ENST00000372727.1 ENST00000414310.1 |
SYS1 |
SYS1 Golgi-localized integral membrane protein homolog (S. cerevisiae) |
| chr11_+_22688150 | 0.00 |
ENST00000454584.2 |
GAS2 |
growth arrest-specific 2 |
| chr10_+_118350522 | 0.00 |
ENST00000530319.1 ENST00000527980.1 ENST00000471549.1 ENST00000534537.1 |
PNLIPRP1 |
pancreatic lipase-related protein 1 |
| chr8_+_12803176 | 0.00 |
ENST00000524591.2 |
KIAA1456 |
KIAA1456 |
| chr4_+_113558612 | 0.00 |
ENST00000505034.1 ENST00000324052.6 |
LARP7 |
La ribonucleoprotein domain family, member 7 |
| chr6_-_154677900 | 0.00 |
ENST00000265198.4 ENST00000520261.1 |
IPCEF1 |
interaction protein for cytohesin exchange factors 1 |
| chr1_-_23504176 | 0.00 |
ENST00000302291.4 |
LUZP1 |
leucine zipper protein 1 |
| chr6_+_149887377 | 0.00 |
ENST00000367419.5 |
GINM1 |
glycoprotein integral membrane 1 |
| chr10_+_118350468 | 0.00 |
ENST00000358834.4 ENST00000528052.1 ENST00000442761.1 |
PNLIPRP1 |
pancreatic lipase-related protein 1 |
| chr9_-_113100088 | 0.00 |
ENST00000374510.4 ENST00000423740.2 ENST00000374511.3 ENST00000374507.4 |
TXNDC8 |
thioredoxin domain containing 8 (spermatozoa) |
| chr8_+_87878640 | 0.00 |
ENST00000518476.1 |
CNBD1 |
cyclic nucleotide binding domain containing 1 |
| chr6_+_146920097 | 0.00 |
ENST00000397944.3 ENST00000522242.1 |
ADGB |
androglobin |
| chr9_+_71820057 | 0.00 |
ENST00000539225.1 |
TJP2 |
tight junction protein 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.2 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.0 | 0.2 | GO:0032571 | response to vitamin K(GO:0032571) bone regeneration(GO:1990523) |
| 0.0 | 0.1 | GO:0043105 | regulation of GTP cyclohydrolase I activity(GO:0043095) negative regulation of GTP cyclohydrolase I activity(GO:0043105) |
| 0.0 | 0.2 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:0008065 | establishment of blood-nerve barrier(GO:0008065) |
| 0.0 | 0.0 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.0 | GO:0016095 | polyprenol catabolic process(GO:0016095) |
| 0.0 | 0.0 | GO:1904425 | negative regulation of GTP binding(GO:1904425) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.0 | 0.1 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044549 | GTP cyclohydrolase binding(GO:0044549) |
| 0.0 | 0.2 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.2 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0005503 | all-trans retinal binding(GO:0005503) |
| 0.0 | 0.1 | GO:0008112 | nicotinamide N-methyltransferase activity(GO:0008112) pyridine N-methyltransferase activity(GO:0030760) |
| 0.0 | 0.0 | GO:0035034 | histone acetyltransferase regulator activity(GO:0035034) |
| 0.0 | 0.1 | GO:0051800 | phosphatidylinositol-3,4-bisphosphate 3-phosphatase activity(GO:0051800) |
| 0.0 | 0.0 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.3 | GO:0004745 | retinol dehydrogenase activity(GO:0004745) |
| 0.0 | 0.0 | GO:0035671 | 3-oxo-5-alpha-steroid 4-dehydrogenase activity(GO:0003865) steroid dehydrogenase activity, acting on the CH-CH group of donors(GO:0033765) enone reductase activity(GO:0035671) cholestenone 5-alpha-reductase activity(GO:0047751) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | PID CONE PATHWAY | Visual signal transduction: Cones |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.5 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |