Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
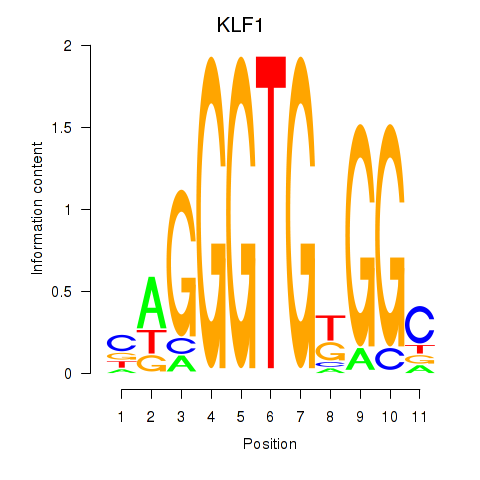
Results for KLF1
Z-value: 2.15
Transcription factors associated with KLF1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF1
|
ENSG00000105610.4 | KLF1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF1 | hg19_v2_chr19_-_12997995_12998021 | 0.51 | 1.9e-01 | Click! |
Activity profile of KLF1 motif
Sorted Z-values of KLF1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr15_+_43885252 | 7.07 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr15_+_43985084 | 6.82 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr19_-_51522955 | 5.69 |
ENST00000358789.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr19_-_36004543 | 4.53 |
ENST00000339686.3 ENST00000447113.2 ENST00000440396.1 |
DMKN |
dermokine |
| chr1_+_209602156 | 4.46 |
ENST00000429156.1 ENST00000366437.3 ENST00000603283.1 ENST00000431096.1 |
MIR205HG |
MIR205 host gene (non-protein coding) |
| chr18_+_29027696 | 4.44 |
ENST00000257189.4 |
DSG3 |
desmoglein 3 |
| chr1_+_44398943 | 3.48 |
ENST00000372359.5 ENST00000414809.3 |
ARTN |
artemin |
| chr19_-_51523412 | 3.47 |
ENST00000391805.1 ENST00000599077.1 |
KLK10 |
kallikrein-related peptidase 10 |
| chr18_+_21452964 | 3.41 |
ENST00000587184.1 |
LAMA3 |
laminin, alpha 3 |
| chr16_-_31147020 | 3.41 |
ENST00000568261.1 ENST00000567797.1 ENST00000317508.6 |
PRSS8 |
protease, serine, 8 |
| chr18_+_21452804 | 3.33 |
ENST00000269217.6 |
LAMA3 |
laminin, alpha 3 |
| chr16_+_67233007 | 3.31 |
ENST00000360833.1 ENST00000393997.2 |
ELMO3 |
engulfment and cell motility 3 |
| chr1_+_44399466 | 3.29 |
ENST00000498139.2 ENST00000491846.1 |
ARTN |
artemin |
| chr19_-_51523275 | 3.25 |
ENST00000309958.3 |
KLK10 |
kallikrein-related peptidase 10 |
| chr10_+_47746929 | 3.18 |
ENST00000340243.6 ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2 AL603965.1 |
annexin A8-like 2 Protein LOC100996760 |
| chr6_+_150464155 | 3.15 |
ENST00000361131.4 |
PPP1R14C |
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr17_-_39507064 | 3.14 |
ENST00000007735.3 |
KRT33A |
keratin 33A |
| chr12_-_8815215 | 3.07 |
ENST00000544889.1 ENST00000543369.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr12_-_8815404 | 3.02 |
ENST00000359478.2 ENST00000396549.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr12_-_8815299 | 2.99 |
ENST00000535336.1 |
MFAP5 |
microfibrillar associated protein 5 |
| chr19_+_35606692 | 2.96 |
ENST00000406242.3 ENST00000454903.2 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
| chr14_-_105635090 | 2.70 |
ENST00000331782.3 ENST00000347004.2 |
JAG2 |
jagged 2 |
| chr17_-_34122596 | 2.65 |
ENST00000250144.8 |
MMP28 |
matrix metallopeptidase 28 |
| chr17_-_7493390 | 2.63 |
ENST00000538513.2 ENST00000570788.1 ENST00000250055.2 |
SOX15 |
SRY (sex determining region Y)-box 15 |
| chr8_-_21988558 | 2.60 |
ENST00000312841.8 |
HR |
hair growth associated |
| chrX_-_153151586 | 2.57 |
ENST00000370060.1 ENST00000370055.1 ENST00000420165.1 |
L1CAM |
L1 cell adhesion molecule |
| chr19_+_35607166 | 2.24 |
ENST00000604255.1 ENST00000346446.5 ENST00000344013.6 ENST00000603449.1 ENST00000406988.1 ENST00000605550.1 ENST00000604804.1 ENST00000605552.1 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
| chr18_-_28682374 | 2.00 |
ENST00000280904.6 |
DSC2 |
desmocollin 2 |
| chr19_+_35606777 | 2.00 |
ENST00000604404.1 ENST00000435734.2 ENST00000603181.1 |
FXYD3 |
FXYD domain containing ion transport regulator 3 |
| chr2_+_173292301 | 1.98 |
ENST00000264106.6 ENST00000375221.2 ENST00000343713.4 |
ITGA6 |
integrin, alpha 6 |
| chr1_-_117210290 | 1.95 |
ENST00000369483.1 ENST00000369486.3 |
IGSF3 |
immunoglobulin superfamily, member 3 |
| chr6_+_30850697 | 1.93 |
ENST00000509639.1 ENST00000412274.2 ENST00000507901.1 ENST00000507046.1 ENST00000437124.2 ENST00000454612.2 ENST00000396342.2 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr2_+_17721920 | 1.93 |
ENST00000295156.4 |
VSNL1 |
visinin-like 1 |
| chr11_+_1860682 | 1.92 |
ENST00000381906.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr11_+_1860832 | 1.84 |
ENST00000252898.7 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr10_-_47173994 | 1.84 |
ENST00000414655.2 ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1 LINC00842 |
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr17_-_46507537 | 1.81 |
ENST00000336915.6 |
SKAP1 |
src kinase associated phosphoprotein 1 |
| chr8_-_75233563 | 1.77 |
ENST00000342232.4 |
JPH1 |
junctophilin 1 |
| chr2_+_182321925 | 1.73 |
ENST00000339307.4 ENST00000397033.2 |
ITGA4 |
integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) |
| chr12_-_85306562 | 1.67 |
ENST00000551612.1 ENST00000450363.3 ENST00000552192.1 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr4_+_48018781 | 1.63 |
ENST00000295461.5 |
NIPAL1 |
NIPA-like domain containing 1 |
| chr6_+_7541808 | 1.63 |
ENST00000379802.3 |
DSP |
desmoplakin |
| chr14_-_23623577 | 1.63 |
ENST00000422941.2 ENST00000453702.1 |
SLC7A8 |
solute carrier family 7 (amino acid transporter light chain, L system), member 8 |
| chr2_-_74667612 | 1.55 |
ENST00000305557.5 ENST00000233330.6 |
RTKN |
rhotekin |
| chr2_+_85360499 | 1.52 |
ENST00000282111.3 |
TCF7L1 |
transcription factor 7-like 1 (T-cell specific, HMG-box) |
| chr17_-_7166500 | 1.51 |
ENST00000575313.1 ENST00000397317.4 |
CLDN7 |
claudin 7 |
| chr2_+_173292390 | 1.50 |
ENST00000442250.1 ENST00000458358.1 ENST00000409080.1 |
ITGA6 |
integrin, alpha 6 |
| chr2_+_173292280 | 1.48 |
ENST00000264107.7 |
ITGA6 |
integrin, alpha 6 |
| chr12_-_8815477 | 1.46 |
ENST00000433590.2 |
MFAP5 |
microfibrillar associated protein 5 |
| chr6_+_106546808 | 1.45 |
ENST00000369089.3 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr5_+_140345820 | 1.45 |
ENST00000289269.5 |
PCDHAC2 |
protocadherin alpha subfamily C, 2 |
| chr12_+_29302119 | 1.43 |
ENST00000536681.3 |
FAR2 |
fatty acyl CoA reductase 2 |
| chr2_-_38604398 | 1.40 |
ENST00000443098.1 ENST00000449130.1 ENST00000378954.4 ENST00000539122.1 ENST00000419554.2 ENST00000451483.1 ENST00000406122.1 |
ATL2 |
atlastin GTPase 2 |
| chr19_+_1077393 | 1.39 |
ENST00000590577.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr1_-_6526192 | 1.35 |
ENST00000377782.3 ENST00000351959.5 ENST00000356876.3 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
| chr1_+_224803995 | 1.35 |
ENST00000272133.3 |
CNIH3 |
cornichon family AMPA receptor auxiliary protein 3 |
| chr12_-_85306594 | 1.33 |
ENST00000266682.5 |
SLC6A15 |
solute carrier family 6 (neutral amino acid transporter), member 15 |
| chr6_-_138428613 | 1.32 |
ENST00000421351.3 |
PERP |
PERP, TP53 apoptosis effector |
| chr11_+_1861399 | 1.32 |
ENST00000381905.3 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr5_+_89770664 | 1.30 |
ENST00000503973.1 ENST00000399107.1 |
POLR3G |
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr11_+_394196 | 1.29 |
ENST00000331563.2 ENST00000531857.1 |
PKP3 |
plakophilin 3 |
| chr17_-_39942940 | 1.28 |
ENST00000310706.5 ENST00000393931.3 ENST00000424457.1 ENST00000591690.1 |
JUP |
junction plakoglobin |
| chr4_-_25865159 | 1.27 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr19_+_41699135 | 1.27 |
ENST00000542619.1 ENST00000600561.1 |
CYP2S1 |
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr5_+_89770696 | 1.24 |
ENST00000504930.1 ENST00000514483.1 |
POLR3G |
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr7_+_86274145 | 1.24 |
ENST00000439827.1 ENST00000394720.2 ENST00000421579.1 |
GRM3 |
glutamate receptor, metabotropic 3 |
| chr1_+_18434240 | 1.14 |
ENST00000251296.1 |
IGSF21 |
immunoglobin superfamily, member 21 |
| chr2_-_26205340 | 1.14 |
ENST00000264712.3 |
KIF3C |
kinesin family member 3C |
| chr4_-_90756769 | 1.13 |
ENST00000345009.4 ENST00000505199.1 ENST00000502987.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr5_-_16617162 | 1.13 |
ENST00000306320.9 |
FAM134B |
family with sequence similarity 134, member B |
| chr5_-_115910091 | 1.12 |
ENST00000257414.8 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chrX_+_17653413 | 1.11 |
ENST00000398097.3 |
NHS |
Nance-Horan syndrome (congenital cataracts and dental anomalies) |
| chr17_-_39661849 | 1.10 |
ENST00000246635.3 ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13 |
keratin 13 |
| chr17_+_30814707 | 1.10 |
ENST00000584792.1 |
CDK5R1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr17_-_2614927 | 1.06 |
ENST00000435359.1 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
| chr18_-_28742813 | 1.05 |
ENST00000257197.3 ENST00000257198.5 |
DSC1 |
desmocollin 1 |
| chr6_+_41606176 | 1.05 |
ENST00000441667.1 ENST00000230321.6 ENST00000373050.4 ENST00000446650.1 ENST00000435476.1 |
MDFI |
MyoD family inhibitor |
| chr17_+_7210921 | 1.03 |
ENST00000573542.1 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr6_+_142622991 | 1.03 |
ENST00000230173.6 ENST00000367608.2 |
GPR126 |
G protein-coupled receptor 126 |
| chr8_+_21912328 | 1.03 |
ENST00000432128.1 ENST00000443491.2 ENST00000517600.1 ENST00000523782.2 |
DMTN |
dematin actin binding protein |
| chr11_-_5248294 | 1.03 |
ENST00000335295.4 |
HBB |
hemoglobin, beta |
| chr17_+_7210898 | 1.02 |
ENST00000572815.1 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr19_+_8117881 | 1.01 |
ENST00000390669.3 |
CCL25 |
chemokine (C-C motif) ligand 25 |
| chr6_+_142623063 | 0.99 |
ENST00000296932.8 ENST00000367609.3 |
GPR126 |
G protein-coupled receptor 126 |
| chr19_+_41698927 | 0.99 |
ENST00000310054.4 |
CYP2S1 |
cytochrome P450, family 2, subfamily S, polypeptide 1 |
| chr3_+_50192537 | 0.98 |
ENST00000002829.3 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr20_-_22565101 | 0.97 |
ENST00000419308.2 |
FOXA2 |
forkhead box A2 |
| chr3_+_50192499 | 0.97 |
ENST00000413852.1 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr6_+_130339710 | 0.93 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr8_-_81083341 | 0.93 |
ENST00000519303.2 |
TPD52 |
tumor protein D52 |
| chr6_+_33589161 | 0.92 |
ENST00000605930.1 |
ITPR3 |
inositol 1,4,5-trisphosphate receptor, type 3 |
| chr1_+_55505184 | 0.90 |
ENST00000302118.5 |
PCSK9 |
proprotein convertase subtilisin/kexin type 9 |
| chr8_+_80523962 | 0.90 |
ENST00000518491.1 |
STMN2 |
stathmin-like 2 |
| chr19_+_39687596 | 0.89 |
ENST00000339852.4 |
NCCRP1 |
non-specific cytotoxic cell receptor protein 1 homolog (zebrafish) |
| chrX_-_152989798 | 0.88 |
ENST00000441714.1 ENST00000442093.1 ENST00000429550.1 ENST00000345046.6 |
BCAP31 |
B-cell receptor-associated protein 31 |
| chr1_+_33207381 | 0.87 |
ENST00000401073.2 |
KIAA1522 |
KIAA1522 |
| chr6_-_20212630 | 0.86 |
ENST00000324607.7 ENST00000541730.1 ENST00000536798.1 |
MBOAT1 |
membrane bound O-acyltransferase domain containing 1 |
| chr8_+_123793633 | 0.84 |
ENST00000314393.4 |
ZHX2 |
zinc fingers and homeoboxes 2 |
| chr12_-_54778471 | 0.84 |
ENST00000550120.1 ENST00000394313.2 ENST00000547210.1 |
ZNF385A |
zinc finger protein 385A |
| chr19_+_8117636 | 0.83 |
ENST00000253451.4 ENST00000315626.4 |
CCL25 |
chemokine (C-C motif) ligand 25 |
| chr9_+_74764278 | 0.83 |
ENST00000238018.4 ENST00000376989.3 |
GDA |
guanine deaminase |
| chr8_+_21911054 | 0.81 |
ENST00000519850.1 ENST00000381470.3 |
DMTN |
dematin actin binding protein |
| chr1_-_6526167 | 0.80 |
ENST00000351748.3 ENST00000348333.3 |
TNFRSF25 |
tumor necrosis factor receptor superfamily, member 25 |
| chr17_-_39023462 | 0.79 |
ENST00000251643.4 |
KRT12 |
keratin 12 |
| chr6_+_106534192 | 0.79 |
ENST00000369091.2 ENST00000369096.4 |
PRDM1 |
PR domain containing 1, with ZNF domain |
| chr15_+_75074385 | 0.79 |
ENST00000220003.9 |
CSK |
c-src tyrosine kinase |
| chr16_-_4466622 | 0.78 |
ENST00000570645.1 ENST00000574025.1 ENST00000572898.1 ENST00000537233.2 ENST00000571059.1 ENST00000251166.4 |
CORO7 |
coronin 7 |
| chrX_+_69672136 | 0.78 |
ENST00000374355.3 |
DLG3 |
discs, large homolog 3 (Drosophila) |
| chr17_+_7210852 | 0.77 |
ENST00000576930.1 |
EIF5A |
eukaryotic translation initiation factor 5A |
| chr10_+_71211212 | 0.76 |
ENST00000373290.2 |
TSPAN15 |
tetraspanin 15 |
| chr7_+_73245193 | 0.76 |
ENST00000340958.2 |
CLDN4 |
claudin 4 |
| chr9_-_100954910 | 0.73 |
ENST00000375077.4 |
CORO2A |
coronin, actin binding protein, 2A |
| chr1_+_33231268 | 0.73 |
ENST00000373480.1 |
KIAA1522 |
KIAA1522 |
| chr19_+_54371114 | 0.73 |
ENST00000448420.1 ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM |
myeloid-associated differentiation marker |
| chr16_+_23847267 | 0.73 |
ENST00000321728.7 |
PRKCB |
protein kinase C, beta |
| chr19_+_42788172 | 0.72 |
ENST00000160740.3 |
CIC |
capicua transcriptional repressor |
| chr1_-_11865982 | 0.70 |
ENST00000418034.1 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr16_-_30125177 | 0.70 |
ENST00000406256.3 |
GDPD3 |
glycerophosphodiester phosphodiesterase domain containing 3 |
| chr17_-_43502987 | 0.70 |
ENST00000376922.2 |
ARHGAP27 |
Rho GTPase activating protein 27 |
| chr1_+_153747746 | 0.69 |
ENST00000368661.3 |
SLC27A3 |
solute carrier family 27 (fatty acid transporter), member 3 |
| chr17_-_56350797 | 0.67 |
ENST00000577220.1 |
MPO |
myeloperoxidase |
| chr2_-_64371546 | 0.66 |
ENST00000358912.4 |
PELI1 |
pellino E3 ubiquitin protein ligase 1 |
| chr20_-_50722183 | 0.66 |
ENST00000371523.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr14_-_21566731 | 0.65 |
ENST00000360947.3 |
ZNF219 |
zinc finger protein 219 |
| chr17_+_38171681 | 0.64 |
ENST00000225474.2 ENST00000331769.2 ENST00000394148.3 ENST00000577675.1 |
CSF3 |
colony stimulating factor 3 (granulocyte) |
| chr7_+_142982023 | 0.64 |
ENST00000359333.3 ENST00000409244.1 ENST00000409541.1 ENST00000410004.1 |
TMEM139 |
transmembrane protein 139 |
| chr2_-_166651191 | 0.64 |
ENST00000392701.3 |
GALNT3 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| chr5_-_115910630 | 0.62 |
ENST00000343348.6 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr2_+_102314161 | 0.62 |
ENST00000425019.1 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr17_+_38171614 | 0.62 |
ENST00000583218.1 ENST00000394149.3 |
CSF3 |
colony stimulating factor 3 (granulocyte) |
| chr3_+_10857885 | 0.61 |
ENST00000254488.2 ENST00000454147.1 |
SLC6A11 |
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr9_-_34048873 | 0.61 |
ENST00000449054.1 ENST00000379239.4 ENST00000539807.1 ENST00000379238.1 ENST00000418786.2 ENST00000360802.1 ENST00000412543.1 |
UBAP2 |
ubiquitin associated protein 2 |
| chr22_-_39639021 | 0.60 |
ENST00000455790.1 |
PDGFB |
platelet-derived growth factor beta polypeptide |
| chr1_-_111743285 | 0.60 |
ENST00000357640.4 |
DENND2D |
DENN/MADD domain containing 2D |
| chrX_+_68835911 | 0.60 |
ENST00000525810.1 ENST00000527388.1 ENST00000374553.2 ENST00000374552.4 ENST00000338901.3 ENST00000524573.1 |
EDA |
ectodysplasin A |
| chr16_+_618837 | 0.60 |
ENST00000409439.2 |
PIGQ |
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr14_-_21979428 | 0.59 |
ENST00000538267.1 ENST00000298717.4 |
METTL3 |
methyltransferase like 3 |
| chr17_-_74023474 | 0.58 |
ENST00000301607.3 |
EVPL |
envoplakin |
| chr13_+_73632897 | 0.58 |
ENST00000377687.4 |
KLF5 |
Kruppel-like factor 5 (intestinal) |
| chr14_+_24783906 | 0.57 |
ENST00000396782.2 |
LTB4R |
leukotriene B4 receptor |
| chr2_+_220306745 | 0.56 |
ENST00000431523.1 ENST00000396698.1 ENST00000396695.2 |
SPEG |
SPEG complex locus |
| chr2_-_220408430 | 0.55 |
ENST00000243776.6 |
CHPF |
chondroitin polymerizing factor |
| chr16_+_46723552 | 0.55 |
ENST00000219097.2 ENST00000568364.2 |
ORC6 |
origin recognition complex, subunit 6 |
| chr16_+_56225248 | 0.55 |
ENST00000262493.6 |
GNAO1 |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide O |
| chr9_-_33264676 | 0.54 |
ENST00000472232.3 ENST00000379704.2 |
BAG1 |
BCL2-associated athanogene |
| chr1_-_11866034 | 0.54 |
ENST00000376590.3 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr12_+_51632666 | 0.53 |
ENST00000604900.1 |
DAZAP2 |
DAZ associated protein 2 |
| chr1_-_205744205 | 0.52 |
ENST00000446390.2 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr17_-_27949911 | 0.52 |
ENST00000492276.2 ENST00000345068.5 ENST00000584602.1 |
CORO6 |
coronin 6 |
| chr22_+_31477296 | 0.52 |
ENST00000426927.1 ENST00000440425.1 ENST00000358743.1 ENST00000347557.2 ENST00000333137.7 |
SMTN |
smoothelin |
| chr1_-_109940550 | 0.51 |
ENST00000256637.6 |
SORT1 |
sortilin 1 |
| chr17_-_7991021 | 0.51 |
ENST00000319144.4 |
ALOX12B |
arachidonate 12-lipoxygenase, 12R type |
| chr1_-_94147385 | 0.50 |
ENST00000260502.6 |
BCAR3 |
breast cancer anti-estrogen resistance 3 |
| chr17_+_65374075 | 0.50 |
ENST00000581322.1 |
PITPNC1 |
phosphatidylinositol transfer protein, cytoplasmic 1 |
| chr11_+_63655987 | 0.50 |
ENST00000509502.2 ENST00000512060.1 |
MARK2 |
MAP/microtubule affinity-regulating kinase 2 |
| chr19_-_44285401 | 0.49 |
ENST00000262888.3 |
KCNN4 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 4 |
| chr1_+_27189631 | 0.49 |
ENST00000339276.4 |
SFN |
stratifin |
| chr11_-_113345995 | 0.49 |
ENST00000355319.2 ENST00000542616.1 |
DRD2 |
dopamine receptor D2 |
| chr1_-_205744574 | 0.49 |
ENST00000367139.3 ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr17_-_39538550 | 0.49 |
ENST00000394001.1 |
KRT34 |
keratin 34 |
| chr6_-_33548006 | 0.49 |
ENST00000374467.3 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr11_+_1074870 | 0.48 |
ENST00000441003.2 ENST00000359061.5 |
MUC2 |
mucin 2, oligomeric mucus/gel-forming |
| chr1_+_11866270 | 0.47 |
ENST00000376497.3 ENST00000376487.3 ENST00000376496.3 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chr1_-_115212696 | 0.47 |
ENST00000393276.3 ENST00000393277.1 |
DENND2C |
DENN/MADD domain containing 2C |
| chr18_-_48723690 | 0.47 |
ENST00000406189.3 |
MEX3C |
mex-3 RNA binding family member C |
| chr11_+_61248583 | 0.47 |
ENST00000432063.2 ENST00000338608.2 |
PPP1R32 |
protein phosphatase 1, regulatory subunit 32 |
| chr1_-_39395165 | 0.47 |
ENST00000372985.3 |
RHBDL2 |
rhomboid, veinlet-like 2 (Drosophila) |
| chr19_-_55791431 | 0.47 |
ENST00000593263.1 ENST00000376343.3 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr15_-_76005170 | 0.46 |
ENST00000308508.5 |
CSPG4 |
chondroitin sulfate proteoglycan 4 |
| chr5_+_141016508 | 0.45 |
ENST00000444782.1 ENST00000521367.1 ENST00000297164.3 |
RELL2 |
RELT-like 2 |
| chr6_-_33547975 | 0.45 |
ENST00000442998.2 ENST00000360661.5 |
BAK1 |
BCL2-antagonist/killer 1 |
| chr20_-_4804244 | 0.45 |
ENST00000379400.3 |
RASSF2 |
Ras association (RalGDS/AF-6) domain family member 2 |
| chr5_-_95297534 | 0.44 |
ENST00000513343.1 ENST00000431061.2 |
ELL2 |
elongation factor, RNA polymerase II, 2 |
| chr10_+_120967072 | 0.43 |
ENST00000392870.2 |
GRK5 |
G protein-coupled receptor kinase 5 |
| chr9_+_74764340 | 0.43 |
ENST00000376986.1 ENST00000358399.3 |
GDA |
guanine deaminase |
| chr1_+_207494853 | 0.43 |
ENST00000367064.3 ENST00000367063.2 ENST00000391921.4 ENST00000367067.4 ENST00000314754.8 ENST00000367065.5 ENST00000391920.4 ENST00000367062.4 ENST00000343420.6 |
CD55 |
CD55 molecule, decay accelerating factor for complement (Cromer blood group) |
| chrX_+_133507389 | 0.42 |
ENST00000370800.4 |
PHF6 |
PHD finger protein 6 |
| chr3_+_8775466 | 0.42 |
ENST00000343849.2 ENST00000397368.2 |
CAV3 |
caveolin 3 |
| chr13_+_24553933 | 0.42 |
ENST00000424834.2 ENST00000439928.2 |
SPATA13 RP11-309I15.1 |
spermatogenesis associated 13 RP11-309I15.1 |
| chr17_-_7518145 | 0.41 |
ENST00000250113.7 ENST00000571597.1 |
FXR2 |
fragile X mental retardation, autosomal homolog 2 |
| chr4_+_146403912 | 0.41 |
ENST00000507367.1 ENST00000394092.2 ENST00000515385.1 |
SMAD1 |
SMAD family member 1 |
| chrX_+_48644962 | 0.41 |
ENST00000376670.3 ENST00000376665.3 |
GATA1 |
GATA binding protein 1 (globin transcription factor 1) |
| chr4_-_926069 | 0.41 |
ENST00000314167.4 ENST00000502656.1 |
GAK |
cyclin G associated kinase |
| chr19_-_55791540 | 0.41 |
ENST00000433386.2 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr22_-_29137771 | 0.40 |
ENST00000439200.1 ENST00000405598.1 ENST00000398017.2 ENST00000425190.2 ENST00000348295.3 ENST00000382578.1 ENST00000382565.1 ENST00000382566.1 ENST00000382580.2 ENST00000328354.6 |
CHEK2 |
checkpoint kinase 2 |
| chr1_+_16090914 | 0.40 |
ENST00000441801.2 |
FBLIM1 |
filamin binding LIM protein 1 |
| chr4_-_165305086 | 0.39 |
ENST00000507270.1 ENST00000514618.1 ENST00000503008.1 |
MARCH1 |
membrane-associated ring finger (C3HC4) 1, E3 ubiquitin protein ligase |
| chrX_+_40944871 | 0.39 |
ENST00000378308.2 ENST00000324545.8 |
USP9X |
ubiquitin specific peptidase 9, X-linked |
| chr3_-_113465065 | 0.39 |
ENST00000497255.1 ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr10_+_104155450 | 0.39 |
ENST00000471698.1 ENST00000189444.6 |
NFKB2 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr15_+_75074410 | 0.39 |
ENST00000439220.2 |
CSK |
c-src tyrosine kinase |
| chr4_-_926161 | 0.38 |
ENST00000511163.1 |
GAK |
cyclin G associated kinase |
| chr17_-_66287350 | 0.38 |
ENST00000580666.1 ENST00000583477.1 |
SLC16A6 |
solute carrier family 16, member 6 |
| chr19_-_55791563 | 0.38 |
ENST00000588971.1 ENST00000255631.5 ENST00000587551.1 |
HSPBP1 |
HSPA (heat shock 70kDa) binding protein, cytoplasmic cochaperone 1 |
| chr14_-_65409502 | 0.38 |
ENST00000389614.5 |
GPX2 |
glutathione peroxidase 2 (gastrointestinal) |
| chr3_-_45838011 | 0.37 |
ENST00000358525.4 ENST00000413781.1 |
SLC6A20 |
solute carrier family 6 (proline IMINO transporter), member 20 |
| chr6_+_158244223 | 0.36 |
ENST00000392185.3 |
SNX9 |
sorting nexin 9 |
| chr16_+_15068916 | 0.36 |
ENST00000455313.2 |
PDXDC1 |
pyridoxal-dependent decarboxylase domain containing 1 |
| chr15_+_42131011 | 0.36 |
ENST00000458483.1 |
PLA2G4B |
phospholipase A2, group IVB (cytosolic) |
| chr20_+_825275 | 0.36 |
ENST00000541082.1 |
FAM110A |
family with sequence similarity 110, member A |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.4 | 6.8 | GO:0061146 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.9 | 2.6 | GO:0048627 | myoblast development(GO:0048627) |
| 0.8 | 4.5 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.7 | 6.8 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.6 | 1.3 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.5 | 4.9 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.5 | 2.7 | GO:0010760 | negative regulation of macrophage chemotaxis(GO:0010760) |
| 0.4 | 2.2 | GO:0033078 | extrathymic T cell differentiation(GO:0033078) |
| 0.4 | 1.7 | GO:0003366 | cell-matrix adhesion involved in ameboidal cell migration(GO:0003366) |
| 0.4 | 11.9 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.4 | 1.1 | GO:0021722 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.4 | 3.0 | GO:0015820 | leucine transport(GO:0015820) proline transmembrane transport(GO:0035524) |
| 0.4 | 3.0 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.4 | 0.7 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.4 | 0.7 | GO:0015824 | proline transport(GO:0015824) |
| 0.3 | 2.7 | GO:0009912 | auditory receptor cell fate commitment(GO:0009912) inner ear receptor cell fate commitment(GO:0060120) |
| 0.3 | 1.0 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.3 | 0.9 | GO:0002352 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) |
| 0.3 | 1.8 | GO:0070560 | protein secretion by platelet(GO:0070560) |
| 0.3 | 0.9 | GO:2000646 | positive regulation of receptor catabolic process(GO:2000646) |
| 0.3 | 1.3 | GO:0002934 | desmosome organization(GO:0002934) |
| 0.3 | 0.8 | GO:1905224 | clathrin-coated pit assembly(GO:1905224) |
| 0.3 | 1.6 | GO:0032185 | septin cytoskeleton organization(GO:0032185) |
| 0.3 | 1.5 | GO:1903237 | negative regulation of leukocyte tethering or rolling(GO:1903237) |
| 0.3 | 1.3 | GO:0006145 | purine nucleobase catabolic process(GO:0006145) |
| 0.2 | 1.2 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.2 | 1.9 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.2 | 1.4 | GO:0010025 | wax biosynthetic process(GO:0010025) wax metabolic process(GO:0010166) |
| 0.2 | 10.2 | GO:0060216 | definitive hemopoiesis(GO:0060216) |
| 0.2 | 1.1 | GO:0051945 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.2 | 0.9 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.2 | 1.9 | GO:0061302 | smooth muscle cell-matrix adhesion(GO:0061302) |
| 0.2 | 0.6 | GO:0006382 | adenosine to inosine editing(GO:0006382) |
| 0.2 | 0.4 | GO:0001300 | chronological cell aging(GO:0001300) |
| 0.2 | 0.6 | GO:1903926 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.2 | 0.9 | GO:0050917 | sensory perception of sweet taste(GO:0050916) sensory perception of umami taste(GO:0050917) |
| 0.2 | 0.7 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.2 | 1.3 | GO:0030223 | neutrophil differentiation(GO:0030223) |
| 0.2 | 0.9 | GO:1904154 | positive regulation of retrograde protein transport, ER to cytosol(GO:1904154) |
| 0.2 | 1.2 | GO:0010989 | negative regulation of low-density lipoprotein particle clearance(GO:0010989) |
| 0.2 | 0.8 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.2 | 1.9 | GO:0032808 | lacrimal gland development(GO:0032808) |
| 0.2 | 0.6 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.2 | 0.6 | GO:1905176 | metanephric glomerular mesangial cell development(GO:0072255) reversible differentiation(GO:0090677) cell dedifferentiation involved in phenotypic switching(GO:0090678) positive regulation of phenotypic switching(GO:1900241) regulation of vascular smooth muscle cell dedifferentiation(GO:1905174) positive regulation of vascular smooth muscle cell dedifferentiation(GO:1905176) vascular smooth muscle cell dedifferentiation(GO:1990936) |
| 0.1 | 0.6 | GO:0030185 | nitric oxide transport(GO:0030185) |
| 0.1 | 0.4 | GO:2000563 | positive regulation of CD4-positive, alpha-beta T cell proliferation(GO:2000563) |
| 0.1 | 2.0 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.1 | 1.8 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.1 | 2.7 | GO:0033169 | histone H3-K9 demethylation(GO:0033169) |
| 0.1 | 0.5 | GO:0044467 | glial cell-derived neurotrophic factor secretion(GO:0044467) regulation of glial cell-derived neurotrophic factor secretion(GO:1900166) positive regulation of glial cell-derived neurotrophic factor secretion(GO:1900168) |
| 0.1 | 0.4 | GO:0060988 | lipid tube assembly(GO:0060988) |
| 0.1 | 0.6 | GO:0061153 | trachea submucosa development(GO:0061152) trachea gland development(GO:0061153) |
| 0.1 | 0.4 | GO:0093001 | glycolysis from storage polysaccharide through glucose-1-phosphate(GO:0093001) |
| 0.1 | 0.5 | GO:0003415 | chondrocyte hypertrophy(GO:0003415) |
| 0.1 | 0.3 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.1 | 14.2 | GO:0070268 | cornification(GO:0070268) |
| 0.1 | 0.7 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.1 | 0.3 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.1 | 1.0 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.1 | 0.6 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) basic amino acid transmembrane transport(GO:1990822) |
| 0.1 | 0.5 | GO:0010481 | epidermal cell division(GO:0010481) regulation of epidermal cell division(GO:0010482) |
| 0.1 | 0.4 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.1 | 0.6 | GO:0061737 | leukotriene signaling pathway(GO:0061737) |
| 0.1 | 2.3 | GO:0019373 | epoxygenase P450 pathway(GO:0019373) |
| 0.1 | 0.3 | GO:1903179 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.1 | 0.4 | GO:1902044 | regulation of Fas signaling pathway(GO:1902044) |
| 0.1 | 0.4 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 1.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.1 | 0.3 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.3 | GO:0061436 | establishment of skin barrier(GO:0061436) |
| 0.1 | 0.2 | GO:2000687 | negative regulation of rubidium ion transport(GO:2000681) regulation of rubidium ion transmembrane transporter activity(GO:2000686) negative regulation of rubidium ion transmembrane transporter activity(GO:2000687) |
| 0.1 | 0.3 | GO:0060666 | dichotomous subdivision of terminal units involved in salivary gland branching(GO:0060666) |
| 0.1 | 0.2 | GO:1990641 | cellular response to bile acid(GO:1903413) negative regulation of intestinal absorption(GO:1904479) response to iron ion starvation(GO:1990641) |
| 0.1 | 5.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.1 | 0.2 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.1 | 0.1 | GO:0003274 | endocardial cushion fusion(GO:0003274) |
| 0.1 | 2.5 | GO:0006359 | regulation of transcription from RNA polymerase III promoter(GO:0006359) |
| 0.1 | 0.3 | GO:0046340 | diacylglycerol catabolic process(GO:0046340) |
| 0.1 | 0.8 | GO:0043249 | erythrocyte maturation(GO:0043249) |
| 0.1 | 0.6 | GO:0032534 | regulation of microvillus assembly(GO:0032534) |
| 0.1 | 0.5 | GO:1903764 | regulation of potassium ion export across plasma membrane(GO:1903764) |
| 0.1 | 0.5 | GO:0051005 | negative regulation of lipoprotein lipase activity(GO:0051005) |
| 0.1 | 1.0 | GO:0060707 | trophoblast giant cell differentiation(GO:0060707) |
| 0.1 | 4.7 | GO:2000649 | regulation of sodium ion transmembrane transporter activity(GO:2000649) |
| 0.1 | 0.2 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.1 | 0.3 | GO:1903830 | magnesium ion transmembrane transport(GO:1903830) |
| 0.1 | 1.3 | GO:2000311 | regulation of alpha-amino-3-hydroxy-5-methyl-4-isoxazole propionate selective glutamate receptor activity(GO:2000311) |
| 0.1 | 0.5 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.1 | 1.5 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.1 | 0.6 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.5 | GO:0001878 | response to yeast(GO:0001878) |
| 0.1 | 0.3 | GO:0042738 | exogenous drug catabolic process(GO:0042738) |
| 0.0 | 1.8 | GO:0060314 | regulation of ryanodine-sensitive calcium-release channel activity(GO:0060314) |
| 0.0 | 0.1 | GO:0006535 | cysteine biosynthetic process from serine(GO:0006535) |
| 0.0 | 1.6 | GO:0015695 | organic cation transport(GO:0015695) |
| 0.0 | 0.4 | GO:2000622 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.3 | GO:0046541 | saliva secretion(GO:0046541) |
| 0.0 | 0.6 | GO:1904896 | ESCRT complex disassembly(GO:1904896) ESCRT III complex disassembly(GO:1904903) |
| 0.0 | 0.3 | GO:0061590 | calcium activated phospholipid scrambling(GO:0061588) calcium activated phosphatidylserine scrambling(GO:0061589) calcium activated phosphatidylcholine scrambling(GO:0061590) calcium activated galactosylceramide scrambling(GO:0061591) |
| 0.0 | 0.3 | GO:0070601 | centromeric sister chromatid cohesion(GO:0070601) |
| 0.0 | 0.4 | GO:0007217 | tachykinin receptor signaling pathway(GO:0007217) |
| 0.0 | 1.9 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.4 | GO:0002051 | osteoblast fate commitment(GO:0002051) |
| 0.0 | 1.2 | GO:0036152 | phosphatidylethanolamine acyl-chain remodeling(GO:0036152) |
| 0.0 | 0.2 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0032639 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.2 | GO:1901165 | positive regulation of trophoblast cell migration(GO:1901165) |
| 0.0 | 0.4 | GO:0033623 | regulation of integrin activation(GO:0033623) |
| 0.0 | 0.2 | GO:2000543 | positive regulation of gastrulation(GO:2000543) |
| 0.0 | 0.4 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.5 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.6 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.3 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.4 | GO:0070389 | chaperone cofactor-dependent protein refolding(GO:0070389) |
| 0.0 | 0.2 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.3 | GO:0097167 | circadian regulation of translation(GO:0097167) |
| 0.0 | 0.2 | GO:0035507 | regulation of myosin-light-chain-phosphatase activity(GO:0035507) |
| 0.0 | 1.5 | GO:1904837 | beta-catenin-TCF complex assembly(GO:1904837) |
| 0.0 | 0.3 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.0 | 0.1 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.3 | GO:0035634 | response to stilbenoid(GO:0035634) |
| 0.0 | 0.4 | GO:0019388 | galactose catabolic process(GO:0019388) |
| 0.0 | 0.6 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.3 | GO:0071787 | endoplasmic reticulum tubular network assembly(GO:0071787) regulation of branching morphogenesis of a nerve(GO:2000172) |
| 0.0 | 0.6 | GO:0000185 | activation of MAPKKK activity(GO:0000185) |
| 0.0 | 0.2 | GO:1902031 | regulation of NADP metabolic process(GO:1902031) |
| 0.0 | 0.3 | GO:0006122 | mitochondrial electron transport, ubiquinol to cytochrome c(GO:0006122) |
| 0.0 | 0.3 | GO:0090168 | Golgi reassembly(GO:0090168) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 1.1 | GO:0007029 | endoplasmic reticulum organization(GO:0007029) |
| 0.0 | 0.7 | GO:0003016 | respiratory system process(GO:0003016) |
| 0.0 | 0.4 | GO:0030277 | maintenance of gastrointestinal epithelium(GO:0030277) |
| 0.0 | 0.5 | GO:0002089 | lens morphogenesis in camera-type eye(GO:0002089) |
| 0.0 | 0.2 | GO:0097428 | protein maturation by iron-sulfur cluster transfer(GO:0097428) |
| 0.0 | 0.1 | GO:0008626 | granzyme-mediated apoptotic signaling pathway(GO:0008626) |
| 0.0 | 1.1 | GO:0035735 | intraciliary transport involved in cilium morphogenesis(GO:0035735) |
| 0.0 | 0.2 | GO:0010993 | regulation of ubiquitin homeostasis(GO:0010993) free ubiquitin chain polymerization(GO:0010994) |
| 0.0 | 0.7 | GO:0046710 | GDP metabolic process(GO:0046710) |
| 0.0 | 0.2 | GO:0006621 | protein retention in ER lumen(GO:0006621) |
| 0.0 | 0.2 | GO:0005981 | regulation of glycogen catabolic process(GO:0005981) |
| 0.0 | 0.7 | GO:0031954 | positive regulation of protein autophosphorylation(GO:0031954) |
| 0.0 | 0.2 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.3 | GO:0034380 | high-density lipoprotein particle assembly(GO:0034380) |
| 0.0 | 0.2 | GO:0002281 | macrophage activation involved in immune response(GO:0002281) |
| 0.0 | 0.2 | GO:1900016 | negative regulation of cytokine production involved in inflammatory response(GO:1900016) |
| 0.0 | 0.3 | GO:0046475 | glycerophospholipid catabolic process(GO:0046475) |
| 0.0 | 0.1 | GO:0007613 | memory(GO:0007613) |
| 0.0 | 0.3 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.3 | GO:0007095 | mitotic G2 DNA damage checkpoint(GO:0007095) |
| 0.0 | 0.0 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.3 | GO:0015937 | coenzyme A biosynthetic process(GO:0015937) |
| 0.0 | 0.2 | GO:0006474 | N-terminal protein amino acid acetylation(GO:0006474) |
| 0.0 | 0.1 | GO:0050819 | negative regulation of coagulation(GO:0050819) |
| 0.0 | 0.1 | GO:0010225 | response to UV-C(GO:0010225) |
| 0.0 | 1.0 | GO:0009301 | snRNA transcription(GO:0009301) snRNA transcription from RNA polymerase II promoter(GO:0042795) |
| 0.0 | 0.5 | GO:0034599 | cellular response to oxidative stress(GO:0034599) |
| 0.0 | 0.5 | GO:0007223 | Wnt signaling pathway, calcium modulating pathway(GO:0007223) |
| 0.0 | 0.5 | GO:0015718 | monocarboxylic acid transport(GO:0015718) |
| 0.0 | 0.2 | GO:0021794 | thalamus development(GO:0021794) |
| 0.0 | 0.1 | GO:1990573 | potassium ion import across plasma membrane(GO:1990573) |
| 0.0 | 0.1 | GO:0006287 | base-excision repair, gap-filling(GO:0006287) |
| 0.0 | 0.1 | GO:0055009 | atrial cardiac muscle tissue development(GO:0003228) atrial cardiac muscle tissue morphogenesis(GO:0055009) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.7 | 5.0 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.8 | 6.7 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.4 | 1.7 | GO:0034669 | integrin alpha4-beta7 complex(GO:0034669) |
| 0.4 | 10.5 | GO:0043205 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.4 | 13.6 | GO:0030057 | desmosome(GO:0030057) |
| 0.3 | 0.9 | GO:1990666 | PCSK9-LDLR complex(GO:1990666) |
| 0.3 | 1.1 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.3 | 1.0 | GO:0031838 | haptoglobin-hemoglobin complex(GO:0031838) |
| 0.2 | 3.0 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.2 | 5.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.2 | 1.8 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.1 | 0.6 | GO:0036396 | MIS complex(GO:0036396) mRNA editing complex(GO:0045293) |
| 0.1 | 0.9 | GO:0005784 | Sec61 translocon complex(GO:0005784) translocon complex(GO:0071256) |
| 0.1 | 2.8 | GO:0031095 | platelet dense tubular network membrane(GO:0031095) |
| 0.1 | 0.4 | GO:0072563 | endothelial microparticle(GO:0072563) |
| 0.1 | 0.3 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.1 | 0.4 | GO:0033257 | Bcl3/NF-kappaB2 complex(GO:0033257) |
| 0.1 | 2.5 | GO:0005666 | DNA-directed RNA polymerase III complex(GO:0005666) |
| 0.1 | 2.4 | GO:0044295 | axonal growth cone(GO:0044295) |
| 0.1 | 2.3 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.4 | GO:0005945 | 6-phosphofructokinase complex(GO:0005945) |
| 0.1 | 0.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.9 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.8 | GO:0000808 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.1 | 0.6 | GO:0097427 | microtubule bundle(GO:0097427) |
| 0.1 | 5.1 | GO:0034707 | chloride channel complex(GO:0034707) |
| 0.0 | 2.3 | GO:0046658 | anchored component of plasma membrane(GO:0046658) |
| 0.0 | 2.0 | GO:0032281 | AMPA glutamate receptor complex(GO:0032281) |
| 0.0 | 0.6 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0031417 | NatC complex(GO:0031417) |
| 0.0 | 0.1 | GO:0036194 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
| 0.0 | 0.5 | GO:0043219 | lateral loop(GO:0043219) |
| 0.0 | 0.3 | GO:0031415 | NatA complex(GO:0031415) |
| 0.0 | 0.3 | GO:0097422 | tubular endosome(GO:0097422) |
| 0.0 | 0.4 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.2 | GO:0032021 | NELF complex(GO:0032021) |
| 0.0 | 0.9 | GO:0031231 | integral component of peroxisomal membrane(GO:0005779) intrinsic component of peroxisomal membrane(GO:0031231) |
| 0.0 | 1.1 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.3 | GO:0070938 | contractile ring(GO:0070938) |
| 0.0 | 0.4 | GO:0008250 | oligosaccharyltransferase complex(GO:0008250) |
| 0.0 | 0.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.5 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.3 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.4 | GO:0071141 | SMAD protein complex(GO:0071141) |
| 0.0 | 0.3 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.0 | 1.7 | GO:0032580 | Golgi cisterna membrane(GO:0032580) |
| 0.0 | 1.2 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.2 | GO:0000164 | protein phosphatase type 1 complex(GO:0000164) |
| 0.0 | 0.8 | GO:0008023 | transcription elongation factor complex(GO:0008023) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.3 | GO:0030014 | CCR4-NOT complex(GO:0030014) |
| 0.0 | 0.4 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0032426 | stereocilium tip(GO:0032426) |
| 0.0 | 0.9 | GO:0005871 | kinesin complex(GO:0005871) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 7.2 | GO:0005743 | mitochondrial inner membrane(GO:0005743) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.1 | 7.4 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 1.0 | 3.0 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.5 | 4.9 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.5 | 1.8 | GO:0031735 | CCR10 chemokine receptor binding(GO:0031735) |
| 0.5 | 2.3 | GO:0008401 | retinoic acid 4-hydroxylase activity(GO:0008401) |
| 0.4 | 5.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.4 | 1.9 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.4 | 1.1 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.4 | 5.1 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.3 | 0.9 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.3 | 1.7 | GO:0019960 | C-X3-C chemokine binding(GO:0019960) |
| 0.3 | 1.4 | GO:0080019 | fatty-acyl-CoA reductase (alcohol-forming) activity(GO:0080019) |
| 0.2 | 1.2 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.2 | 1.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.2 | 0.7 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.2 | 0.9 | GO:0000822 | inositol hexakisphosphate binding(GO:0000822) inositol 1,4,5-trisphosphate-sensitive calcium-release channel activity(GO:0005220) |
| 0.2 | 1.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.2 | 3.2 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.2 | 0.6 | GO:0016422 | mRNA (2'-O-methyladenosine-N6-)-methyltransferase activity(GO:0016422) |
| 0.2 | 0.6 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.2 | 1.0 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.2 | 3.0 | GO:0017070 | U6 snRNA binding(GO:0017070) |
| 0.1 | 0.4 | GO:0047696 | beta-adrenergic receptor kinase activity(GO:0047696) |
| 0.1 | 9.1 | GO:0017080 | sodium channel regulator activity(GO:0017080) |
| 0.1 | 2.7 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.1 | 0.6 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.1 | 1.6 | GO:0019534 | toxin transporter activity(GO:0019534) |
| 0.1 | 0.5 | GO:0004052 | arachidonate 12-lipoxygenase activity(GO:0004052) |
| 0.1 | 0.5 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.1 | 1.9 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.1 | 1.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.1 | 2.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.1 | 0.3 | GO:0045155 | electron transporter, transferring electrons from CoQH2-cytochrome c reductase complex and cytochrome c oxidase complex activity(GO:0045155) |
| 0.1 | 0.3 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.1 | 0.6 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 0.5 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.1 | 0.5 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.1 | 0.4 | GO:0003974 | UDP-N-acetylglucosamine 4-epimerase activity(GO:0003974) UDP-glucose 4-epimerase activity(GO:0003978) |
| 0.1 | 0.6 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
| 0.1 | 0.3 | GO:0097158 | pre-mRNA intronic pyrimidine-rich binding(GO:0097158) |
| 0.1 | 0.3 | GO:0004142 | diacylglycerol cholinephosphotransferase activity(GO:0004142) ethanolaminephosphotransferase activity(GO:0004307) |
| 0.1 | 0.3 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.1 | 0.2 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.1 | 8.4 | GO:0005201 | extracellular matrix structural constituent(GO:0005201) |
| 0.1 | 0.5 | GO:0001591 | dopamine neurotransmitter receptor activity, coupled via Gi/Go(GO:0001591) |
| 0.1 | 1.6 | GO:0017049 | GTP-Rho binding(GO:0017049) |
| 0.1 | 0.6 | GO:0048407 | platelet-derived growth factor binding(GO:0048407) |
| 0.1 | 0.9 | GO:0047144 | 2-acylglycerol-3-phosphate O-acyltransferase activity(GO:0047144) |
| 0.1 | 0.7 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.1 | 0.4 | GO:0003872 | 6-phosphofructokinase activity(GO:0003872) |
| 0.1 | 0.3 | GO:0016361 | activin receptor activity, type I(GO:0016361) |
| 0.1 | 2.3 | GO:0005035 | tumor necrosis factor-activated receptor activity(GO:0005031) death receptor activity(GO:0005035) |
| 0.1 | 2.2 | GO:1990841 | promoter-specific chromatin binding(GO:1990841) |
| 0.1 | 0.5 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.6 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.5 | GO:0051430 | corticotropin-releasing hormone receptor 1 binding(GO:0051430) |
| 0.0 | 1.0 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.8 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0098809 | cystathionine beta-synthase activity(GO:0004122) oxidoreductase activity, acting on other nitrogenous compounds as donors, cytochrome as acceptor(GO:0016662) nitrite reductase (NO-forming) activity(GO:0050421) carbon monoxide binding(GO:0070025) nitrite reductase activity(GO:0098809) |
| 0.0 | 0.4 | GO:0005534 | galactose binding(GO:0005534) |
| 0.0 | 0.5 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.3 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.0 | 0.9 | GO:0051400 | BH domain binding(GO:0051400) |
| 0.0 | 0.7 | GO:0005328 | neurotransmitter:sodium symporter activity(GO:0005328) |
| 0.0 | 2.5 | GO:0003899 | DNA-directed RNA polymerase activity(GO:0003899) |
| 0.0 | 0.6 | GO:0015174 | basic amino acid transmembrane transporter activity(GO:0015174) |
| 0.0 | 0.2 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.3 | GO:0031748 | D1 dopamine receptor binding(GO:0031748) |
| 0.0 | 0.3 | GO:0071532 | ankyrin repeat binding(GO:0071532) |
| 0.0 | 0.7 | GO:0031957 | very long-chain fatty acid-CoA ligase activity(GO:0031957) |
| 0.0 | 0.3 | GO:0071253 | connexin binding(GO:0071253) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 7.9 | GO:0008083 | growth factor activity(GO:0008083) |
| 0.0 | 0.5 | GO:0004596 | peptide alpha-N-acetyltransferase activity(GO:0004596) |
| 0.0 | 0.4 | GO:0047498 | calcium-dependent phospholipase A2 activity(GO:0047498) |
| 0.0 | 0.2 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.4 | GO:0070878 | primary miRNA binding(GO:0070878) |
| 0.0 | 0.5 | GO:0097109 | neuroligin family protein binding(GO:0097109) |
| 0.0 | 0.4 | GO:0000774 | adenyl-nucleotide exchange factor activity(GO:0000774) |
| 0.0 | 8.8 | GO:0004252 | serine-type endopeptidase activity(GO:0004252) |
| 0.0 | 0.2 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.9 | GO:0042288 | MHC class I protein binding(GO:0042288) |
| 0.0 | 1.3 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.2 | GO:0000405 | bubble DNA binding(GO:0000405) |
| 0.0 | 1.3 | GO:0019239 | deaminase activity(GO:0019239) |
| 0.0 | 0.4 | GO:0042287 | MHC protein binding(GO:0042287) |
| 0.0 | 0.6 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 2.7 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
| 0.0 | 0.3 | GO:0030215 | semaphorin receptor binding(GO:0030215) |
| 0.0 | 0.1 | GO:0046912 | transferase activity, transferring acyl groups, acyl groups converted into alkyl on transfer(GO:0046912) |
| 0.0 | 0.7 | GO:0004385 | guanylate kinase activity(GO:0004385) |
| 0.0 | 0.4 | GO:0008494 | translation activator activity(GO:0008494) |
| 0.0 | 0.1 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.3 | GO:0017128 | phospholipid scramblase activity(GO:0017128) |
| 0.0 | 1.5 | GO:0043621 | protein self-association(GO:0043621) |
| 0.0 | 0.3 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.8 | GO:0048306 | calcium-dependent protein binding(GO:0048306) |
| 0.0 | 1.9 | GO:0008307 | structural constituent of muscle(GO:0008307) |
| 0.0 | 0.2 | GO:0009922 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.4 | GO:0031005 | filamin binding(GO:0031005) |
| 0.0 | 0.1 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 1.1 | GO:0017112 | Rab guanyl-nucleotide exchange factor activity(GO:0017112) |
| 0.0 | 1.2 | GO:0004715 | non-membrane spanning protein tyrosine kinase activity(GO:0004715) |
| 0.0 | 0.3 | GO:0048018 | receptor agonist activity(GO:0048018) |
| 0.0 | 1.1 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.4 | GO:0030676 | Rac guanyl-nucleotide exchange factor activity(GO:0030676) |
| 0.0 | 0.4 | GO:0070742 | C2H2 zinc finger domain binding(GO:0070742) |
| 0.0 | 0.4 | GO:0019902 | phosphatase binding(GO:0019902) |
| 0.0 | 0.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.0 | GO:0070984 | SET domain binding(GO:0070984) |
| 0.0 | 0.1 | GO:0086089 | voltage-gated potassium channel activity involved in atrial cardiac muscle cell action potential repolarization(GO:0086089) |
| 0.0 | 0.4 | GO:0005123 | death receptor binding(GO:0005123) |
| 0.0 | 0.2 | GO:0071889 | 14-3-3 protein binding(GO:0071889) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.3 | GO:0019706 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.2 | GO:0008190 | eukaryotic initiation factor 4E binding(GO:0008190) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 11.7 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.1 | 13.9 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.1 | 1.9 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 3.0 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 3.5 | PID DELTA NP63 PATHWAY | Validated transcriptional targets of deltaNp63 isoforms |
| 0.0 | 1.2 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 3.1 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.9 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 1.4 | ST MYOCYTE AD PATHWAY | Myocyte Adrenergic Pathway is a specific case of the generalized Adrenergic Pathway. |
| 0.0 | 0.6 | PID ALK2 PATHWAY | ALK2 signaling events |
| 0.0 | 0.5 | ST G ALPHA I PATHWAY | G alpha i Pathway |
| 0.0 | 0.4 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 1.6 | ST B CELL ANTIGEN RECEPTOR | B Cell Antigen Receptor |
| 0.0 | 0.5 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 9.3 | NABA SECRETED FACTORS | Genes encoding secreted soluble factors |
| 0.0 | 0.3 | PID SYNDECAN 2 PATHWAY | Syndecan-2-mediated signaling events |
| 0.0 | 4.8 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 1.0 | PID CERAMIDE PATHWAY | Ceramide signaling pathway |
| 0.0 | 0.7 | PID FOXM1 PATHWAY | FOXM1 transcription factor network |
| 0.0 | 0.8 | PID MAPK TRK PATHWAY | Trk receptor signaling mediated by the MAPK pathway |
| 0.0 | 1.2 | PID HNF3A PATHWAY | FOXA1 transcription factor network |
| 0.0 | 0.9 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 0.4 | PID TCR JNK PATHWAY | JNK signaling in the CD4+ TCR pathway |
| 0.0 | 0.6 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
| 0.0 | 0.2 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.6 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.4 | ST PHOSPHOINOSITIDE 3 KINASE PATHWAY | PI3K Pathway |
| 0.0 | 0.2 | PID FAK PATHWAY | Signaling events mediated by focal adhesion kinase |
| 0.0 | 0.4 | PID HES HEY PATHWAY | Notch-mediated HES/HEY network |
| 0.0 | 0.3 | PID P38 MKK3 6PATHWAY | p38 MAPK signaling pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 6.0 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.2 | 2.7 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.2 | 4.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.1 | 4.8 | REACTOME BASIGIN INTERACTIONS | Genes involved in Basigin interactions |
| 0.1 | 6.3 | REACTOME NCAM1 INTERACTIONS | Genes involved in NCAM1 interactions |
| 0.1 | 10.8 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.1 | 1.9 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.1 | 1.3 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.1 | 0.9 | REACTOME ELEVATION OF CYTOSOLIC CA2 LEVELS | Genes involved in Elevation of cytosolic Ca2+ levels |
| 0.1 | 3.0 | REACTOME PTM GAMMA CARBOXYLATION HYPUSINE FORMATION AND ARYLSULFATASE ACTIVATION | Genes involved in PTM: gamma carboxylation, hypusine formation and arylsulfatase activation |
| 0.1 | 0.8 | REACTOME CDC6 ASSOCIATION WITH THE ORC ORIGIN COMPLEX | Genes involved in CDC6 association with the ORC:origin complex |
| 0.1 | 1.2 | REACTOME CLASS C 3 METABOTROPIC GLUTAMATE PHEROMONE RECEPTORS | Genes involved in Class C/3 (Metabotropic glutamate/pheromone receptors) |
| 0.1 | 0.6 | REACTOME P53 INDEPENDENT G1 S DNA DAMAGE CHECKPOINT | Genes involved in p53-Independent G1/S DNA damage checkpoint |
| 0.0 | 0.9 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.1 | REACTOME IONOTROPIC ACTIVITY OF KAINATE RECEPTORS | Genes involved in Ionotropic activity of Kainate Receptors |
| 0.0 | 1.2 | REACTOME PHOSPHORYLATION OF CD3 AND TCR ZETA CHAINS | Genes involved in Phosphorylation of CD3 and TCR zeta chains |
| 0.0 | 0.6 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 2.9 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 2.6 | REACTOME CYTOCHROME P450 ARRANGED BY SUBSTRATE TYPE | Genes involved in Cytochrome P450 - arranged by substrate type |
| 0.0 | 12.5 | REACTOME METABOLISM OF AMINO ACIDS AND DERIVATIVES | Genes involved in Metabolism of amino acids and derivatives |
| 0.0 | 0.6 | REACTOME EICOSANOID LIGAND BINDING RECEPTORS | Genes involved in Eicosanoid ligand-binding receptors |
| 0.0 | 0.1 | REACTOME PLATELET SENSITIZATION BY LDL | Genes involved in Platelet sensitization by LDL |
| 0.0 | 0.5 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.7 | REACTOME ACTIVATION OF NF KAPPAB IN B CELLS | Genes involved in Activation of NF-kappaB in B Cells |
| 0.0 | 0.3 | REACTOME PECAM1 INTERACTIONS | Genes involved in PECAM1 interactions |
| 0.0 | 0.9 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.8 | REACTOME REGULATION OF GENE EXPRESSION IN BETA CELLS | Genes involved in Regulation of gene expression in beta cells |
| 0.0 | 0.4 | REACTOME DESTABILIZATION OF MRNA BY AUF1 HNRNP D0 | Genes involved in Destabilization of mRNA by AUF1 (hnRNP D0) |
| 0.0 | 0.9 | REACTOME KINESINS | Genes involved in Kinesins |
| 0.0 | 0.6 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.6 | REACTOME CRMPS IN SEMA3A SIGNALING | Genes involved in CRMPs in Sema3A signaling |
| 0.0 | 1.5 | REACTOME CHEMOKINE RECEPTORS BIND CHEMOKINES | Genes involved in Chemokine receptors bind chemokines |
| 0.0 | 0.4 | REACTOME SIGNALING BY BMP | Genes involved in Signaling by BMP |
| 0.0 | 0.6 | REACTOME ENDOSOMAL SORTING COMPLEX REQUIRED FOR TRANSPORT ESCRT | Genes involved in Endosomal Sorting Complex Required For Transport (ESCRT) |
| 0.0 | 2.0 | REACTOME GOLGI ASSOCIATED VESICLE BIOGENESIS | Genes involved in Golgi Associated Vesicle Biogenesis |
| 0.0 | 0.3 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME RIP MEDIATED NFKB ACTIVATION VIA DAI | Genes involved in RIP-mediated NFkB activation via DAI |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 1.0 | REACTOME O LINKED GLYCOSYLATION OF MUCINS | Genes involved in O-linked glycosylation of mucins |
| 0.0 | 0.8 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.3 | REACTOME DEADENYLATION OF MRNA | Genes involved in Deadenylation of mRNA |
| 0.0 | 0.3 | REACTOME DESTABILIZATION OF MRNA BY KSRP | Genes involved in Destabilization of mRNA by KSRP |