Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
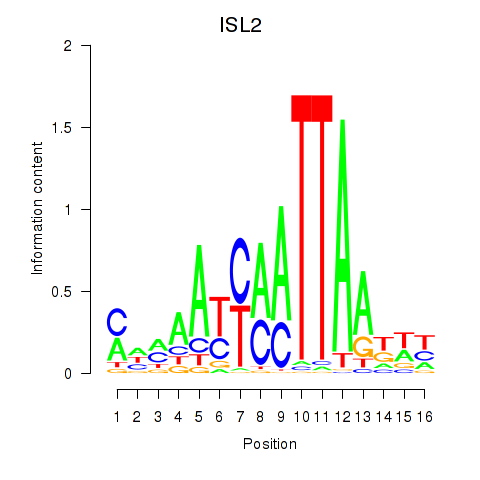
Results for ISL2
Z-value: 0.24
Transcription factors associated with ISL2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISL2
|
ENSG00000159556.5 | ISL2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ISL2 | hg19_v2_chr15_+_76629064_76629073 | -0.11 | 7.9e-01 | Click! |
Activity profile of ISL2 motif
Sorted Z-values of ISL2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ISL2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr13_-_20806440 | 0.14 |
ENST00000400066.3 ENST00000400065.3 ENST00000356192.6 |
GJB6 |
gap junction protein, beta 6, 30kDa |
| chr6_-_155776966 | 0.14 |
ENST00000159060.2 |
NOX3 |
NADPH oxidase 3 |
| chr19_-_36001113 | 0.14 |
ENST00000434389.1 |
DMKN |
dermokine |
| chr7_-_111032971 | 0.13 |
ENST00000450877.1 |
IMMP2L |
IMP2 inner mitochondrial membrane peptidase-like (S. cerevisiae) |
| chr1_+_160370344 | 0.12 |
ENST00000368061.2 |
VANGL2 |
VANGL planar cell polarity protein 2 |
| chr3_-_141747950 | 0.12 |
ENST00000497579.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr12_+_122356488 | 0.10 |
ENST00000397454.2 |
WDR66 |
WD repeat domain 66 |
| chr4_-_90756769 | 0.09 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr9_+_12693336 | 0.08 |
ENST00000381137.2 ENST00000388918.5 |
TYRP1 |
tyrosinase-related protein 1 |
| chr20_-_50722183 | 0.08 |
ENST00000371523.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr8_-_125577940 | 0.08 |
ENST00000519168.1 ENST00000395508.2 |
MTSS1 |
metastasis suppressor 1 |
| chr1_+_77333117 | 0.08 |
ENST00000477717.1 |
ST6GALNAC5 |
ST6 (alpha-N-acetyl-neuraminyl-2,3-beta-galactosyl-1,3)-N-acetylgalactosaminide alpha-2,6-sialyltransferase 5 |
| chrX_-_32173579 | 0.07 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chr12_-_12715266 | 0.06 |
ENST00000228862.2 |
DUSP16 |
dual specificity phosphatase 16 |
| chr6_+_31105426 | 0.06 |
ENST00000547221.1 |
PSORS1C1 |
psoriasis susceptibility 1 candidate 1 |
| chr22_-_29107919 | 0.05 |
ENST00000434810.1 ENST00000456369.1 |
CHEK2 |
checkpoint kinase 2 |
| chr4_+_128702969 | 0.05 |
ENST00000508776.1 ENST00000439123.2 |
HSPA4L |
heat shock 70kDa protein 4-like |
| chr14_+_102276132 | 0.04 |
ENST00000350249.3 ENST00000557621.1 ENST00000556946.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chr6_-_24358264 | 0.04 |
ENST00000378454.3 |
DCDC2 |
doublecortin domain containing 2 |
| chr1_+_149871171 | 0.04 |
ENST00000369150.1 |
BOLA1 |
bolA family member 1 |
| chr3_-_191000172 | 0.04 |
ENST00000427544.2 |
UTS2B |
urotensin 2B |
| chr2_-_85108363 | 0.04 |
ENST00000335459.5 |
TRABD2A |
TraB domain containing 2A |
| chr14_-_67878917 | 0.04 |
ENST00000216446.4 |
PLEK2 |
pleckstrin 2 |
| chr17_-_10372875 | 0.04 |
ENST00000255381.2 |
MYH4 |
myosin, heavy chain 4, skeletal muscle |
| chr7_-_45151272 | 0.04 |
ENST00000461363.1 ENST00000495078.1 ENST00000494076.1 ENST00000478532.1 ENST00000258770.3 ENST00000361278.3 |
TBRG4 |
transforming growth factor beta regulator 4 |
| chr2_+_113825547 | 0.04 |
ENST00000341010.2 ENST00000337569.3 |
IL1F10 |
interleukin 1 family, member 10 (theta) |
| chr11_-_13517565 | 0.03 |
ENST00000282091.1 ENST00000529816.1 |
PTH |
parathyroid hormone |
| chr1_+_149871135 | 0.03 |
ENST00000369152.5 |
BOLA1 |
bolA family member 1 |
| chr4_-_120243545 | 0.03 |
ENST00000274024.3 |
FABP2 |
fatty acid binding protein 2, intestinal |
| chr11_-_5255861 | 0.03 |
ENST00000380299.3 |
HBD |
hemoglobin, delta |
| chrX_-_84634737 | 0.03 |
ENST00000262753.4 |
POF1B |
premature ovarian failure, 1B |
| chr18_-_3845293 | 0.03 |
ENST00000400145.2 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr12_-_95945246 | 0.03 |
ENST00000258499.3 |
USP44 |
ubiquitin specific peptidase 44 |
| chr21_+_39644305 | 0.03 |
ENST00000398930.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr11_-_62609281 | 0.03 |
ENST00000525239.1 ENST00000538098.2 |
WDR74 |
WD repeat domain 74 |
| chr11_+_86085778 | 0.03 |
ENST00000354755.1 ENST00000278487.3 ENST00000531271.1 ENST00000445632.2 |
CCDC81 |
coiled-coil domain containing 81 |
| chr2_+_234668894 | 0.03 |
ENST00000305208.5 ENST00000608383.1 ENST00000360418.3 |
UGT1A8 UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A1 UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr13_-_46716969 | 0.03 |
ENST00000435666.2 |
LCP1 |
lymphocyte cytosolic protein 1 (L-plastin) |
| chr2_+_86669118 | 0.03 |
ENST00000427678.1 ENST00000542128.1 |
KDM3A |
lysine (K)-specific demethylase 3A |
| chr5_+_40909354 | 0.03 |
ENST00000313164.9 |
C7 |
complement component 7 |
| chr4_+_88532028 | 0.03 |
ENST00000282478.7 |
DSPP |
dentin sialophosphoprotein |
| chr1_-_17766198 | 0.03 |
ENST00000375436.4 |
RCC2 |
regulator of chromosome condensation 2 |
| chr21_+_39644395 | 0.03 |
ENST00000398934.1 |
KCNJ15 |
potassium inwardly-rectifying channel, subfamily J, member 15 |
| chr11_-_124190184 | 0.03 |
ENST00000357438.2 |
OR8D2 |
olfactory receptor, family 8, subfamily D, member 2 |
| chr10_-_5046042 | 0.03 |
ENST00000421196.3 ENST00000455190.1 |
AKR1C2 |
aldo-keto reductase family 1, member C2 |
| chr7_-_16844611 | 0.03 |
ENST00000401412.1 ENST00000419304.2 |
AGR2 |
anterior gradient 2 |
| chr15_-_83378638 | 0.03 |
ENST00000261722.3 |
AP3B2 |
adaptor-related protein complex 3, beta 2 subunit |
| chr18_-_64271316 | 0.03 |
ENST00000540086.1 ENST00000580157.1 |
CDH19 |
cadherin 19, type 2 |
| chr17_+_3118915 | 0.03 |
ENST00000304094.1 |
OR1A1 |
olfactory receptor, family 1, subfamily A, member 1 |
| chrX_+_65384182 | 0.02 |
ENST00000441993.2 ENST00000419594.1 |
HEPH |
hephaestin |
| chr1_+_63063152 | 0.02 |
ENST00000371129.3 |
ANGPTL3 |
angiopoietin-like 3 |
| chr9_+_35042205 | 0.02 |
ENST00000312292.5 ENST00000378745.3 |
C9orf131 |
chromosome 9 open reading frame 131 |
| chr17_-_7307358 | 0.02 |
ENST00000576017.1 ENST00000302422.3 ENST00000535512.1 |
TMEM256 TMEM256-PLSCR3 |
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr17_-_48785216 | 0.02 |
ENST00000285243.6 |
ANKRD40 |
ankyrin repeat domain 40 |
| chr7_-_37488834 | 0.02 |
ENST00000310758.4 |
ELMO1 |
engulfment and cell motility 1 |
| chr2_-_163008903 | 0.02 |
ENST00000418842.2 ENST00000375497.3 |
GCG |
glucagon |
| chrX_-_15332665 | 0.02 |
ENST00000537676.1 ENST00000344384.4 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr11_+_71544246 | 0.02 |
ENST00000328698.1 |
DEFB108B |
defensin, beta 108B |
| chrX_-_84634708 | 0.02 |
ENST00000373145.3 |
POF1B |
premature ovarian failure, 1B |
| chr4_+_169013666 | 0.02 |
ENST00000359299.3 |
ANXA10 |
annexin A10 |
| chr6_+_130339710 | 0.02 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr11_-_327537 | 0.02 |
ENST00000602735.1 |
IFITM3 |
interferon induced transmembrane protein 3 |
| chr17_-_4938712 | 0.02 |
ENST00000254853.5 ENST00000424747.1 |
SLC52A1 |
solute carrier family 52 (riboflavin transporter), member 1 |
| chr11_+_73498898 | 0.02 |
ENST00000535529.1 ENST00000497094.2 ENST00000411840.2 ENST00000535277.1 ENST00000398483.3 ENST00000542303.1 |
MRPL48 |
mitochondrial ribosomal protein L48 |
| chr14_-_106781017 | 0.02 |
ENST00000390612.2 |
IGHV4-28 |
immunoglobulin heavy variable 4-28 |
| chr8_-_121457608 | 0.02 |
ENST00000306185.3 |
MRPL13 |
mitochondrial ribosomal protein L13 |
| chr18_-_64271363 | 0.02 |
ENST00000262150.2 |
CDH19 |
cadherin 19, type 2 |
| chr18_-_53303123 | 0.02 |
ENST00000569357.1 ENST00000565124.1 ENST00000398339.1 |
TCF4 |
transcription factor 4 |
| chr11_+_28129795 | 0.02 |
ENST00000406787.3 ENST00000342303.5 ENST00000403099.1 ENST00000407364.3 |
METTL15 |
methyltransferase like 15 |
| chr1_+_15668240 | 0.02 |
ENST00000444385.1 |
FHAD1 |
forkhead-associated (FHA) phosphopeptide binding domain 1 |
| chr19_-_46234119 | 0.02 |
ENST00000317683.3 |
FBXO46 |
F-box protein 46 |
| chr9_-_28670283 | 0.02 |
ENST00000379992.2 |
LINGO2 |
leucine rich repeat and Ig domain containing 2 |
| chr11_+_101983176 | 0.02 |
ENST00000524575.1 |
YAP1 |
Yes-associated protein 1 |
| chr5_+_140201183 | 0.02 |
ENST00000529619.1 ENST00000529859.1 ENST00000378126.3 |
PCDHA5 |
protocadherin alpha 5 |
| chr17_+_4710391 | 0.02 |
ENST00000263088.6 ENST00000572940.1 |
PLD2 |
phospholipase D2 |
| chr2_-_209010874 | 0.02 |
ENST00000260988.4 |
CRYGB |
crystallin, gamma B |
| chrX_-_119077695 | 0.01 |
ENST00000371410.3 |
NKAP |
NFKB activating protein |
| chr1_-_178840157 | 0.01 |
ENST00000367629.1 ENST00000234816.2 |
ANGPTL1 |
angiopoietin-like 1 |
| chrX_+_11311533 | 0.01 |
ENST00000380714.3 ENST00000380712.3 ENST00000348912.4 |
AMELX |
amelogenin, X-linked |
| chr19_+_29456034 | 0.01 |
ENST00000589921.1 |
LINC00906 |
long intergenic non-protein coding RNA 906 |
| chr12_-_118628350 | 0.01 |
ENST00000537952.1 ENST00000537822.1 |
TAOK3 |
TAO kinase 3 |
| chr2_+_234580525 | 0.01 |
ENST00000609637.1 |
UGT1A1 |
UDP glucuronosyltransferase 1 family, polypeptide A8 |
| chr5_-_139726181 | 0.01 |
ENST00000507104.1 ENST00000230990.6 |
HBEGF |
heparin-binding EGF-like growth factor |
| chr14_+_92789498 | 0.01 |
ENST00000531433.1 |
SLC24A4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr5_+_110427983 | 0.01 |
ENST00000513710.2 ENST00000505303.1 |
WDR36 |
WD repeat domain 36 |
| chr6_+_26440700 | 0.01 |
ENST00000494393.1 ENST00000482451.1 ENST00000244519.2 ENST00000339789.4 ENST00000471353.1 ENST00000361232.3 ENST00000487627.1 ENST00000496719.1 ENST00000490254.1 ENST00000487272.1 |
BTN3A3 |
butyrophilin, subfamily 3, member A3 |
| chr17_+_7155819 | 0.01 |
ENST00000570322.1 ENST00000576496.1 ENST00000574841.2 |
ELP5 |
elongator acetyltransferase complex subunit 5 |
| chr2_+_234580499 | 0.01 |
ENST00000354728.4 |
UGT1A9 |
UDP glucuronosyltransferase 1 family, polypeptide A9 |
| chr4_+_15471489 | 0.01 |
ENST00000424120.1 ENST00000413206.1 ENST00000438599.2 ENST00000511544.1 ENST00000512702.1 ENST00000507954.1 ENST00000515124.1 ENST00000503292.1 ENST00000503658.1 |
CC2D2A |
coiled-coil and C2 domain containing 2A |
| chr2_-_231860596 | 0.01 |
ENST00000441063.1 ENST00000434094.1 ENST00000418330.1 ENST00000457803.1 ENST00000414876.1 ENST00000446741.1 ENST00000426904.1 |
AC105344.2 |
SPATA3 antisense RNA 1 (head to head) |
| chr10_-_50396407 | 0.01 |
ENST00000374153.2 ENST00000374151.3 |
C10orf128 |
chromosome 10 open reading frame 128 |
| chr17_-_64225508 | 0.01 |
ENST00000205948.6 |
APOH |
apolipoprotein H (beta-2-glycoprotein I) |
| chr2_-_169887827 | 0.01 |
ENST00000263817.6 |
ABCB11 |
ATP-binding cassette, sub-family B (MDR/TAP), member 11 |
| chr3_+_139063372 | 0.01 |
ENST00000478464.1 |
MRPS22 |
mitochondrial ribosomal protein S22 |
| chr1_-_174992544 | 0.01 |
ENST00000476371.1 |
MRPS14 |
mitochondrial ribosomal protein S14 |
| chr11_-_107729887 | 0.01 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chr16_+_28565230 | 0.01 |
ENST00000317058.3 |
CCDC101 |
coiled-coil domain containing 101 |
| chrX_+_65382433 | 0.01 |
ENST00000374727.3 |
HEPH |
hephaestin |
| chr15_-_75748115 | 0.01 |
ENST00000360439.4 |
SIN3A |
SIN3 transcription regulator family member A |
| chr2_+_74685413 | 0.01 |
ENST00000233615.2 |
WBP1 |
WW domain binding protein 1 |
| chr6_-_32095968 | 0.01 |
ENST00000375203.3 ENST00000375201.4 |
ATF6B |
activating transcription factor 6 beta |
| chr19_+_4402659 | 0.01 |
ENST00000301280.5 ENST00000585854.1 |
CHAF1A |
chromatin assembly factor 1, subunit A (p150) |
| chr21_-_30445886 | 0.01 |
ENST00000431234.1 ENST00000540844.1 ENST00000286788.4 |
CCT8 |
chaperonin containing TCP1, subunit 8 (theta) |
| chrX_+_96138907 | 0.01 |
ENST00000373040.3 |
RPA4 |
replication protein A4, 30kDa |
| chr1_-_168464875 | 0.01 |
ENST00000422253.1 |
RP5-968D22.3 |
RP5-968D22.3 |
| chr4_-_185275104 | 0.01 |
ENST00000317596.3 |
RP11-290F5.2 |
RP11-290F5.2 |
| chr6_+_26087509 | 0.01 |
ENST00000397022.3 ENST00000353147.5 ENST00000352392.4 ENST00000349999.4 ENST00000317896.7 ENST00000357618.5 ENST00000470149.1 ENST00000336625.8 ENST00000461397.1 ENST00000488199.1 |
HFE |
hemochromatosis |
| chr2_+_234826016 | 0.01 |
ENST00000324695.4 ENST00000433712.2 |
TRPM8 |
transient receptor potential cation channel, subfamily M, member 8 |
| chr17_-_3496171 | 0.01 |
ENST00000399756.4 |
TRPV1 |
transient receptor potential cation channel, subfamily V, member 1 |
| chr8_-_116504448 | 0.01 |
ENST00000518018.1 |
TRPS1 |
trichorhinophalangeal syndrome I |
| chr3_+_136649311 | 0.01 |
ENST00000469404.1 ENST00000467911.1 |
NCK1 |
NCK adaptor protein 1 |
| chr19_+_49199209 | 0.01 |
ENST00000522966.1 ENST00000425340.2 ENST00000391876.4 |
FUT2 |
fucosyltransferase 2 (secretor status included) |
| chr12_-_10282681 | 0.01 |
ENST00000533022.1 |
CLEC7A |
C-type lectin domain family 7, member A |
| chr1_+_234765057 | 0.01 |
ENST00000429269.1 |
LINC00184 |
long intergenic non-protein coding RNA 184 |
| chr10_-_128359074 | 0.00 |
ENST00000544758.1 |
C10orf90 |
chromosome 10 open reading frame 90 |
| chr20_-_29978286 | 0.00 |
ENST00000376315.2 |
DEFB119 |
defensin, beta 119 |
| chr1_-_202897724 | 0.00 |
ENST00000435533.3 ENST00000367258.1 |
KLHL12 |
kelch-like family member 12 |
| chr18_-_3845321 | 0.00 |
ENST00000539435.1 ENST00000400147.2 |
DLGAP1 |
discs, large (Drosophila) homolog-associated protein 1 |
| chr3_-_193096600 | 0.00 |
ENST00000446087.1 ENST00000342358.4 |
ATP13A5 |
ATPase type 13A5 |
| chr14_-_90798418 | 0.00 |
ENST00000354366.3 |
NRDE2 |
NRDE-2, necessary for RNA interference, domain containing |
| chr15_+_58702742 | 0.00 |
ENST00000356113.6 ENST00000414170.3 |
LIPC |
lipase, hepatic |
| chr4_+_169418255 | 0.00 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr1_+_13516066 | 0.00 |
ENST00000332192.6 |
PRAMEF21 |
PRAME family member 21 |
| chr6_+_29555683 | 0.00 |
ENST00000383640.2 |
OR2H2 |
olfactory receptor, family 2, subfamily H, member 2 |
| chr14_+_45605157 | 0.00 |
ENST00000542564.2 |
FANCM |
Fanconi anemia, complementation group M |
| chr3_+_134514093 | 0.00 |
ENST00000398015.3 |
EPHB1 |
EPH receptor B1 |
| chr10_-_29923893 | 0.00 |
ENST00000355867.4 |
SVIL |
supervillin |
| chr3_-_20053741 | 0.00 |
ENST00000389050.4 |
PP2D1 |
protein phosphatase 2C-like domain containing 1 |
| chr15_+_79166065 | 0.00 |
ENST00000559690.1 ENST00000559158.1 |
MORF4L1 |
mortality factor 4 like 1 |
| chr10_+_35484793 | 0.00 |
ENST00000488741.1 ENST00000474931.1 ENST00000468236.1 ENST00000344351.5 ENST00000490511.1 |
CREM |
cAMP responsive element modulator |
| chr12_+_104337515 | 0.00 |
ENST00000550595.1 |
HSP90B1 |
heat shock protein 90kDa beta (Grp94), member 1 |
| chr1_-_21377383 | 0.00 |
ENST00000374935.3 |
EIF4G3 |
eukaryotic translation initiation factor 4 gamma, 3 |
| chr7_-_115670804 | 0.00 |
ENST00000320239.7 |
TFEC |
transcription factor EC |
| chr3_-_157221380 | 0.00 |
ENST00000468233.1 |
VEPH1 |
ventricular zone expressed PH domain-containing 1 |
| chr4_+_169418195 | 0.00 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr18_+_61575200 | 0.00 |
ENST00000238508.3 |
SERPINB10 |
serpin peptidase inhibitor, clade B (ovalbumin), member 10 |
| chr15_-_72563585 | 0.00 |
ENST00000287196.9 ENST00000260376.7 |
PARP6 |
poly (ADP-ribose) polymerase family, member 6 |
| chr1_-_13390765 | 0.00 |
ENST00000357367.2 |
PRAMEF8 |
PRAME family member 8 |
| chr10_+_63422695 | 0.00 |
ENST00000330194.2 ENST00000389639.3 |
C10orf107 |
chromosome 10 open reading frame 107 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0061300 | cerebellum vasculature development(GO:0061300) |
| 0.0 | 0.1 | GO:0061346 | orthogonal dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060488) planar dichotomous subdivision of terminal units involved in lung branching morphogenesis(GO:0060489) lateral sprouting involved in lung morphogenesis(GO:0060490) non-canonical Wnt signaling pathway involved in heart development(GO:0061341) planar cell polarity pathway involved in heart morphogenesis(GO:0061346) |
| 0.0 | 0.1 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.0 | 0.1 | GO:0051945 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.0 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.1 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.0 | GO:1904806 | regulation of protein oxidation(GO:1904806) positive regulation of protein oxidation(GO:1904808) |
| 0.0 | 0.1 | GO:0030035 | microspike assembly(GO:0030035) |
| 0.0 | 0.1 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.0 | GO:0090290 | positive regulation of osteoclast proliferation(GO:0090290) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042720 | mitochondrial inner membrane peptidase complex(GO:0042720) |
| 0.0 | 0.1 | GO:0001534 | radial spoke(GO:0001534) |
| 0.0 | 0.1 | GO:0060187 | cell pole(GO:0060187) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.1 | GO:0001665 | alpha-N-acetylgalactosaminide alpha-2,6-sialyltransferase activity(GO:0001665) |