Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
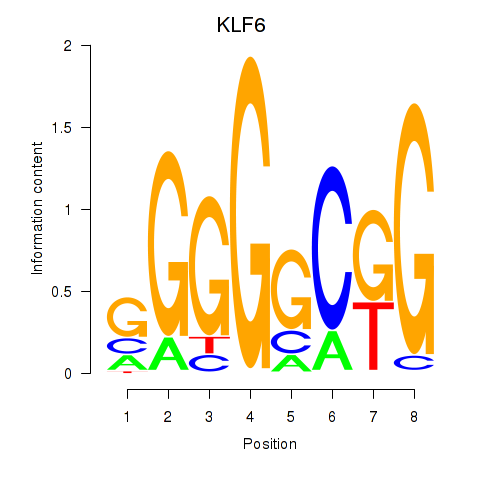
Results for KLF6
Z-value: 0.61
Transcription factors associated with KLF6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
KLF6
|
ENSG00000067082.10 | KLF6 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| KLF6 | hg19_v2_chr10_-_3827417_3827473 | -0.18 | 6.7e-01 | Click! |
Activity profile of KLF6 motif
Sorted Z-values of KLF6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of KLF6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_72596604 | 0.32 |
ENST00000348717.5 ENST00000517960.1 ENST00000518273.1 ENST00000522291.1 ENST00000521978.1 ENST00000520567.1 ENST00000264839.7 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr1_-_6321035 | 0.30 |
ENST00000377893.2 |
GPR153 |
G protein-coupled receptor 153 |
| chr4_-_36246060 | 0.29 |
ENST00000303965.4 |
ARAP2 |
ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| chr19_+_54372639 | 0.25 |
ENST00000391769.2 |
MYADM |
myeloid-associated differentiation marker |
| chr6_-_82462425 | 0.24 |
ENST00000369754.3 ENST00000320172.6 ENST00000369756.3 |
FAM46A |
family with sequence similarity 46, member A |
| chr7_-_143105941 | 0.22 |
ENST00000275815.3 |
EPHA1 |
EPH receptor A1 |
| chr11_+_57365150 | 0.21 |
ENST00000457869.1 ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1 |
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr13_+_88324870 | 0.20 |
ENST00000325089.6 |
SLITRK5 |
SLIT and NTRK-like family, member 5 |
| chr20_+_30555805 | 0.17 |
ENST00000562532.2 |
XKR7 |
XK, Kell blood group complex subunit-related family, member 7 |
| chr1_-_44497118 | 0.16 |
ENST00000537678.1 ENST00000466926.1 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr1_+_156698234 | 0.15 |
ENST00000368218.4 ENST00000368216.4 |
RRNAD1 |
ribosomal RNA adenine dimethylase domain containing 1 |
| chr2_+_42275153 | 0.15 |
ENST00000294964.5 |
PKDCC |
protein kinase domain containing, cytoplasmic |
| chr11_-_108464465 | 0.14 |
ENST00000525344.1 |
EXPH5 |
exophilin 5 |
| chr16_+_68678739 | 0.14 |
ENST00000264012.4 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr6_+_72596406 | 0.14 |
ENST00000491071.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr20_-_48099182 | 0.14 |
ENST00000371741.4 |
KCNB1 |
potassium voltage-gated channel, Shab-related subfamily, member 1 |
| chr17_+_54671047 | 0.13 |
ENST00000332822.4 |
NOG |
noggin |
| chr16_+_68678892 | 0.13 |
ENST00000429102.2 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr5_-_94620239 | 0.13 |
ENST00000515393.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
| chr1_-_22263790 | 0.13 |
ENST00000374695.3 |
HSPG2 |
heparan sulfate proteoglycan 2 |
| chr3_+_38495333 | 0.13 |
ENST00000352511.4 |
ACVR2B |
activin A receptor, type IIB |
| chr16_+_68679193 | 0.13 |
ENST00000581171.1 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr1_-_44497024 | 0.13 |
ENST00000372306.3 ENST00000372310.3 ENST00000475075.2 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr2_+_203499901 | 0.12 |
ENST00000303116.6 ENST00000392238.2 |
FAM117B |
family with sequence similarity 117, member B |
| chr8_-_110656995 | 0.12 |
ENST00000276646.9 ENST00000533065.1 |
SYBU |
syntabulin (syntaxin-interacting) |
| chr7_+_130131907 | 0.12 |
ENST00000223215.4 ENST00000437945.1 |
MEST |
mesoderm specific transcript |
| chr19_-_15311713 | 0.12 |
ENST00000601011.1 ENST00000263388.2 |
NOTCH3 |
notch 3 |
| chr7_-_27213893 | 0.12 |
ENST00000283921.4 |
HOXA10 |
homeobox A10 |
| chr12_+_79258547 | 0.11 |
ENST00000457153.2 |
SYT1 |
synaptotagmin I |
| chr19_+_45504688 | 0.11 |
ENST00000221452.8 ENST00000540120.1 ENST00000505236.1 |
RELB |
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr12_+_79258444 | 0.11 |
ENST00000261205.4 |
SYT1 |
synaptotagmin I |
| chr11_-_108464321 | 0.11 |
ENST00000265843.4 |
EXPH5 |
exophilin 5 |
| chr11_-_94964354 | 0.11 |
ENST00000536441.1 |
SESN3 |
sestrin 3 |
| chr14_-_37051798 | 0.10 |
ENST00000258829.5 |
NKX2-8 |
NK2 homeobox 8 |
| chr4_+_155665123 | 0.10 |
ENST00000336356.3 |
LRAT |
lecithin retinol acyltransferase (phosphatidylcholine--retinol O-acyltransferase) |
| chr17_-_42277203 | 0.10 |
ENST00000587097.1 |
ATXN7L3 |
ataxin 7-like 3 |
| chr17_-_73874654 | 0.10 |
ENST00000254816.2 |
TRIM47 |
tripartite motif containing 47 |
| chr11_-_2160180 | 0.10 |
ENST00000381406.4 |
IGF2 |
insulin-like growth factor 2 (somatomedin A) |
| chr15_-_72612470 | 0.09 |
ENST00000287202.5 |
CELF6 |
CUGBP, Elav-like family member 6 |
| chr9_-_130517522 | 0.09 |
ENST00000373274.3 ENST00000420366.1 |
SH2D3C |
SH2 domain containing 3C |
| chr10_+_72972281 | 0.09 |
ENST00000335350.6 |
UNC5B |
unc-5 homolog B (C. elegans) |
| chr11_-_46940074 | 0.09 |
ENST00000378623.1 ENST00000534404.1 |
LRP4 |
low density lipoprotein receptor-related protein 4 |
| chr11_+_100558384 | 0.09 |
ENST00000524892.2 ENST00000298815.8 |
ARHGAP42 |
Rho GTPase activating protein 42 |
| chr2_+_70142189 | 0.09 |
ENST00000264444.2 |
MXD1 |
MAX dimerization protein 1 |
| chr2_-_27603582 | 0.09 |
ENST00000323703.6 ENST00000436006.1 |
ZNF513 |
zinc finger protein 513 |
| chr4_-_6202291 | 0.09 |
ENST00000282924.5 |
JAKMIP1 |
janus kinase and microtubule interacting protein 1 |
| chr1_-_156698181 | 0.09 |
ENST00000313146.6 |
ISG20L2 |
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr16_-_30102547 | 0.09 |
ENST00000279386.2 |
TBX6 |
T-box 6 |
| chr17_-_7145475 | 0.08 |
ENST00000571129.1 ENST00000571253.1 ENST00000573928.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr19_+_34287751 | 0.08 |
ENST00000590771.1 ENST00000589786.1 ENST00000284006.6 ENST00000588881.1 |
KCTD15 |
potassium channel tetramerization domain containing 15 |
| chr12_-_51611477 | 0.08 |
ENST00000389243.4 |
POU6F1 |
POU class 6 homeobox 1 |
| chr12_-_112450915 | 0.08 |
ENST00000437003.2 ENST00000552374.2 ENST00000550831.3 ENST00000354825.3 ENST00000549537.2 ENST00000355445.3 |
TMEM116 |
transmembrane protein 116 |
| chr10_+_23983671 | 0.08 |
ENST00000376462.1 |
KIAA1217 |
KIAA1217 |
| chr4_-_6202247 | 0.08 |
ENST00000409021.3 ENST00000409371.3 |
JAKMIP1 |
janus kinase and microtubule interacting protein 1 |
| chr6_+_24495067 | 0.08 |
ENST00000357578.3 ENST00000546278.1 ENST00000491546.1 |
ALDH5A1 |
aldehyde dehydrogenase 5 family, member A1 |
| chr2_+_220408724 | 0.08 |
ENST00000421791.1 ENST00000373883.3 ENST00000451952.1 |
TMEM198 |
transmembrane protein 198 |
| chr1_+_38273818 | 0.08 |
ENST00000373042.4 |
C1orf122 |
chromosome 1 open reading frame 122 |
| chr10_-_75255668 | 0.08 |
ENST00000545874.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr3_-_171178157 | 0.08 |
ENST00000465393.1 ENST00000436636.2 ENST00000369326.5 ENST00000538048.1 ENST00000341852.6 |
TNIK |
TRAF2 and NCK interacting kinase |
| chr6_+_107811162 | 0.08 |
ENST00000317357.5 |
SOBP |
sine oculis binding protein homolog (Drosophila) |
| chr11_+_71238313 | 0.08 |
ENST00000398536.4 |
KRTAP5-7 |
keratin associated protein 5-7 |
| chr19_+_10400615 | 0.08 |
ENST00000221980.4 |
ICAM5 |
intercellular adhesion molecule 5, telencephalin |
| chr11_+_117070037 | 0.08 |
ENST00000392951.4 ENST00000525531.1 ENST00000278968.6 |
TAGLN |
transgelin |
| chr11_+_64008525 | 0.08 |
ENST00000449942.2 |
FKBP2 |
FK506 binding protein 2, 13kDa |
| chr20_-_41818536 | 0.07 |
ENST00000373193.3 ENST00000373198.4 ENST00000373201.1 |
PTPRT |
protein tyrosine phosphatase, receptor type, T |
| chr9_-_95896550 | 0.07 |
ENST00000375446.4 |
NINJ1 |
ninjurin 1 |
| chr1_+_160336851 | 0.07 |
ENST00000302101.5 |
NHLH1 |
nescient helix loop helix 1 |
| chrX_+_150863596 | 0.07 |
ENST00000448726.1 ENST00000538575.1 |
PRRG3 |
proline rich Gla (G-carboxyglutamic acid) 3 (transmembrane) |
| chr8_-_57123815 | 0.07 |
ENST00000316981.3 ENST00000423799.2 ENST00000429357.2 |
PLAG1 |
pleiomorphic adenoma gene 1 |
| chr1_-_154531095 | 0.07 |
ENST00000292211.4 |
UBE2Q1 |
ubiquitin-conjugating enzyme E2Q family member 1 |
| chr1_-_165325449 | 0.07 |
ENST00000294816.2 |
LMX1A |
LIM homeobox transcription factor 1, alpha |
| chr20_-_31172598 | 0.07 |
ENST00000201961.2 |
C20orf112 |
chromosome 20 open reading frame 112 |
| chr20_-_41818373 | 0.07 |
ENST00000373187.1 ENST00000356100.2 ENST00000373184.1 ENST00000373190.1 |
PTPRT |
protein tyrosine phosphatase, receptor type, T |
| chr16_+_75032901 | 0.07 |
ENST00000335325.4 ENST00000320619.6 |
ZNRF1 |
zinc and ring finger 1, E3 ubiquitin protein ligase |
| chr12_+_10365404 | 0.07 |
ENST00000266458.5 ENST00000421801.2 ENST00000544284.1 ENST00000545047.1 ENST00000543602.1 ENST00000545887.1 |
GABARAPL1 |
GABA(A) receptor-associated protein like 1 |
| chr17_-_36762095 | 0.07 |
ENST00000578925.1 ENST00000264659.7 |
SRCIN1 |
SRC kinase signaling inhibitor 1 |
| chr8_-_145559943 | 0.07 |
ENST00000332135.4 |
SCRT1 |
scratch family zinc finger 1 |
| chr15_-_83316254 | 0.07 |
ENST00000567678.1 ENST00000450751.2 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr7_-_150864635 | 0.07 |
ENST00000297537.4 |
GBX1 |
gastrulation brain homeobox 1 |
| chr20_+_2673383 | 0.07 |
ENST00000380648.4 ENST00000342725.5 |
EBF4 |
early B-cell factor 4 |
| chr2_-_220408430 | 0.07 |
ENST00000243776.6 |
CHPF |
chondroitin polymerizing factor |
| chr12_+_53773944 | 0.06 |
ENST00000551969.1 ENST00000327443.4 |
SP1 |
Sp1 transcription factor |
| chr3_+_50192499 | 0.06 |
ENST00000413852.1 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr19_+_10196981 | 0.06 |
ENST00000591813.1 |
C19orf66 |
chromosome 19 open reading frame 66 |
| chr15_+_41136586 | 0.06 |
ENST00000431806.1 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr1_-_226924980 | 0.06 |
ENST00000272117.3 |
ITPKB |
inositol-trisphosphate 3-kinase B |
| chr8_+_28196157 | 0.06 |
ENST00000522209.1 |
PNOC |
prepronociceptin |
| chr1_+_107682629 | 0.06 |
ENST00000370074.4 ENST00000370073.2 ENST00000370071.2 ENST00000542803.1 ENST00000370061.3 ENST00000370072.3 ENST00000370070.2 |
NTNG1 |
netrin G1 |
| chr1_+_6845578 | 0.06 |
ENST00000467404.2 ENST00000439411.2 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr14_-_92572894 | 0.06 |
ENST00000532032.1 ENST00000506466.1 ENST00000555381.1 ENST00000557311.1 ENST00000554592.1 ENST00000554672.1 ENST00000553491.1 ENST00000556220.1 ENST00000502250.1 ENST00000503767.1 ENST00000393287.5 ENST00000340660.6 ENST00000545170.1 ENST00000429774.2 |
ATXN3 |
ataxin 3 |
| chr16_+_67465016 | 0.06 |
ENST00000326152.5 |
HSD11B2 |
hydroxysteroid (11-beta) dehydrogenase 2 |
| chr1_+_107683436 | 0.06 |
ENST00000370068.1 |
NTNG1 |
netrin G1 |
| chr1_-_221915418 | 0.06 |
ENST00000323825.3 ENST00000366899.3 |
DUSP10 |
dual specificity phosphatase 10 |
| chr6_+_19837592 | 0.06 |
ENST00000378700.3 |
ID4 |
inhibitor of DNA binding 4, dominant negative helix-loop-helix protein |
| chr2_-_30144432 | 0.06 |
ENST00000389048.3 |
ALK |
anaplastic lymphoma receptor tyrosine kinase |
| chr2_+_220306745 | 0.06 |
ENST00000431523.1 ENST00000396698.1 ENST00000396695.2 |
SPEG |
SPEG complex locus |
| chr4_+_88928777 | 0.06 |
ENST00000237596.2 |
PKD2 |
polycystic kidney disease 2 (autosomal dominant) |
| chr11_-_133826852 | 0.06 |
ENST00000533871.2 ENST00000321016.8 |
IGSF9B |
immunoglobulin superfamily, member 9B |
| chr11_-_72492903 | 0.06 |
ENST00000537947.1 |
STARD10 |
StAR-related lipid transfer (START) domain containing 10 |
| chr2_-_219850277 | 0.06 |
ENST00000295727.1 |
FEV |
FEV (ETS oncogene family) |
| chr17_-_79008373 | 0.06 |
ENST00000577066.1 ENST00000573167.1 |
BAIAP2-AS1 |
BAIAP2 antisense RNA 1 (head to head) |
| chr5_+_42423872 | 0.06 |
ENST00000230882.4 ENST00000357703.3 |
GHR |
growth hormone receptor |
| chr1_+_207226574 | 0.06 |
ENST00000367080.3 ENST00000367079.2 |
PFKFB2 |
6-phosphofructo-2-kinase/fructose-2,6-biphosphatase 2 |
| chr3_+_50192537 | 0.06 |
ENST00000002829.3 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr19_-_41196534 | 0.06 |
ENST00000252891.4 |
NUMBL |
numb homolog (Drosophila)-like |
| chr12_+_110152033 | 0.06 |
ENST00000538780.1 |
FAM222A |
family with sequence similarity 222, member A |
| chr14_+_23340822 | 0.06 |
ENST00000359591.4 |
LRP10 |
low density lipoprotein receptor-related protein 10 |
| chr1_+_36023035 | 0.06 |
ENST00000373253.3 |
NCDN |
neurochondrin |
| chr1_-_205649580 | 0.06 |
ENST00000367145.3 |
SLC45A3 |
solute carrier family 45, member 3 |
| chr2_-_211036051 | 0.05 |
ENST00000418791.1 ENST00000452086.1 ENST00000281772.9 |
KANSL1L |
KAT8 regulatory NSL complex subunit 1-like |
| chr10_+_76871229 | 0.05 |
ENST00000372690.3 |
SAMD8 |
sterile alpha motif domain containing 8 |
| chr1_+_6845497 | 0.05 |
ENST00000473578.1 ENST00000557126.1 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chrX_-_49056635 | 0.05 |
ENST00000472598.1 ENST00000538567.1 ENST00000479808.1 ENST00000263233.4 |
SYP |
synaptophysin |
| chr15_+_74422585 | 0.05 |
ENST00000561740.1 ENST00000435464.1 ENST00000453268.2 |
ISLR2 |
immunoglobulin superfamily containing leucine-rich repeat 2 |
| chr17_+_36508111 | 0.05 |
ENST00000331159.5 ENST00000577233.1 |
SOCS7 |
suppressor of cytokine signaling 7 |
| chr12_+_57610562 | 0.05 |
ENST00000349394.5 |
NXPH4 |
neurexophilin 4 |
| chr1_+_33722080 | 0.05 |
ENST00000483388.1 ENST00000539719.1 |
ZNF362 |
zinc finger protein 362 |
| chr1_-_87380002 | 0.05 |
ENST00000331835.5 |
SEP15 |
Homo sapiens 15 kDa selenoprotein (SEP15), transcript variant 2, mRNA. |
| chr1_-_117664317 | 0.05 |
ENST00000256649.4 ENST00000369464.3 ENST00000485032.1 |
TRIM45 |
tripartite motif containing 45 |
| chr12_+_56473628 | 0.05 |
ENST00000549282.1 ENST00000549061.1 ENST00000267101.3 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr1_-_156698591 | 0.05 |
ENST00000368219.1 |
ISG20L2 |
interferon stimulated exonuclease gene 20kDa-like 2 |
| chr7_+_36192855 | 0.05 |
ENST00000534978.1 |
EEPD1 |
endonuclease/exonuclease/phosphatase family domain containing 1 |
| chr7_-_27239703 | 0.05 |
ENST00000222753.4 |
HOXA13 |
homeobox A13 |
| chr8_-_101322132 | 0.05 |
ENST00000523481.1 |
RNF19A |
ring finger protein 19A, RBR E3 ubiquitin protein ligase |
| chr22_+_31608219 | 0.05 |
ENST00000406516.1 ENST00000444929.2 ENST00000331728.4 |
LIMK2 |
LIM domain kinase 2 |
| chr3_-_24536253 | 0.05 |
ENST00000428492.1 ENST00000396671.2 ENST00000431815.1 ENST00000418247.1 ENST00000416420.1 ENST00000356447.4 |
THRB |
thyroid hormone receptor, beta |
| chr12_+_112451120 | 0.05 |
ENST00000261735.3 ENST00000455836.1 |
ERP29 |
endoplasmic reticulum protein 29 |
| chr14_+_100705322 | 0.05 |
ENST00000262238.4 |
YY1 |
YY1 transcription factor |
| chr11_+_129939811 | 0.05 |
ENST00000345598.5 ENST00000338167.5 |
APLP2 |
amyloid beta (A4) precursor-like protein 2 |
| chr3_+_52280173 | 0.05 |
ENST00000296487.4 |
PPM1M |
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr19_+_50084561 | 0.05 |
ENST00000246794.5 |
PRRG2 |
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr3_-_121740969 | 0.05 |
ENST00000393631.1 ENST00000273691.3 ENST00000344209.5 |
ILDR1 |
immunoglobulin-like domain containing receptor 1 |
| chr10_-_124768300 | 0.05 |
ENST00000368886.5 |
IKZF5 |
IKAROS family zinc finger 5 (Pegasus) |
| chr1_+_87797351 | 0.05 |
ENST00000370542.1 |
LMO4 |
LIM domain only 4 |
| chr17_-_27278304 | 0.05 |
ENST00000577226.1 |
PHF12 |
PHD finger protein 12 |
| chr1_+_200993071 | 0.05 |
ENST00000446333.1 ENST00000458003.1 |
RP11-168O16.1 |
RP11-168O16.1 |
| chr3_+_52280220 | 0.05 |
ENST00000409502.3 ENST00000323588.4 |
PPM1M |
protein phosphatase, Mg2+/Mn2+ dependent, 1M |
| chr1_-_36022979 | 0.05 |
ENST00000469892.1 ENST00000325722.3 |
KIAA0319L |
KIAA0319-like |
| chr11_+_22688150 | 0.05 |
ENST00000454584.2 |
GAS2 |
growth arrest-specific 2 |
| chr10_+_11060004 | 0.05 |
ENST00000542579.1 ENST00000399850.3 ENST00000417956.2 |
CELF2 |
CUGBP, Elav-like family member 2 |
| chr1_-_37499726 | 0.05 |
ENST00000373091.3 ENST00000373093.4 |
GRIK3 |
glutamate receptor, ionotropic, kainate 3 |
| chr3_-_48700310 | 0.05 |
ENST00000164024.4 ENST00000544264.1 |
CELSR3 |
cadherin, EGF LAG seven-pass G-type receptor 3 |
| chr5_+_109025067 | 0.05 |
ENST00000261483.4 |
MAN2A1 |
mannosidase, alpha, class 2A, member 1 |
| chrX_+_152990302 | 0.05 |
ENST00000218104.3 |
ABCD1 |
ATP-binding cassette, sub-family D (ALD), member 1 |
| chr11_+_129939779 | 0.05 |
ENST00000533195.1 ENST00000533713.1 ENST00000528499.1 ENST00000539648.1 ENST00000263574.5 |
APLP2 |
amyloid beta (A4) precursor-like protein 2 |
| chr19_+_42788172 | 0.05 |
ENST00000160740.3 |
CIC |
capicua transcriptional repressor |
| chr11_-_115375107 | 0.05 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chr12_-_50222187 | 0.04 |
ENST00000335999.6 |
NCKAP5L |
NCK-associated protein 5-like |
| chr15_+_41136216 | 0.04 |
ENST00000562057.1 ENST00000344051.4 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr5_-_81046841 | 0.04 |
ENST00000509013.2 ENST00000505980.1 ENST00000509053.1 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr14_-_23451845 | 0.04 |
ENST00000262713.2 |
AJUBA |
ajuba LIM protein |
| chr8_-_103668114 | 0.04 |
ENST00000285407.6 |
KLF10 |
Kruppel-like factor 10 |
| chr16_+_69600209 | 0.04 |
ENST00000566899.1 |
NFAT5 |
nuclear factor of activated T-cells 5, tonicity-responsive |
| chr17_+_42264556 | 0.04 |
ENST00000319511.6 ENST00000589785.1 ENST00000592825.1 ENST00000589184.1 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr8_+_26240414 | 0.04 |
ENST00000380629.2 |
BNIP3L |
BCL2/adenovirus E1B 19kDa interacting protein 3-like |
| chr8_+_125486939 | 0.04 |
ENST00000303545.3 |
RNF139 |
ring finger protein 139 |
| chr17_+_40932610 | 0.04 |
ENST00000246914.5 |
WNK4 |
WNK lysine deficient protein kinase 4 |
| chr11_-_1629693 | 0.04 |
ENST00000399685.1 |
KRTAP5-3 |
keratin associated protein 5-3 |
| chr4_-_129208940 | 0.04 |
ENST00000296425.5 |
PGRMC2 |
progesterone receptor membrane component 2 |
| chr6_+_32811861 | 0.04 |
ENST00000453426.1 |
TAPSAR1 |
TAP1 and PSMB8 antisense RNA 1 |
| chr10_+_70587279 | 0.04 |
ENST00000298596.6 ENST00000399169.4 ENST00000399165.4 ENST00000399162.2 |
STOX1 |
storkhead box 1 |
| chr1_+_6845384 | 0.04 |
ENST00000303635.7 |
CAMTA1 |
calmodulin binding transcription activator 1 |
| chr6_+_21593972 | 0.04 |
ENST00000244745.1 ENST00000543472.1 |
SOX4 |
SRY (sex determining region Y)-box 4 |
| chr3_-_53080047 | 0.04 |
ENST00000482396.1 ENST00000358080.2 ENST00000296295.6 ENST00000394752.3 |
SFMBT1 |
Scm-like with four mbt domains 1 |
| chr8_-_8243968 | 0.04 |
ENST00000520004.1 |
SGK223 |
Tyrosine-protein kinase SgK223 |
| chr2_+_210636697 | 0.04 |
ENST00000439458.1 ENST00000272845.6 |
UNC80 |
unc-80 homolog (C. elegans) |
| chr10_-_103603523 | 0.04 |
ENST00000370046.1 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr10_-_103603568 | 0.04 |
ENST00000356640.2 |
KCNIP2 |
Kv channel interacting protein 2 |
| chr19_+_4153598 | 0.04 |
ENST00000078445.2 ENST00000252587.3 ENST00000595923.1 ENST00000602257.1 ENST00000602147.1 |
CREB3L3 |
cAMP responsive element binding protein 3-like 3 |
| chr15_+_44084040 | 0.04 |
ENST00000249786.4 |
SERF2 |
small EDRK-rich factor 2 |
| chr13_-_45151259 | 0.04 |
ENST00000493016.1 |
TSC22D1 |
TSC22 domain family, member 1 |
| chr1_-_160040038 | 0.04 |
ENST00000368089.3 |
KCNJ10 |
potassium inwardly-rectifying channel, subfamily J, member 10 |
| chr4_+_52709229 | 0.04 |
ENST00000334635.5 ENST00000381441.3 ENST00000381437.4 |
DCUN1D4 |
DCN1, defective in cullin neddylation 1, domain containing 4 |
| chr17_+_9548845 | 0.04 |
ENST00000570475.1 ENST00000285199.7 |
USP43 |
ubiquitin specific peptidase 43 |
| chr17_-_39942940 | 0.04 |
ENST00000310706.5 ENST00000393931.3 ENST00000424457.1 ENST00000591690.1 |
JUP |
junction plakoglobin |
| chr14_+_94640633 | 0.04 |
ENST00000304338.3 |
PPP4R4 |
protein phosphatase 4, regulatory subunit 4 |
| chr17_-_10101868 | 0.04 |
ENST00000432992.2 ENST00000540214.1 |
GAS7 |
growth arrest-specific 7 |
| chr2_-_97405775 | 0.04 |
ENST00000264963.4 ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L |
lectin, mannose-binding 2-like |
| chr20_+_48807351 | 0.04 |
ENST00000303004.3 |
CEBPB |
CCAAT/enhancer binding protein (C/EBP), beta |
| chr5_-_81046904 | 0.04 |
ENST00000515395.1 |
SSBP2 |
single-stranded DNA binding protein 2 |
| chr4_+_26862313 | 0.04 |
ENST00000467087.1 ENST00000382009.3 ENST00000237364.5 |
STIM2 |
stromal interaction molecule 2 |
| chr1_-_231004220 | 0.04 |
ENST00000366663.5 |
C1orf198 |
chromosome 1 open reading frame 198 |
| chr2_-_135476552 | 0.04 |
ENST00000281924.6 |
TMEM163 |
transmembrane protein 163 |
| chr4_+_183370146 | 0.04 |
ENST00000510504.1 |
TENM3 |
teneurin transmembrane protein 3 |
| chrX_-_47479246 | 0.04 |
ENST00000295987.7 ENST00000340666.4 |
SYN1 |
synapsin I |
| chr6_+_32811885 | 0.04 |
ENST00000458296.1 ENST00000413039.1 ENST00000429600.1 ENST00000412095.1 ENST00000415067.1 ENST00000395330.1 |
TAPSAR1 PSMB9 |
TAP1 and PSMB8 antisense RNA 1 proteasome (prosome, macropain) subunit, beta type, 9 |
| chr4_-_153456153 | 0.04 |
ENST00000603548.1 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr2_+_241375069 | 0.04 |
ENST00000264039.2 |
GPC1 |
glypican 1 |
| chr18_-_24129367 | 0.04 |
ENST00000408011.3 |
KCTD1 |
potassium channel tetramerization domain containing 1 |
| chr17_+_59477233 | 0.04 |
ENST00000240328.3 |
TBX2 |
T-box 2 |
| chr17_+_10600894 | 0.04 |
ENST00000379774.4 |
ADPRM |
ADP-ribose/CDP-alcohol diphosphatase, manganese-dependent |
| chr1_+_186344883 | 0.04 |
ENST00000367470.3 |
C1orf27 |
chromosome 1 open reading frame 27 |
| chr2_+_32502952 | 0.04 |
ENST00000238831.4 |
YIPF4 |
Yip1 domain family, member 4 |
| chr12_+_70760056 | 0.04 |
ENST00000258111.4 |
KCNMB4 |
potassium large conductance calcium-activated channel, subfamily M, beta member 4 |
| chr3_+_49449636 | 0.04 |
ENST00000273590.3 |
TCTA |
T-cell leukemia translocation altered |
| chr3_-_141868357 | 0.04 |
ENST00000489671.1 ENST00000475734.1 ENST00000467072.1 ENST00000499676.2 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr4_+_26862431 | 0.04 |
ENST00000465503.1 |
STIM2 |
stromal interaction molecule 2 |
| chr12_-_54785074 | 0.04 |
ENST00000338010.5 ENST00000550774.1 |
ZNF385A |
zinc finger protein 385A |
| chr1_+_43148059 | 0.04 |
ENST00000321358.7 ENST00000332220.6 |
YBX1 |
Y box binding protein 1 |
| chr5_-_153857819 | 0.04 |
ENST00000231121.2 |
HAND1 |
heart and neural crest derivatives expressed 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0060901 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.1 | 0.3 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.1 | 0.2 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.1 | GO:0097374 | sensory neuron axon guidance(GO:0097374) |
| 0.1 | 0.3 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0009450 | gamma-aminobutyric acid catabolic process(GO:0009450) |
| 0.0 | 0.1 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.0 | 0.1 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) |
| 0.0 | 0.5 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.0 | 0.1 | GO:0021849 | neuroblast division in subventricular zone(GO:0021849) |
| 0.0 | 0.1 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.0 | 0.1 | GO:0009726 | detection of endogenous stimulus(GO:0009726) |
| 0.0 | 0.1 | GO:0015770 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.0 | 0.1 | GO:0042997 | negative regulation of Golgi to plasma membrane protein transport(GO:0042997) |
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0014043 | negative regulation of neuron maturation(GO:0014043) |
| 0.0 | 0.1 | GO:0032237 | activation of store-operated calcium channel activity(GO:0032237) positive regulation of store-operated calcium channel activity(GO:1901341) |
| 0.0 | 0.2 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.1 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.0 | 0.1 | GO:0036269 | swimming behavior(GO:0036269) |
| 0.0 | 0.1 | GO:0061438 | renal system vasculature morphogenesis(GO:0061438) kidney vasculature morphogenesis(GO:0061439) glomerulus vasculature morphogenesis(GO:0072103) glomerular capillary formation(GO:0072104) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.0 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.0 | 0.1 | GO:1902164 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902162) positive regulation of DNA damage response, signal transduction by p53 class mediator resulting in transcription of p21 class mediator(GO:1902164) |
| 0.0 | 0.0 | GO:1901207 | regulation of heart looping(GO:1901207) |
| 0.0 | 0.1 | GO:0033564 | anterior/posterior axon guidance(GO:0033564) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.0 | GO:0003218 | cardiac left ventricle formation(GO:0003218) |
| 0.0 | 0.0 | GO:2000820 | negative regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000820) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) |
| 0.0 | 0.1 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.0 | GO:0042996 | regulation of Golgi to plasma membrane protein transport(GO:0042996) |
| 0.0 | 0.1 | GO:0061107 | prostate gland stromal morphogenesis(GO:0060741) seminal vesicle development(GO:0061107) |
| 0.0 | 0.1 | GO:0014826 | vein smooth muscle contraction(GO:0014826) |
| 0.0 | 0.0 | GO:1902336 | VEGF-activated platelet-derived growth factor receptor signaling pathway(GO:0038086) positive regulation of cell proliferation by VEGF-activated platelet derived growth factor receptor signaling pathway(GO:0038091) neuropilin signaling pathway(GO:0038189) VEGF-activated neuropilin signaling pathway(GO:0038190) positive regulation of retinal ganglion cell axon guidance(GO:1902336) regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.0 | GO:0015910 | peroxisomal long-chain fatty acid import(GO:0015910) |
| 0.0 | 0.1 | GO:1903301 | positive regulation of glucokinase activity(GO:0033133) positive regulation of hexokinase activity(GO:1903301) |
| 0.0 | 0.0 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.0 | 0.0 | GO:0043397 | corticotropin-releasing hormone secretion(GO:0043396) regulation of corticotropin-releasing hormone secretion(GO:0043397) |
| 0.0 | 0.0 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.1 | GO:0032927 | positive regulation of activin receptor signaling pathway(GO:0032927) |
| 0.0 | 0.2 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.0 | GO:0071500 | cellular response to nitrosative stress(GO:0071500) |
| 0.0 | 0.0 | GO:0035910 | N-terminal peptidyl-lysine acetylation(GO:0018076) ascending aorta development(GO:0035905) ascending aorta morphogenesis(GO:0035910) |
| 0.0 | 0.1 | GO:0038028 | insulin receptor signaling pathway via phosphatidylinositol 3-kinase(GO:0038028) |
| 0.0 | 0.0 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.0 | 0.1 | GO:0070195 | growth hormone receptor complex(GO:0070195) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.0 | GO:0060187 | cell pole(GO:0060187) |
| 0.0 | 0.0 | GO:0036195 | muscle cell projection(GO:0036194) muscle cell projection membrane(GO:0036195) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0030348 | syntaxin-3 binding(GO:0030348) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.3 | GO:0045294 | alpha-catenin binding(GO:0045294) |
| 0.0 | 0.1 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.0 | 0.0 | GO:0070506 | high-density lipoprotein particle receptor activity(GO:0070506) |
| 0.0 | 0.1 | GO:0050510 | N-acetylgalactosaminyl-proteoglycan 3-beta-glucuronosyltransferase activity(GO:0050510) |
| 0.0 | 0.1 | GO:0017002 | activin-activated receptor activity(GO:0017002) |
| 0.0 | 0.1 | GO:0048763 | HLH domain binding(GO:0043398) calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.1 | GO:0016416 | O-palmitoyltransferase activity(GO:0016416) |
| 0.0 | 0.0 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.0 | 0.0 | GO:0050698 | proteoglycan sulfotransferase activity(GO:0050698) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0031708 | endothelin B receptor binding(GO:0031708) |
| 0.0 | 0.1 | GO:0008420 | CTD phosphatase activity(GO:0008420) |
| 0.0 | 0.1 | GO:0033188 | sphingomyelin synthase activity(GO:0033188) ceramide cholinephosphotransferase activity(GO:0047493) |
| 0.0 | 0.0 | GO:0008184 | purine nucleobase binding(GO:0002060) glycogen phosphorylase activity(GO:0008184) |
| 0.0 | 0.3 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.0 | GO:0003863 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME DOPAMINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Dopamine Neurotransmitter Release Cycle |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.2 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |