Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
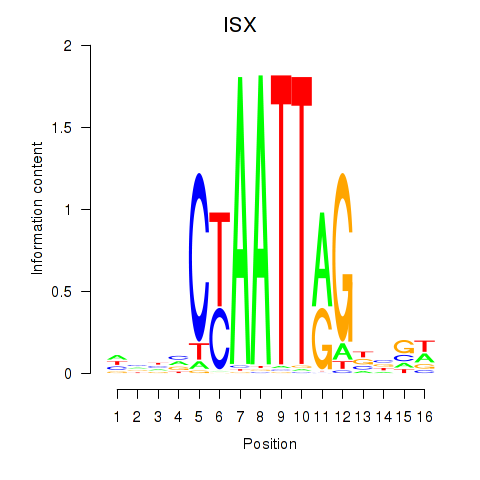
Results for ISX
Z-value: 0.25
Transcription factors associated with ISX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ISX
|
ENSG00000175329.8 | ISX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ISX | hg19_v2_chr22_+_35462129_35462156 | -0.11 | 7.9e-01 | Click! |
Activity profile of ISX motif
Sorted Z-values of ISX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ISX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_138453606 | 0.17 |
ENST00000412923.2 ENST00000344876.4 ENST00000507846.1 ENST00000510305.1 |
PCDH18 |
protocadherin 18 |
| chr6_+_73076432 | 0.13 |
ENST00000414192.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr10_-_28571015 | 0.13 |
ENST00000375719.3 ENST00000375732.1 |
MPP7 |
membrane protein, palmitoylated 7 (MAGUK p55 subfamily member 7) |
| chr17_-_43045439 | 0.13 |
ENST00000253407.3 |
C1QL1 |
complement component 1, q subcomponent-like 1 |
| chr2_+_217524323 | 0.12 |
ENST00000456764.1 |
IGFBP2 |
insulin-like growth factor binding protein 2, 36kDa |
| chr7_+_120628731 | 0.11 |
ENST00000310396.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr11_-_63381925 | 0.11 |
ENST00000415826.1 |
PLA2G16 |
phospholipase A2, group XVI |
| chr2_-_79315112 | 0.09 |
ENST00000305089.3 |
REG1B |
regenerating islet-derived 1 beta |
| chr1_+_151735431 | 0.08 |
ENST00000321531.5 ENST00000315067.8 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr11_+_77532155 | 0.07 |
ENST00000532481.1 ENST00000526415.1 ENST00000393427.2 ENST00000527134.1 ENST00000304716.8 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr19_-_14945933 | 0.07 |
ENST00000322301.3 |
OR7A5 |
olfactory receptor, family 7, subfamily A, member 5 |
| chr3_-_105588231 | 0.07 |
ENST00000545639.1 ENST00000394027.3 ENST00000438603.1 ENST00000447441.1 ENST00000443752.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr4_-_138453559 | 0.07 |
ENST00000511115.1 |
PCDH18 |
protocadherin 18 |
| chr14_-_51027838 | 0.07 |
ENST00000555216.1 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr6_+_34204642 | 0.07 |
ENST00000347617.6 ENST00000401473.3 ENST00000311487.5 ENST00000447654.1 ENST00000395004.3 |
HMGA1 |
high mobility group AT-hook 1 |
| chr7_-_122339162 | 0.07 |
ENST00000340112.2 |
RNF133 |
ring finger protein 133 |
| chr22_+_17956618 | 0.07 |
ENST00000262608.8 |
CECR2 |
cat eye syndrome chromosome region, candidate 2 |
| chr17_-_42277203 | 0.06 |
ENST00000587097.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chr11_+_77532233 | 0.06 |
ENST00000525409.1 |
AAMDC |
adipogenesis associated, Mth938 domain containing |
| chr2_+_204571375 | 0.06 |
ENST00000374478.4 |
CD28 |
CD28 molecule |
| chr15_-_42500351 | 0.06 |
ENST00000348544.4 ENST00000318006.5 |
VPS39 |
vacuolar protein sorting 39 homolog (S. cerevisiae) |
| chr14_-_53258314 | 0.06 |
ENST00000216410.3 ENST00000557604.1 |
GNPNAT1 |
glucosamine-phosphate N-acetyltransferase 1 |
| chr17_+_59489112 | 0.05 |
ENST00000335108.2 |
C17orf82 |
chromosome 17 open reading frame 82 |
| chr2_-_227050079 | 0.05 |
ENST00000423838.1 |
AC068138.1 |
AC068138.1 |
| chr11_+_75526212 | 0.05 |
ENST00000356136.3 |
UVRAG |
UV radiation resistance associated |
| chr22_-_24316648 | 0.05 |
ENST00000403754.3 ENST00000430101.2 ENST00000398344.4 |
DDT |
D-dopachrome tautomerase |
| chr12_+_3000073 | 0.05 |
ENST00000397132.2 |
TULP3 |
tubby like protein 3 |
| chr5_-_147286065 | 0.05 |
ENST00000318315.4 ENST00000515291.1 |
C5orf46 |
chromosome 5 open reading frame 46 |
| chr5_-_35938674 | 0.05 |
ENST00000397366.1 ENST00000513623.1 ENST00000514524.1 ENST00000397367.2 |
CAPSL |
calcyphosine-like |
| chr22_+_24309089 | 0.05 |
ENST00000215770.5 |
DDTL |
D-dopachrome tautomerase-like |
| chr1_-_150849174 | 0.05 |
ENST00000515192.1 |
ARNT |
aryl hydrocarbon receptor nuclear translocator |
| chr8_-_82395461 | 0.05 |
ENST00000256104.4 |
FABP4 |
fatty acid binding protein 4, adipocyte |
| chr6_+_127898312 | 0.05 |
ENST00000329722.7 |
C6orf58 |
chromosome 6 open reading frame 58 |
| chr12_+_26348246 | 0.05 |
ENST00000422622.2 |
SSPN |
sarcospan |
| chr12_+_7013897 | 0.04 |
ENST00000007969.8 ENST00000323702.5 |
LRRC23 |
leucine rich repeat containing 23 |
| chr12_+_7014064 | 0.04 |
ENST00000443597.2 |
LRRC23 |
leucine rich repeat containing 23 |
| chr1_-_150849047 | 0.04 |
ENST00000354396.2 ENST00000505755.1 |
ARNT |
aryl hydrocarbon receptor nuclear translocator |
| chr10_+_135160844 | 0.04 |
ENST00000423766.1 ENST00000458230.1 |
PRAP1 |
proline-rich acidic protein 1 |
| chr11_-_63381823 | 0.04 |
ENST00000323646.5 |
PLA2G16 |
phospholipase A2, group XVI |
| chr8_+_105235572 | 0.04 |
ENST00000523362.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr17_-_18266765 | 0.04 |
ENST00000354098.3 |
SHMT1 |
serine hydroxymethyltransferase 1 (soluble) |
| chr2_-_3521518 | 0.04 |
ENST00000382093.5 |
ADI1 |
acireductone dioxygenase 1 |
| chr22_-_24303340 | 0.04 |
ENST00000404172.3 ENST00000290765.4 |
GSTT2B |
glutathione S-transferase theta 2B (gene/pseudogene) |
| chr8_-_12612962 | 0.04 |
ENST00000398246.3 |
LONRF1 |
LON peptidase N-terminal domain and ring finger 1 |
| chr22_+_24322322 | 0.04 |
ENST00000215780.5 ENST00000402588.3 |
GSTT2 |
glutathione S-transferase theta 2 |
| chr6_+_160542821 | 0.04 |
ENST00000366963.4 |
SLC22A1 |
solute carrier family 22 (organic cation transporter), member 1 |
| chr15_+_55700741 | 0.04 |
ENST00000569691.1 |
C15orf65 |
chromosome 15 open reading frame 65 |
| chr3_-_15563229 | 0.04 |
ENST00000383786.5 ENST00000383787.2 ENST00000383785.2 ENST00000383788.5 ENST00000603808.1 |
COLQ |
collagen-like tail subunit (single strand of homotrimer) of asymmetric acetylcholinesterase |
| chr6_+_148663729 | 0.04 |
ENST00000367467.3 |
SASH1 |
SAM and SH3 domain containing 1 |
| chr8_-_7243080 | 0.04 |
ENST00000400156.4 |
ZNF705G |
zinc finger protein 705G |
| chr19_-_40931891 | 0.04 |
ENST00000357949.4 |
SERTAD1 |
SERTA domain containing 1 |
| chr6_-_85474219 | 0.04 |
ENST00000369663.5 |
TBX18 |
T-box 18 |
| chr12_-_120966943 | 0.04 |
ENST00000552443.1 ENST00000547736.1 ENST00000445328.2 ENST00000547943.1 ENST00000288532.6 |
COQ5 |
coenzyme Q5 homolog, methyltransferase (S. cerevisiae) |
| chr6_+_36164487 | 0.04 |
ENST00000357641.6 |
BRPF3 |
bromodomain and PHD finger containing, 3 |
| chr1_-_150849208 | 0.03 |
ENST00000358595.5 |
ARNT |
aryl hydrocarbon receptor nuclear translocator |
| chr1_-_36916066 | 0.03 |
ENST00000315643.9 |
OSCP1 |
organic solute carrier partner 1 |
| chr10_-_4285923 | 0.03 |
ENST00000418372.1 ENST00000608792.1 |
LINC00702 |
long intergenic non-protein coding RNA 702 |
| chr11_-_124981475 | 0.03 |
ENST00000532156.1 ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218 |
transmembrane protein 218 |
| chr6_+_160542870 | 0.03 |
ENST00000324965.4 ENST00000457470.2 |
SLC22A1 |
solute carrier family 22 (organic cation transporter), member 1 |
| chr9_+_99212403 | 0.03 |
ENST00000375251.3 ENST00000375249.4 |
HABP4 |
hyaluronan binding protein 4 |
| chr12_+_69201923 | 0.03 |
ENST00000462284.1 ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2 |
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr12_+_3000037 | 0.03 |
ENST00000544943.1 ENST00000448120.2 |
TULP3 |
tubby like protein 3 |
| chr6_+_39760129 | 0.03 |
ENST00000274867.4 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr1_-_68698197 | 0.03 |
ENST00000370973.2 ENST00000370971.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr1_-_36915880 | 0.03 |
ENST00000445843.3 |
OSCP1 |
organic solute carrier partner 1 |
| chr3_-_105587879 | 0.03 |
ENST00000264122.4 ENST00000403724.1 ENST00000405772.1 |
CBLB |
Cbl proto-oncogene B, E3 ubiquitin protein ligase |
| chr12_+_104680659 | 0.03 |
ENST00000526691.1 ENST00000531691.1 ENST00000388854.3 ENST00000354940.6 ENST00000526390.1 ENST00000531689.1 |
TXNRD1 |
thioredoxin reductase 1 |
| chr1_+_32666188 | 0.03 |
ENST00000421922.2 |
CCDC28B |
coiled-coil domain containing 28B |
| chr17_-_57229155 | 0.03 |
ENST00000584089.1 |
SKA2 |
spindle and kinetochore associated complex subunit 2 |
| chr12_-_53730147 | 0.03 |
ENST00000536324.2 |
SP7 |
Sp7 transcription factor |
| chr17_-_40337470 | 0.03 |
ENST00000293330.1 |
HCRT |
hypocretin (orexin) neuropeptide precursor |
| chr14_+_22964877 | 0.03 |
ENST00000390494.1 |
TRAJ43 |
T cell receptor alpha joining 43 |
| chr1_-_36916011 | 0.03 |
ENST00000356637.5 ENST00000354267.3 ENST00000235532.5 |
OSCP1 |
organic solute carrier partner 1 |
| chr5_-_177580855 | 0.03 |
ENST00000514354.1 ENST00000511078.1 |
NHP2 |
NHP2 ribonucleoprotein |
| chr19_-_46285736 | 0.03 |
ENST00000291270.4 ENST00000447742.2 ENST00000354227.5 |
DMPK |
dystrophia myotonica-protein kinase |
| chrX_-_153363188 | 0.03 |
ENST00000303391.6 |
MECP2 |
methyl CpG binding protein 2 (Rett syndrome) |
| chr20_+_44441215 | 0.03 |
ENST00000356455.4 ENST00000405520.1 |
UBE2C |
ubiquitin-conjugating enzyme E2C |
| chr4_+_66536248 | 0.03 |
ENST00000514260.1 ENST00000507117.1 |
RP11-807H7.1 |
RP11-807H7.1 |
| chr11_+_33061543 | 0.03 |
ENST00000432887.1 ENST00000528898.1 ENST00000531632.2 |
TCP11L1 |
t-complex 11, testis-specific-like 1 |
| chrX_-_8139308 | 0.03 |
ENST00000317103.4 |
VCX2 |
variable charge, X-linked 2 |
| chr4_-_66536057 | 0.03 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr22_-_32766972 | 0.03 |
ENST00000382084.4 ENST00000382086.2 |
RFPL3S |
RFPL3 antisense |
| chr16_-_31105870 | 0.03 |
ENST00000394971.3 |
VKORC1 |
vitamin K epoxide reductase complex, subunit 1 |
| chr5_-_177580777 | 0.03 |
ENST00000314397.4 |
NHP2 |
NHP2 ribonucleoprotein |
| chr13_+_38923959 | 0.03 |
ENST00000379649.1 ENST00000239878.4 ENST00000437952.1 ENST00000379641.1 |
UFM1 |
ubiquitin-fold modifier 1 |
| chr17_+_78965624 | 0.03 |
ENST00000325167.5 |
CHMP6 |
charged multivesicular body protein 6 |
| chr19_-_51071302 | 0.03 |
ENST00000389201.3 ENST00000600381.1 |
LRRC4B |
leucine rich repeat containing 4B |
| chr20_-_35580104 | 0.03 |
ENST00000373694.5 |
SAMHD1 |
SAM domain and HD domain 1 |
| chr10_-_99447024 | 0.03 |
ENST00000370626.3 |
AVPI1 |
arginine vasopressin-induced 1 |
| chr5_-_177580933 | 0.03 |
ENST00000274606.3 |
NHP2 |
NHP2 ribonucleoprotein |
| chr17_-_18266797 | 0.03 |
ENST00000316694.3 ENST00000539052.1 |
SHMT1 |
serine hydroxymethyltransferase 1 (soluble) |
| chr5_+_159848807 | 0.02 |
ENST00000352433.5 |
PTTG1 |
pituitary tumor-transforming 1 |
| chr3_+_115342349 | 0.02 |
ENST00000393780.3 |
GAP43 |
growth associated protein 43 |
| chr14_+_57671888 | 0.02 |
ENST00000391612.1 |
AL391152.1 |
AL391152.1 |
| chr19_-_46285646 | 0.02 |
ENST00000458663.2 |
DMPK |
dystrophia myotonica-protein kinase |
| chr7_+_142985308 | 0.02 |
ENST00000310447.5 |
CASP2 |
caspase 2, apoptosis-related cysteine peptidase |
| chr3_+_148709128 | 0.02 |
ENST00000345003.4 ENST00000296048.6 ENST00000483267.1 |
GYG1 |
glycogenin 1 |
| chr17_-_40828969 | 0.02 |
ENST00000591022.1 ENST00000587627.1 ENST00000293349.6 |
PLEKHH3 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 3 |
| chr10_-_99205607 | 0.02 |
ENST00000477692.2 ENST00000485122.2 ENST00000370886.5 ENST00000370885.4 ENST00000370902.3 ENST00000370884.5 |
EXOSC1 |
exosome component 1 |
| chr3_-_164796269 | 0.02 |
ENST00000264382.3 |
SI |
sucrase-isomaltase (alpha-glucosidase) |
| chr16_+_12059050 | 0.02 |
ENST00000396495.3 |
TNFRSF17 |
tumor necrosis factor receptor superfamily, member 17 |
| chr10_+_114206956 | 0.02 |
ENST00000432306.1 ENST00000393077.2 |
VTI1A |
vesicle transport through interaction with t-SNAREs 1A |
| chr16_-_31106048 | 0.02 |
ENST00000300851.6 |
VKORC1 |
vitamin K epoxide reductase complex, subunit 1 |
| chr17_-_7307358 | 0.02 |
ENST00000576017.1 ENST00000302422.3 ENST00000535512.1 |
TMEM256 TMEM256-PLSCR3 |
transmembrane protein 256 TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr4_+_108911036 | 0.02 |
ENST00000505878.1 |
HADH |
hydroxyacyl-CoA dehydrogenase |
| chrX_+_108779004 | 0.02 |
ENST00000218004.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr14_+_104182061 | 0.02 |
ENST00000216602.6 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr20_+_44441304 | 0.02 |
ENST00000352551.5 |
UBE2C |
ubiquitin-conjugating enzyme E2C |
| chr16_-_31106211 | 0.02 |
ENST00000532364.1 ENST00000529564.1 ENST00000319788.7 ENST00000354895.4 ENST00000394975.2 |
RP11-196G11.1 VKORC1 |
Uncharacterized protein vitamin K epoxide reductase complex, subunit 1 |
| chr17_-_79650818 | 0.02 |
ENST00000397498.4 |
ARL16 |
ADP-ribosylation factor-like 16 |
| chr14_+_32798462 | 0.02 |
ENST00000280979.4 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr15_-_75199213 | 0.02 |
ENST00000562698.1 |
FAM219B |
family with sequence similarity 219, member B |
| chr22_-_29137771 | 0.02 |
ENST00000439200.1 ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2 |
checkpoint kinase 2 |
| chr16_+_89160217 | 0.02 |
ENST00000317447.4 ENST00000537290.1 |
ACSF3 |
acyl-CoA synthetase family member 3 |
| chr10_-_105110831 | 0.02 |
ENST00000337211.4 |
PCGF6 |
polycomb group ring finger 6 |
| chr22_-_32767017 | 0.02 |
ENST00000400234.1 |
RFPL3S |
RFPL3 antisense |
| chrX_-_153363125 | 0.02 |
ENST00000407218.1 ENST00000453960.2 |
MECP2 |
methyl CpG binding protein 2 (Rett syndrome) |
| chr1_-_150669500 | 0.02 |
ENST00000271732.3 |
GOLPH3L |
golgi phosphoprotein 3-like |
| chr10_-_1095050 | 0.02 |
ENST00000381344.3 |
IDI1 |
isopentenyl-diphosphate delta isomerase 1 |
| chr20_+_44441271 | 0.02 |
ENST00000335046.3 ENST00000243893.6 |
UBE2C |
ubiquitin-conjugating enzyme E2C |
| chr11_-_62521614 | 0.02 |
ENST00000527994.1 ENST00000394807.3 |
ZBTB3 |
zinc finger and BTB domain containing 3 |
| chr20_+_31805131 | 0.02 |
ENST00000375454.3 ENST00000375452.3 |
BPIFA3 |
BPI fold containing family A, member 3 |
| chr16_+_31724552 | 0.02 |
ENST00000539915.1 ENST00000316491.9 ENST00000399681.3 ENST00000398696.3 ENST00000534369.1 |
ZNF720 |
zinc finger protein 720 |
| chr16_+_1359511 | 0.02 |
ENST00000397514.3 ENST00000397515.2 ENST00000567383.1 ENST00000403747.2 ENST00000566587.1 |
UBE2I |
ubiquitin-conjugating enzyme E2I |
| chr3_-_186288097 | 0.02 |
ENST00000446782.1 |
TBCCD1 |
TBCC domain containing 1 |
| chr12_+_7014126 | 0.02 |
ENST00000415834.1 ENST00000436789.1 |
LRRC23 |
leucine rich repeat containing 23 |
| chr10_+_1095416 | 0.02 |
ENST00000358220.1 |
WDR37 |
WD repeat domain 37 |
| chr17_-_79650689 | 0.02 |
ENST00000574938.1 ENST00000570561.1 ENST00000573392.1 ENST00000576135.1 |
ARL16 |
ADP-ribosylation factor-like 16 |
| chr11_+_95523621 | 0.02 |
ENST00000325542.5 ENST00000325486.5 ENST00000544522.1 ENST00000541365.1 |
CEP57 |
centrosomal protein 57kDa |
| chrX_+_77154935 | 0.02 |
ENST00000481445.1 |
COX7B |
cytochrome c oxidase subunit VIIb |
| chr14_+_104182105 | 0.02 |
ENST00000311141.2 |
ZFYVE21 |
zinc finger, FYVE domain containing 21 |
| chr9_+_136501478 | 0.02 |
ENST00000393056.2 ENST00000263611.2 |
DBH |
dopamine beta-hydroxylase (dopamine beta-monooxygenase) |
| chr1_+_155278539 | 0.02 |
ENST00000447866.1 |
FDPS |
farnesyl diphosphate synthase |
| chr2_+_242089833 | 0.02 |
ENST00000404405.3 ENST00000439916.1 ENST00000406106.3 ENST00000401987.1 |
PPP1R7 |
protein phosphatase 1, regulatory subunit 7 |
| chr1_+_155278625 | 0.02 |
ENST00000368356.4 ENST00000356657.6 |
FDPS |
farnesyl diphosphate synthase |
| chrX_-_47509887 | 0.02 |
ENST00000247161.3 ENST00000592066.1 ENST00000376983.3 |
ELK1 |
ELK1, member of ETS oncogene family |
| chr14_-_24711470 | 0.02 |
ENST00000559969.1 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr4_-_70080449 | 0.02 |
ENST00000446444.1 |
UGT2B11 |
UDP glucuronosyltransferase 2 family, polypeptide B11 |
| chr2_+_86947296 | 0.02 |
ENST00000283632.4 |
RMND5A |
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr14_+_32798547 | 0.02 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr1_-_161208013 | 0.02 |
ENST00000515452.1 ENST00000367983.4 |
NR1I3 |
nuclear receptor subfamily 1, group I, member 3 |
| chr4_-_186733363 | 0.02 |
ENST00000393523.2 ENST00000393528.3 ENST00000449407.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr3_+_111393501 | 0.02 |
ENST00000393934.3 |
PLCXD2 |
phosphatidylinositol-specific phospholipase C, X domain containing 2 |
| chr5_+_159848854 | 0.02 |
ENST00000517480.1 ENST00000520452.1 ENST00000393964.1 |
PTTG1 |
pituitary tumor-transforming 1 |
| chr8_+_92261516 | 0.01 |
ENST00000276609.3 ENST00000309536.2 |
SLC26A7 |
solute carrier family 26 (anion exchanger), member 7 |
| chr10_-_48416849 | 0.01 |
ENST00000249598.1 |
GDF2 |
growth differentiation factor 2 |
| chr7_-_36764004 | 0.01 |
ENST00000431169.1 |
AOAH |
acyloxyacyl hydrolase (neutrophil) |
| chr4_-_177190364 | 0.01 |
ENST00000296525.3 |
ASB5 |
ankyrin repeat and SOCS box containing 5 |
| chr6_-_30080863 | 0.01 |
ENST00000540829.1 |
TRIM31 |
tripartite motif containing 31 |
| chr4_-_122686261 | 0.01 |
ENST00000337677.5 |
TMEM155 |
transmembrane protein 155 |
| chr17_-_18266660 | 0.01 |
ENST00000582653.1 ENST00000352886.6 |
SHMT1 |
serine hydroxymethyltransferase 1 (soluble) |
| chrX_+_8433376 | 0.01 |
ENST00000440654.2 ENST00000381029.4 |
VCX3B |
variable charge, X-linked 3B |
| chr17_+_46970134 | 0.01 |
ENST00000503641.1 ENST00000514808.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr17_-_60883993 | 0.01 |
ENST00000583803.1 ENST00000456609.2 |
MARCH10 |
membrane-associated ring finger (C3HC4) 10, E3 ubiquitin protein ligase |
| chr1_-_20250110 | 0.01 |
ENST00000375116.3 |
PLA2G2E |
phospholipase A2, group IIE |
| chr19_-_6424783 | 0.01 |
ENST00000398148.3 |
KHSRP |
KH-type splicing regulatory protein |
| chr20_-_50722183 | 0.01 |
ENST00000371523.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr8_+_9413410 | 0.01 |
ENST00000520408.1 ENST00000310430.6 ENST00000522110.1 |
TNKS |
tankyrase, TRF1-interacting ankyrin-related ADP-ribose polymerase |
| chr6_-_10115007 | 0.01 |
ENST00000485268.1 |
OFCC1 |
orofacial cleft 1 candidate 1 |
| chr19_+_50270219 | 0.01 |
ENST00000354293.5 ENST00000359032.5 |
AP2A1 |
adaptor-related protein complex 2, alpha 1 subunit |
| chrX_+_43515467 | 0.01 |
ENST00000338702.3 ENST00000542639.1 |
MAOA |
monoamine oxidase A |
| chr11_-_121986923 | 0.01 |
ENST00000560104.1 |
BLID |
BH3-like motif containing, cell death inducer |
| chr20_+_24449821 | 0.01 |
ENST00000376862.3 |
SYNDIG1 |
synapse differentiation inducing 1 |
| chr11_-_77532050 | 0.01 |
ENST00000308488.6 |
RSF1 |
remodeling and spacing factor 1 |
| chr6_-_31125850 | 0.01 |
ENST00000507751.1 ENST00000448162.2 ENST00000502557.1 ENST00000503420.1 ENST00000507892.1 ENST00000507226.1 ENST00000513222.1 ENST00000503934.1 ENST00000396263.2 ENST00000508683.1 ENST00000428174.1 ENST00000448141.2 ENST00000507829.1 ENST00000455279.2 ENST00000376266.5 |
CCHCR1 |
coiled-coil alpha-helical rod protein 1 |
| chr22_-_50523760 | 0.01 |
ENST00000395876.2 |
MLC1 |
megalencephalic leukoencephalopathy with subcortical cysts 1 |
| chr1_+_40713573 | 0.01 |
ENST00000372766.3 |
TMCO2 |
transmembrane and coiled-coil domains 2 |
| chrX_+_77166172 | 0.01 |
ENST00000343533.5 ENST00000350425.4 ENST00000341514.6 |
ATP7A |
ATPase, Cu++ transporting, alpha polypeptide |
| chr7_+_50348268 | 0.01 |
ENST00000438033.1 ENST00000439701.1 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
| chr4_-_69434245 | 0.01 |
ENST00000317746.2 |
UGT2B17 |
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr22_+_40322595 | 0.01 |
ENST00000420971.1 ENST00000544756.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr17_+_46970178 | 0.01 |
ENST00000393366.2 ENST00000506855.1 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr5_+_177631497 | 0.01 |
ENST00000358344.3 |
HNRNPAB |
heterogeneous nuclear ribonucleoprotein A/B |
| chr22_+_40322623 | 0.01 |
ENST00000399090.2 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr3_-_46069223 | 0.01 |
ENST00000309285.3 |
XCR1 |
chemokine (C motif) receptor 1 |
| chr5_+_177631523 | 0.01 |
ENST00000506339.1 ENST00000355836.5 ENST00000514633.1 ENST00000515193.1 ENST00000506259.1 ENST00000504898.1 |
HNRNPAB |
heterogeneous nuclear ribonucleoprotein A/B |
| chr17_+_46970127 | 0.01 |
ENST00000355938.5 |
ATP5G1 |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit C1 (subunit 9) |
| chr14_-_24711806 | 0.01 |
ENST00000540705.1 ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr19_+_13001840 | 0.01 |
ENST00000222214.5 ENST00000589039.1 ENST00000591470.1 ENST00000457854.1 ENST00000422947.2 ENST00000588905.1 ENST00000587072.1 |
GCDH |
glutaryl-CoA dehydrogenase |
| chr19_-_10227503 | 0.01 |
ENST00000593054.1 |
EIF3G |
eukaryotic translation initiation factor 3, subunit G |
| chr3_+_133292574 | 0.01 |
ENST00000264993.3 |
CDV3 |
CDV3 homolog (mouse) |
| chr6_-_49681235 | 0.01 |
ENST00000339139.4 |
CRISP2 |
cysteine-rich secretory protein 2 |
| chr7_-_100493482 | 0.01 |
ENST00000411582.1 ENST00000419336.2 ENST00000241069.5 ENST00000302913.4 |
ACHE |
acetylcholinesterase (Yt blood group) |
| chr6_+_36165133 | 0.01 |
ENST00000446974.1 ENST00000454960.1 |
BRPF3 |
bromodomain and PHD finger containing, 3 |
| chr14_-_24711865 | 0.01 |
ENST00000399423.4 ENST00000267415.7 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chrX_+_37639302 | 0.01 |
ENST00000545017.1 ENST00000536160.1 |
CYBB |
cytochrome b-245, beta polypeptide |
| chr15_+_80351910 | 0.01 |
ENST00000261749.6 ENST00000561060.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chrX_+_8432871 | 0.01 |
ENST00000381032.1 ENST00000453306.1 ENST00000444481.1 |
VCX3B |
variable charge, X-linked 3B |
| chr4_+_144303093 | 0.01 |
ENST00000505913.1 |
GAB1 |
GRB2-associated binding protein 1 |
| chr1_+_23345930 | 0.01 |
ENST00000356634.3 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr7_-_86849883 | 0.01 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr12_-_57039739 | 0.01 |
ENST00000552959.1 ENST00000551020.1 ENST00000553007.2 ENST00000552919.1 ENST00000552104.1 ENST00000262030.3 |
ATP5B |
ATP synthase, H+ transporting, mitochondrial F1 complex, beta polypeptide |
| chr1_-_68698222 | 0.01 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr8_+_50824233 | 0.01 |
ENST00000522124.1 |
SNTG1 |
syntrophin, gamma 1 |
| chr16_+_89160269 | 0.01 |
ENST00000540697.1 ENST00000406948.3 ENST00000378345.4 ENST00000541755.2 |
ACSF3 |
acyl-CoA synthetase family member 3 |
| chr1_+_23345943 | 0.01 |
ENST00000400181.4 ENST00000542151.1 |
KDM1A |
lysine (K)-specific demethylase 1A |
| chr12_+_15125954 | 0.01 |
ENST00000266395.2 |
PDE6H |
phosphodiesterase 6H, cGMP-specific, cone, gamma |
| chr19_-_47137942 | 0.01 |
ENST00000300873.4 |
GNG8 |
guanine nucleotide binding protein (G protein), gamma 8 |
| chr14_-_20801427 | 0.01 |
ENST00000557665.1 ENST00000358932.4 ENST00000353689.4 |
CCNB1IP1 |
cyclin B1 interacting protein 1, E3 ubiquitin protein ligase |
| chr14_-_73925225 | 0.01 |
ENST00000356296.4 ENST00000355058.3 ENST00000359560.3 ENST00000557597.1 ENST00000554394.1 ENST00000555238.1 ENST00000535282.1 ENST00000555987.1 ENST00000555394.1 ENST00000554546.1 |
NUMB |
numb homolog (Drosophila) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.0 | 0.1 | GO:0060434 | bronchus morphogenesis(GO:0060434) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 0.1 | GO:0046485 | ether lipid metabolic process(GO:0046485) |
| 0.0 | 0.1 | GO:0071896 | protein localization to adherens junction(GO:0071896) |
| 0.0 | 0.1 | GO:0090402 | oncogene-induced cell senescence(GO:0090402) |
| 0.0 | 0.0 | GO:0060823 | canonical Wnt signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060823) |
| 0.0 | 0.1 | GO:0051684 | maintenance of Golgi location(GO:0051684) |
| 0.0 | 0.0 | GO:1904404 | cellular response to vitamin B1(GO:0071301) response to formaldehyde(GO:1904404) |
| 0.0 | 0.1 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.1 | GO:0048241 | epinephrine transport(GO:0048241) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0044301 | climbing fiber(GO:0044301) |
| 0.0 | 0.1 | GO:0090661 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0052740 | 1-acyl-2-lysophosphatidylserine acylhydrolase activity(GO:0052740) |
| 0.0 | 0.1 | GO:0005277 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.1 | GO:0070905 | serine binding(GO:0070905) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0047057 | oxidoreductase activity, acting on the CH-OH group of donors, disulfide as acceptor(GO:0016900) vitamin-K-epoxide reductase (warfarin-sensitive) activity(GO:0047057) |
| 0.0 | 0.0 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.0 | GO:0033981 | D-dopachrome decarboxylase activity(GO:0033981) |
| 0.0 | 0.1 | GO:0031995 | insulin-like growth factor II binding(GO:0031995) |
| 0.0 | 0.0 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |