Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for MAF_NRL
Z-value: 0.53
Transcription factors associated with MAF_NRL
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
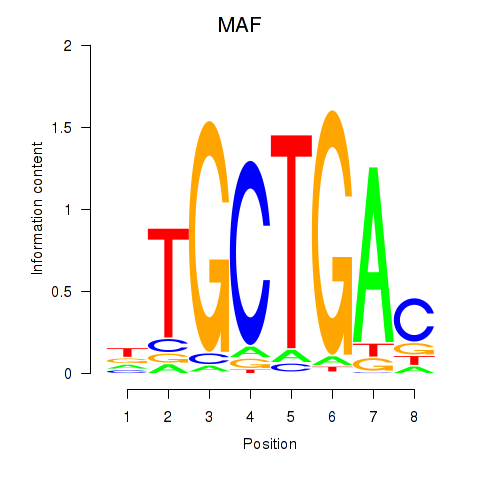
MAF
|
ENSG00000178573.6 | MAF |
|
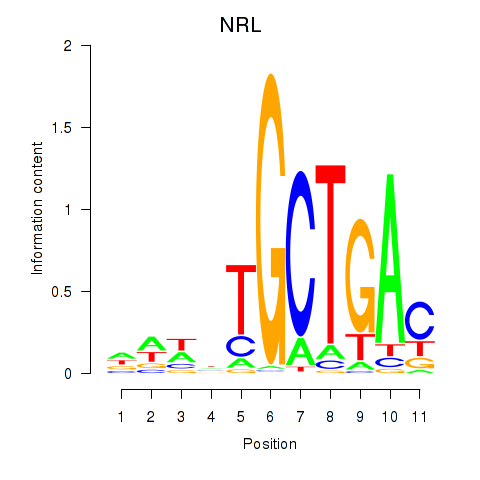
NRL
|
ENSG00000129535.8 | NRL |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NRL | hg19_v2_chr14_-_24551137_24551178, hg19_v2_chr14_-_24551195_24551214 | -0.66 | 7.4e-02 | Click! |
| MAF | hg19_v2_chr16_-_79633799_79633872 | -0.50 | 2.1e-01 | Click! |
Activity profile of MAF_NRL motif
Sorted Z-values of MAF_NRL motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MAF_NRL
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_233749739 | 0.65 |
ENST00000366621.3 |
KCNK1 |
potassium channel, subfamily K, member 1 |
| chr18_+_61554932 | 0.62 |
ENST00000299502.4 ENST00000457692.1 ENST00000413956.1 |
SERPINB2 |
serpin peptidase inhibitor, clade B (ovalbumin), member 2 |
| chr1_-_201368653 | 0.60 |
ENST00000367313.3 |
LAD1 |
ladinin 1 |
| chr1_-_201368707 | 0.59 |
ENST00000391967.2 |
LAD1 |
ladinin 1 |
| chr1_+_24646002 | 0.57 |
ENST00000356046.2 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr1_+_24645807 | 0.55 |
ENST00000361548.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr1_+_24645865 | 0.54 |
ENST00000342072.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr17_+_48610074 | 0.43 |
ENST00000503690.1 ENST00000514874.1 ENST00000537145.1 ENST00000541226.1 |
EPN3 |
epsin 3 |
| chr14_+_67999999 | 0.41 |
ENST00000329153.5 |
PLEKHH1 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr17_-_39769005 | 0.41 |
ENST00000301653.4 ENST00000593067.1 |
KRT16 |
keratin 16 |
| chr7_-_41742697 | 0.38 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr8_+_32579341 | 0.38 |
ENST00000519240.1 ENST00000539990.1 |
NRG1 |
neuregulin 1 |
| chr17_-_39674668 | 0.37 |
ENST00000393981.3 |
KRT15 |
keratin 15 |
| chr6_+_80129989 | 0.32 |
ENST00000429444.1 |
RP1-232L24.3 |
RP1-232L24.3 |
| chr1_+_2004901 | 0.30 |
ENST00000400921.2 |
PRKCZ |
protein kinase C, zeta |
| chr7_+_26331541 | 0.30 |
ENST00000416246.1 ENST00000338523.4 ENST00000412416.1 |
SNX10 |
sorting nexin 10 |
| chr6_-_33041378 | 0.30 |
ENST00000428995.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr1_+_2005425 | 0.27 |
ENST00000461106.2 |
PRKCZ |
protein kinase C, zeta |
| chr19_-_51456321 | 0.26 |
ENST00000391809.2 |
KLK5 |
kallikrein-related peptidase 5 |
| chr19_-_51456344 | 0.25 |
ENST00000336334.3 ENST00000593428.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr17_-_56494713 | 0.25 |
ENST00000407977.2 |
RNF43 |
ring finger protein 43 |
| chr1_+_62208091 | 0.25 |
ENST00000316485.6 ENST00000371158.2 |
INADL |
InaD-like (Drosophila) |
| chr1_+_22333943 | 0.25 |
ENST00000400271.2 |
CELA3A |
chymotrypsin-like elastase family, member 3A |
| chr17_-_3595181 | 0.25 |
ENST00000552050.1 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chrX_-_15683147 | 0.25 |
ENST00000380342.3 |
TMEM27 |
transmembrane protein 27 |
| chr5_+_148206156 | 0.24 |
ENST00000305988.4 |
ADRB2 |
adrenoceptor beta 2, surface |
| chr11_-_118122996 | 0.24 |
ENST00000525386.1 ENST00000527472.1 ENST00000278949.4 |
MPZL3 |
myelin protein zero-like 3 |
| chr19_+_15052301 | 0.24 |
ENST00000248072.3 |
OR7C2 |
olfactory receptor, family 7, subfamily C, member 2 |
| chr17_+_30771279 | 0.23 |
ENST00000261712.3 ENST00000578213.1 ENST00000457654.2 ENST00000579451.1 |
PSMD11 |
proteasome (prosome, macropain) 26S subunit, non-ATPase, 11 |
| chr14_+_23842018 | 0.23 |
ENST00000397242.2 ENST00000329715.2 |
IL25 |
interleukin 25 |
| chr17_-_56494882 | 0.22 |
ENST00000584437.1 |
RNF43 |
ring finger protein 43 |
| chr17_-_56494908 | 0.22 |
ENST00000577716.1 |
RNF43 |
ring finger protein 43 |
| chr22_+_45148432 | 0.22 |
ENST00000389774.2 ENST00000396119.2 ENST00000336963.4 ENST00000356099.6 ENST00000412433.1 |
ARHGAP8 |
Rho GTPase activating protein 8 |
| chr9_+_93589734 | 0.22 |
ENST00000375746.1 |
SYK |
spleen tyrosine kinase |
| chr2_+_207804278 | 0.22 |
ENST00000272852.3 |
CPO |
carboxypeptidase O |
| chr11_-_119599794 | 0.21 |
ENST00000264025.3 |
PVRL1 |
poliovirus receptor-related 1 (herpesvirus entry mediator C) |
| chr16_+_67233412 | 0.20 |
ENST00000477898.1 |
ELMO3 |
engulfment and cell motility 3 |
| chr6_+_121756809 | 0.20 |
ENST00000282561.3 |
GJA1 |
gap junction protein, alpha 1, 43kDa |
| chr17_-_73874654 | 0.20 |
ENST00000254816.2 |
TRIM47 |
tripartite motif containing 47 |
| chr1_+_152956549 | 0.20 |
ENST00000307122.2 |
SPRR1A |
small proline-rich protein 1A |
| chr6_+_143381979 | 0.20 |
ENST00000367598.5 ENST00000447498.1 ENST00000357847.4 ENST00000344492.5 ENST00000367596.1 ENST00000494282.2 ENST00000275235.4 |
AIG1 |
androgen-induced 1 |
| chr14_+_23654525 | 0.20 |
ENST00000399910.1 ENST00000492621.1 |
C14orf164 |
chromosome 14 open reading frame 164 |
| chr17_+_55183261 | 0.20 |
ENST00000576295.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr22_+_42196666 | 0.20 |
ENST00000402061.3 ENST00000255784.5 |
CCDC134 |
coiled-coil domain containing 134 |
| chr10_-_123357598 | 0.20 |
ENST00000358487.5 ENST00000369058.3 ENST00000369060.4 ENST00000359354.2 |
FGFR2 |
fibroblast growth factor receptor 2 |
| chr1_-_109935819 | 0.19 |
ENST00000538502.1 |
SORT1 |
sortilin 1 |
| chr3_+_189349162 | 0.19 |
ENST00000264731.3 ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63 |
tumor protein p63 |
| chr3_+_50192499 | 0.18 |
ENST00000413852.1 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr2_-_129076151 | 0.18 |
ENST00000259241.6 |
HS6ST1 |
heparan sulfate 6-O-sulfotransferase 1 |
| chr10_+_71389983 | 0.18 |
ENST00000373279.4 |
C10orf35 |
chromosome 10 open reading frame 35 |
| chr17_-_54893250 | 0.17 |
ENST00000397862.2 |
C17orf67 |
chromosome 17 open reading frame 67 |
| chr7_-_22234381 | 0.17 |
ENST00000458533.1 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr19_-_49567124 | 0.17 |
ENST00000301411.3 |
NTF4 |
neurotrophin 4 |
| chrX_+_105969893 | 0.17 |
ENST00000255499.2 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr2_-_74757066 | 0.17 |
ENST00000377526.3 |
AUP1 |
ancient ubiquitous protein 1 |
| chr3_-_69435428 | 0.16 |
ENST00000542259.1 |
FRMD4B |
FERM domain containing 4B |
| chr5_+_89770696 | 0.16 |
ENST00000504930.1 ENST00000514483.1 |
POLR3G |
polymerase (RNA) III (DNA directed) polypeptide G (32kD) |
| chr16_+_2510081 | 0.16 |
ENST00000361837.4 ENST00000569496.1 ENST00000567489.1 ENST00000563531.1 ENST00000483320.1 |
C16orf59 |
chromosome 16 open reading frame 59 |
| chr1_-_85514120 | 0.16 |
ENST00000370589.2 ENST00000341115.4 ENST00000370587.1 |
MCOLN3 |
mucolipin 3 |
| chr21_+_44589118 | 0.16 |
ENST00000291554.2 |
CRYAA |
crystallin, alpha A |
| chr18_-_53089723 | 0.15 |
ENST00000561992.1 ENST00000562512.2 |
TCF4 |
transcription factor 4 |
| chr17_+_635786 | 0.15 |
ENST00000572018.1 ENST00000301324.8 |
FAM57A |
family with sequence similarity 57, member A |
| chr17_+_55173933 | 0.15 |
ENST00000539273.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr19_-_51530916 | 0.15 |
ENST00000594768.1 |
KLK11 |
kallikrein-related peptidase 11 |
| chr5_-_16936340 | 0.14 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr10_-_5446786 | 0.14 |
ENST00000479328.1 ENST00000380419.3 |
TUBAL3 |
tubulin, alpha-like 3 |
| chr17_+_635652 | 0.14 |
ENST00000308278.8 |
FAM57A |
family with sequence similarity 57, member A |
| chr14_-_77787198 | 0.14 |
ENST00000261534.4 |
POMT2 |
protein-O-mannosyltransferase 2 |
| chr22_+_31489344 | 0.14 |
ENST00000404574.1 |
SMTN |
smoothelin |
| chr8_-_145018905 | 0.14 |
ENST00000398774.2 |
PLEC |
plectin |
| chr14_-_106237742 | 0.14 |
ENST00000390551.2 |
IGHG3 |
immunoglobulin heavy constant gamma 3 (G3m marker) |
| chr13_+_113656022 | 0.14 |
ENST00000423482.2 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chr18_-_53253323 | 0.14 |
ENST00000540999.1 ENST00000563888.2 |
TCF4 |
transcription factor 4 |
| chr20_-_18774614 | 0.14 |
ENST00000412553.1 |
LINC00652 |
long intergenic non-protein coding RNA 652 |
| chr19_-_55660561 | 0.13 |
ENST00000587758.1 ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr15_-_68497657 | 0.13 |
ENST00000448060.2 ENST00000467889.1 |
CALML4 |
calmodulin-like 4 |
| chr1_-_12677714 | 0.13 |
ENST00000376223.2 |
DHRS3 |
dehydrogenase/reductase (SDR family) member 3 |
| chr22_+_45072925 | 0.13 |
ENST00000006251.7 |
PRR5 |
proline rich 5 (renal) |
| chr12_-_52828147 | 0.13 |
ENST00000252245.5 |
KRT75 |
keratin 75 |
| chr14_+_21510385 | 0.13 |
ENST00000298690.4 |
RNASE7 |
ribonuclease, RNase A family, 7 |
| chr11_-_119991589 | 0.13 |
ENST00000526881.1 |
TRIM29 |
tripartite motif containing 29 |
| chr22_+_45072958 | 0.13 |
ENST00000403581.1 |
PRR5 |
proline rich 5 (renal) |
| chr8_+_104831472 | 0.13 |
ENST00000262231.10 ENST00000507740.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr11_+_10471836 | 0.13 |
ENST00000444303.2 |
AMPD3 |
adenosine monophosphate deaminase 3 |
| chr11_+_6866883 | 0.13 |
ENST00000299454.4 ENST00000379831.2 |
OR10A5 |
olfactory receptor, family 10, subfamily A, member 5 |
| chr18_-_52969844 | 0.13 |
ENST00000561831.3 |
TCF4 |
transcription factor 4 |
| chr1_+_161691353 | 0.13 |
ENST00000367948.2 |
FCRLB |
Fc receptor-like B |
| chr3_+_50192537 | 0.13 |
ENST00000002829.3 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr1_+_3388181 | 0.12 |
ENST00000418137.1 ENST00000413250.2 |
ARHGEF16 |
Rho guanine nucleotide exchange factor (GEF) 16 |
| chr2_-_166651191 | 0.12 |
ENST00000392701.3 |
GALNT3 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 3 (GalNAc-T3) |
| chrX_-_153141302 | 0.12 |
ENST00000361699.4 ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM |
L1 cell adhesion molecule |
| chrX_+_128913906 | 0.12 |
ENST00000356892.3 |
SASH3 |
SAM and SH3 domain containing 3 |
| chr3_-_123339418 | 0.12 |
ENST00000583087.1 |
MYLK |
myosin light chain kinase |
| chr12_+_53662073 | 0.12 |
ENST00000553219.1 ENST00000257934.4 |
ESPL1 |
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr8_+_80523321 | 0.12 |
ENST00000518111.1 |
STMN2 |
stathmin-like 2 |
| chr17_-_57784755 | 0.12 |
ENST00000537860.1 ENST00000393038.2 ENST00000409433.2 |
PTRH2 |
peptidyl-tRNA hydrolase 2 |
| chr6_-_11779840 | 0.12 |
ENST00000506810.1 |
ADTRP |
androgen-dependent TFPI-regulating protein |
| chr15_-_59665062 | 0.12 |
ENST00000288235.4 |
MYO1E |
myosin IE |
| chr7_-_32111009 | 0.12 |
ENST00000396184.3 ENST00000396189.2 ENST00000321453.7 |
PDE1C |
phosphodiesterase 1C, calmodulin-dependent 70kDa |
| chr15_-_52587945 | 0.12 |
ENST00000443683.2 ENST00000558479.1 ENST00000261839.7 |
MYO5C |
myosin VC |
| chr3_-_98241358 | 0.12 |
ENST00000503004.1 ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1 |
claudin domain containing 1 |
| chr17_-_4448361 | 0.12 |
ENST00000572759.1 |
MYBBP1A |
MYB binding protein (P160) 1a |
| chr14_+_93118813 | 0.11 |
ENST00000556418.1 |
RIN3 |
Ras and Rab interactor 3 |
| chr9_+_27109392 | 0.11 |
ENST00000406359.4 |
TEK |
TEK tyrosine kinase, endothelial |
| chr3_+_98699880 | 0.11 |
ENST00000473756.1 |
LINC00973 |
long intergenic non-protein coding RNA 973 |
| chr9_-_117111222 | 0.11 |
ENST00000374079.4 |
AKNA |
AT-hook transcription factor |
| chr12_+_53662110 | 0.11 |
ENST00000552462.1 |
ESPL1 |
extra spindle pole bodies homolog 1 (S. cerevisiae) |
| chr12_+_20848282 | 0.11 |
ENST00000545604.1 |
SLCO1C1 |
solute carrier organic anion transporter family, member 1C1 |
| chr12_+_20848377 | 0.11 |
ENST00000540354.1 ENST00000266509.2 ENST00000381552.1 |
SLCO1C1 |
solute carrier organic anion transporter family, member 1C1 |
| chr18_-_53253112 | 0.11 |
ENST00000568673.1 ENST00000562847.1 ENST00000568147.1 |
TCF4 |
transcription factor 4 |
| chr19_-_51471362 | 0.11 |
ENST00000376853.4 ENST00000424910.2 |
KLK6 |
kallikrein-related peptidase 6 |
| chr3_-_50340996 | 0.11 |
ENST00000266031.4 ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1 |
hyaluronoglucosaminidase 1 |
| chr3_-_190167571 | 0.11 |
ENST00000354905.2 |
TMEM207 |
transmembrane protein 207 |
| chr2_+_113885138 | 0.10 |
ENST00000409930.3 |
IL1RN |
interleukin 1 receptor antagonist |
| chr17_-_27503770 | 0.10 |
ENST00000533112.1 |
MYO18A |
myosin XVIIIA |
| chr17_-_76128488 | 0.10 |
ENST00000322914.3 |
TMC6 |
transmembrane channel-like 6 |
| chr6_+_31582961 | 0.10 |
ENST00000376059.3 ENST00000337917.7 |
AIF1 |
allograft inflammatory factor 1 |
| chr4_+_128554081 | 0.10 |
ENST00000335251.6 ENST00000296461.5 |
INTU |
inturned planar cell polarity protein |
| chr12_-_14721283 | 0.10 |
ENST00000240617.5 |
PLBD1 |
phospholipase B domain containing 1 |
| chr14_+_76776957 | 0.10 |
ENST00000512784.1 |
ESRRB |
estrogen-related receptor beta |
| chr6_-_52109335 | 0.10 |
ENST00000336123.4 |
IL17F |
interleukin 17F |
| chr9_-_112260531 | 0.10 |
ENST00000374541.2 ENST00000262539.3 |
PTPN3 |
protein tyrosine phosphatase, non-receptor type 3 |
| chr6_-_33048483 | 0.10 |
ENST00000419277.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr15_-_34502197 | 0.10 |
ENST00000557877.1 |
KATNBL1 |
katanin p80 subunit B-like 1 |
| chr20_-_4721314 | 0.10 |
ENST00000418528.1 ENST00000326539.2 ENST00000423718.2 |
PRNT |
prion protein (testis specific) |
| chr6_-_42419649 | 0.10 |
ENST00000372922.4 ENST00000541110.1 ENST00000372917.4 |
TRERF1 |
transcriptional regulating factor 1 |
| chr2_-_20425158 | 0.10 |
ENST00000381150.1 |
SDC1 |
syndecan 1 |
| chr2_+_85661918 | 0.10 |
ENST00000340326.2 |
SH2D6 |
SH2 domain containing 6 |
| chr13_-_44735393 | 0.10 |
ENST00000400419.1 |
SMIM2 |
small integral membrane protein 2 |
| chr10_+_24755416 | 0.10 |
ENST00000396446.1 ENST00000396445.1 ENST00000376451.2 |
KIAA1217 |
KIAA1217 |
| chr16_-_84538218 | 0.10 |
ENST00000562447.1 ENST00000565765.1 ENST00000535580.1 ENST00000343629.6 |
TLDC1 |
TBC/LysM-associated domain containing 1 |
| chr11_-_116708302 | 0.10 |
ENST00000375320.1 ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1 |
apolipoprotein A-I |
| chr2_-_136678123 | 0.10 |
ENST00000422708.1 |
DARS |
aspartyl-tRNA synthetase |
| chr3_-_123339343 | 0.09 |
ENST00000578202.1 |
MYLK |
myosin light chain kinase |
| chr12_+_7282795 | 0.09 |
ENST00000266546.6 |
CLSTN3 |
calsyntenin 3 |
| chr5_+_55149150 | 0.09 |
ENST00000297015.3 |
IL31RA |
interleukin 31 receptor A |
| chr22_+_21128167 | 0.09 |
ENST00000215727.5 |
SERPIND1 |
serpin peptidase inhibitor, clade D (heparin cofactor), member 1 |
| chr3_+_130569429 | 0.09 |
ENST00000505330.1 ENST00000504381.1 ENST00000507488.2 ENST00000393221.4 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr14_-_23834411 | 0.09 |
ENST00000429593.2 |
EFS |
embryonal Fyn-associated substrate |
| chr9_+_139839686 | 0.09 |
ENST00000371634.2 |
C8G |
complement component 8, gamma polypeptide |
| chr11_-_124311054 | 0.09 |
ENST00000328064.2 |
OR8B8 |
olfactory receptor, family 8, subfamily B, member 8 |
| chr22_+_29279552 | 0.09 |
ENST00000544604.2 |
ZNRF3 |
zinc and ring finger 3 |
| chrX_+_41306575 | 0.09 |
ENST00000342595.2 ENST00000378220.1 |
NYX |
nyctalopin |
| chr1_-_112046289 | 0.09 |
ENST00000241356.4 |
ADORA3 |
adenosine A3 receptor |
| chr1_-_197115818 | 0.09 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr5_+_80529104 | 0.09 |
ENST00000254035.4 ENST00000511719.1 ENST00000437669.1 ENST00000424301.2 ENST00000505060.1 |
CKMT2 |
creatine kinase, mitochondrial 2 (sarcomeric) |
| chr5_-_176056974 | 0.09 |
ENST00000510387.1 ENST00000506696.1 |
SNCB |
synuclein, beta |
| chr10_+_5566916 | 0.08 |
ENST00000315238.1 |
CALML3 |
calmodulin-like 3 |
| chr18_-_74839891 | 0.08 |
ENST00000581878.1 |
MBP |
myelin basic protein |
| chr5_-_77844974 | 0.08 |
ENST00000515007.2 |
LHFPL2 |
lipoma HMGIC fusion partner-like 2 |
| chr15_-_68498376 | 0.08 |
ENST00000540479.1 ENST00000395465.3 |
CALML4 |
calmodulin-like 4 |
| chr1_+_100111479 | 0.08 |
ENST00000263174.4 |
PALMD |
palmdelphin |
| chr10_+_81272287 | 0.08 |
ENST00000520547.2 |
EIF5AL1 |
eukaryotic translation initiation factor 5A-like 1 |
| chr12_-_91451758 | 0.08 |
ENST00000266719.3 |
KERA |
keratocan |
| chr3_+_97851542 | 0.08 |
ENST00000354565.2 |
OR5H1 |
olfactory receptor, family 5, subfamily H, member 1 |
| chrX_-_100662881 | 0.08 |
ENST00000218516.3 |
GLA |
galactosidase, alpha |
| chr7_+_20370300 | 0.08 |
ENST00000537992.1 |
ITGB8 |
integrin, beta 8 |
| chr6_+_32407619 | 0.08 |
ENST00000395388.2 ENST00000374982.5 |
HLA-DRA |
major histocompatibility complex, class II, DR alpha |
| chr12_+_19358228 | 0.08 |
ENST00000424268.1 ENST00000543806.1 |
PLEKHA5 |
pleckstrin homology domain containing, family A member 5 |
| chr1_-_161015752 | 0.08 |
ENST00000435396.1 ENST00000368021.3 |
USF1 |
upstream transcription factor 1 |
| chr5_+_135364584 | 0.08 |
ENST00000442011.2 ENST00000305126.8 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr5_+_110427983 | 0.08 |
ENST00000513710.2 ENST00000505303.1 |
WDR36 |
WD repeat domain 36 |
| chr11_-_123756334 | 0.08 |
ENST00000528595.1 ENST00000375026.2 |
TMEM225 |
transmembrane protein 225 |
| chr17_-_39023462 | 0.08 |
ENST00000251643.4 |
KRT12 |
keratin 12 |
| chr4_+_95972822 | 0.08 |
ENST00000509540.1 ENST00000440890.2 |
BMPR1B |
bone morphogenetic protein receptor, type IB |
| chr19_+_50094866 | 0.08 |
ENST00000418929.2 |
PRR12 |
proline rich 12 |
| chr19_-_38746979 | 0.08 |
ENST00000591291.1 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr11_+_2323236 | 0.08 |
ENST00000182290.4 |
TSPAN32 |
tetraspanin 32 |
| chr14_+_24600484 | 0.07 |
ENST00000267426.5 |
FITM1 |
fat storage-inducing transmembrane protein 1 |
| chr17_-_39280419 | 0.07 |
ENST00000394014.1 |
KRTAP4-12 |
keratin associated protein 4-12 |
| chr3_+_184097905 | 0.07 |
ENST00000450923.1 |
CHRD |
chordin |
| chr19_-_54804173 | 0.07 |
ENST00000391744.3 ENST00000251390.3 |
LILRA3 |
leukocyte immunoglobulin-like receptor, subfamily A (without TM domain), member 3 |
| chr2_+_196521845 | 0.07 |
ENST00000359634.5 ENST00000412905.1 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
| chr2_+_196521458 | 0.07 |
ENST00000409086.3 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
| chr1_-_25256368 | 0.07 |
ENST00000308873.6 |
RUNX3 |
runt-related transcription factor 3 |
| chr9_-_131486367 | 0.07 |
ENST00000372663.4 ENST00000406904.2 ENST00000452105.1 ENST00000372672.2 ENST00000372667.5 |
ZDHHC12 |
zinc finger, DHHC-type containing 12 |
| chr10_-_50970322 | 0.07 |
ENST00000374103.4 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr16_-_10868853 | 0.07 |
ENST00000572428.1 |
TVP23A |
trans-golgi network vesicle protein 23 homolog A (S. cerevisiae) |
| chr17_-_38520067 | 0.07 |
ENST00000337376.4 ENST00000578689.1 |
GJD3 |
gap junction protein, delta 3, 31.9kDa |
| chr7_+_39125365 | 0.07 |
ENST00000559001.1 ENST00000464276.2 |
POU6F2 |
POU class 6 homeobox 2 |
| chr19_-_38747172 | 0.07 |
ENST00000347262.4 ENST00000591585.1 ENST00000301242.4 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr8_-_20040638 | 0.07 |
ENST00000519026.1 ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr8_-_20040601 | 0.07 |
ENST00000265808.7 ENST00000522513.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr2_-_64371546 | 0.07 |
ENST00000358912.4 |
PELI1 |
pellino E3 ubiquitin protein ligase 1 |
| chr17_+_34430980 | 0.07 |
ENST00000250151.4 |
CCL4 |
chemokine (C-C motif) ligand 4 |
| chr6_+_33048222 | 0.07 |
ENST00000428835.1 |
HLA-DPB1 |
major histocompatibility complex, class II, DP beta 1 |
| chr2_+_113735575 | 0.07 |
ENST00000376489.2 ENST00000259205.4 |
IL36G |
interleukin 36, gamma |
| chr19_+_39138320 | 0.07 |
ENST00000424234.2 ENST00000390009.3 ENST00000589528.1 |
ACTN4 |
actinin, alpha 4 |
| chr1_-_16539094 | 0.07 |
ENST00000270747.3 |
ARHGEF19 |
Rho guanine nucleotide exchange factor (GEF) 19 |
| chrX_-_54824673 | 0.07 |
ENST00000218436.6 |
ITIH6 |
inter-alpha-trypsin inhibitor heavy chain family, member 6 |
| chr2_+_101869262 | 0.07 |
ENST00000289382.3 |
CNOT11 |
CCR4-NOT transcription complex, subunit 11 |
| chr2_-_47798044 | 0.07 |
ENST00000327876.4 |
KCNK12 |
potassium channel, subfamily K, member 12 |
| chr11_+_60609537 | 0.07 |
ENST00000227520.5 |
CCDC86 |
coiled-coil domain containing 86 |
| chr11_+_118826999 | 0.07 |
ENST00000264031.2 |
UPK2 |
uroplakin 2 |
| chr2_-_86333244 | 0.07 |
ENST00000263857.6 ENST00000409681.1 |
POLR1A |
polymerase (RNA) I polypeptide A, 194kDa |
| chrX_-_51239425 | 0.07 |
ENST00000375992.3 |
NUDT11 |
nudix (nucleoside diphosphate linked moiety X)-type motif 11 |
| chr10_-_98031265 | 0.07 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr8_+_104831554 | 0.07 |
ENST00000408894.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr11_+_62186498 | 0.07 |
ENST00000278282.2 |
SCGB1A1 |
secretoglobin, family 1A, member 1 (uteroglobin) |
| chrX_-_154563889 | 0.07 |
ENST00000369449.2 ENST00000321926.4 |
CLIC2 |
chloride intracellular channel 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.8 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.1 | 0.4 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.1 | 0.3 | GO:0002025 | vasodilation by norepinephrine-epinephrine involved in regulation of systemic arterial blood pressure(GO:0002025) |
| 0.1 | 0.5 | GO:0002225 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 1.6 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.5 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 0.2 | GO:0010652 | regulation of cell communication by chemical coupling(GO:0010645) positive regulation of cell communication by chemical coupling(GO:0010652) |
| 0.1 | 0.2 | GO:0021919 | BMP signaling pathway involved in spinal cord dorsal/ventral patterning(GO:0021919) |
| 0.1 | 0.2 | GO:0035604 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.1 | 0.2 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.1 | 0.6 | GO:0060075 | regulation of resting membrane potential(GO:0060075) |
| 0.1 | 0.2 | GO:0045875 | negative regulation of sister chromatid cohesion(GO:0045875) |
| 0.1 | 0.2 | GO:0002238 | response to molecule of fungal origin(GO:0002238) cellular response to molecule of fungal origin(GO:0071226) |
| 0.1 | 0.4 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.2 | GO:0060529 | ectoderm and mesoderm interaction(GO:0007499) squamous basal epithelial stem cell differentiation involved in prostate gland acinus development(GO:0060529) |
| 0.0 | 0.1 | GO:1902396 | protein localization to bicellular tight junction(GO:1902396) |
| 0.0 | 0.2 | GO:1990535 | neuron projection maintenance(GO:1990535) |
| 0.0 | 0.3 | GO:0036486 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.0 | 0.2 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.1 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.1 | GO:0009107 | lipoate biosynthetic process(GO:0009107) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.0 | 0.1 | GO:0031117 | positive regulation of microtubule depolymerization(GO:0031117) |
| 0.0 | 0.1 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:1904100 | regulation of protein O-linked glycosylation(GO:1904098) positive regulation of protein O-linked glycosylation(GO:1904100) |
| 0.0 | 0.1 | GO:1900106 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:0061669 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.0 | 0.3 | GO:0035542 | regulation of SNARE complex assembly(GO:0035542) |
| 0.0 | 0.2 | GO:0060414 | aorta smooth muscle tissue morphogenesis(GO:0060414) |
| 0.0 | 0.4 | GO:0051546 | keratinocyte migration(GO:0051546) |
| 0.0 | 0.1 | GO:0000430 | regulation of transcription from RNA polymerase II promoter by glucose(GO:0000430) positive regulation of transcription from RNA polymerase II promoter by glucose(GO:0000432) |
| 0.0 | 0.1 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.0 | 0.3 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0006196 | AMP catabolic process(GO:0006196) |
| 0.0 | 0.1 | GO:0046900 | tetrahydrofolylpolyglutamate metabolic process(GO:0046900) |
| 0.0 | 0.0 | GO:0042214 | terpene metabolic process(GO:0042214) |
| 0.0 | 0.1 | GO:0002503 | peptide antigen assembly with MHC class II protein complex(GO:0002503) |
| 0.0 | 0.1 | GO:0035166 | post-embryonic hemopoiesis(GO:0035166) |
| 0.0 | 0.1 | GO:0006624 | vacuolar protein processing(GO:0006624) |
| 0.0 | 0.1 | GO:1904924 | negative regulation of mitophagy in response to mitochondrial depolarization(GO:1904924) |
| 0.0 | 0.1 | GO:1990928 | response to amino acid starvation(GO:1990928) |
| 0.0 | 0.1 | GO:0045065 | cytotoxic T cell differentiation(GO:0045065) |
| 0.0 | 0.1 | GO:0002728 | negative regulation of natural killer cell cytokine production(GO:0002728) |
| 0.0 | 0.1 | GO:0014738 | regulation of muscle hyperplasia(GO:0014738) |
| 0.0 | 0.3 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.0 | 0.0 | GO:0051088 | PMA-inducible membrane protein ectodomain proteolysis(GO:0051088) |
| 0.0 | 0.3 | GO:0070587 | negative regulation of heterotypic cell-cell adhesion(GO:0034115) regulation of cell-cell adhesion involved in gastrulation(GO:0070587) |
| 0.0 | 0.1 | GO:0030886 | negative regulation of myeloid dendritic cell activation(GO:0030886) |
| 0.0 | 0.1 | GO:0051673 | membrane disruption in other organism(GO:0051673) |
| 0.0 | 0.0 | GO:0002309 | T cell proliferation involved in immune response(GO:0002309) |
| 0.0 | 0.0 | GO:0021589 | hindbrain structural organization(GO:0021577) cerebellum structural organization(GO:0021589) |
| 0.0 | 0.0 | GO:0045608 | negative regulation of auditory receptor cell differentiation(GO:0045608) |
| 0.0 | 0.0 | GO:0044254 | angiotensin catabolic process in blood(GO:0002005) multicellular organismal protein catabolic process(GO:0044254) protein digestion(GO:0044256) multicellular organismal macromolecule catabolic process(GO:0044266) |
| 0.0 | 0.2 | GO:0070327 | thyroid hormone transport(GO:0070327) |
| 0.0 | 0.0 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.1 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.0 | 0.1 | GO:0048165 | ovarian cumulus expansion(GO:0001550) fused antrum stage(GO:0048165) |
| 0.0 | 0.0 | GO:1903984 | regulation of transcription from RNA polymerase II promoter in response to arsenic-containing substance(GO:0061394) positive regulation of TRAIL-activated apoptotic signaling pathway(GO:1903984) |
| 0.0 | 0.5 | GO:0042730 | fibrinolysis(GO:0042730) |
| 0.0 | 0.2 | GO:0048227 | plasma membrane to endosome transport(GO:0048227) |
| 0.0 | 0.1 | GO:0048706 | embryonic skeletal system development(GO:0048706) |
| 0.0 | 0.1 | GO:0035745 | T-helper 2 cell cytokine production(GO:0035745) |
| 0.0 | 0.1 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.2 | GO:0043248 | proteasome assembly(GO:0043248) |
| 0.0 | 0.0 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.0 | 0.1 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0051045 | negative regulation of membrane protein ectodomain proteolysis(GO:0051045) |
| 0.0 | 0.1 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0001927 | exocyst assembly(GO:0001927) |
| 0.0 | 0.1 | GO:1902474 | positive regulation of protein localization to synapse(GO:1902474) |
| 0.0 | 0.1 | GO:0008063 | Toll signaling pathway(GO:0008063) |
| 0.0 | 0.1 | GO:0001757 | somite specification(GO:0001757) |
| 0.0 | 0.1 | GO:1904749 | regulation of protein localization to nucleolus(GO:1904749) |
| 0.0 | 0.0 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.0 | 0.0 | GO:0051596 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.2 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.0 | GO:0019056 | modulation by virus of host transcription(GO:0019056) positive regulation of sprouting of injured axon(GO:0048687) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:0045905 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.0 | GO:0042851 | L-alanine metabolic process(GO:0042851) L-alanine catabolic process(GO:0042853) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:1904995 | negative regulation of leukocyte tethering or rolling(GO:1903237) negative regulation of leukocyte adhesion to vascular endothelial cell(GO:1904995) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.1 | 0.4 | GO:0043512 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.1 | 0.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.0 | 0.6 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.2 | GO:0019815 | B cell receptor complex(GO:0019815) |
| 0.0 | 0.1 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.0 | 0.5 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.3 | GO:0008541 | proteasome regulatory particle, lid subcomplex(GO:0008541) |
| 0.0 | 0.1 | GO:0031074 | nucleocytoplasmic shuttling complex(GO:0031074) |
| 0.0 | 0.1 | GO:0005850 | eukaryotic translation initiation factor 2 complex(GO:0005850) |
| 0.0 | 0.1 | GO:0070081 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.0 | 0.0 | GO:1990622 | CHOP-ATF3 complex(GO:1990622) |
| 0.0 | 0.0 | GO:0043159 | acrosomal matrix(GO:0043159) |
| 0.0 | 0.2 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:0034388 | Pwp2p-containing subcomplex of 90S preribosome(GO:0034388) |
| 0.0 | 0.0 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.1 | GO:0016942 | insulin-like growth factor binding protein complex(GO:0016942) growth factor complex(GO:0036454) |
| 0.0 | 0.1 | GO:0045252 | oxoglutarate dehydrogenase complex(GO:0045252) |
| 0.0 | 0.3 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0043527 | tRNA methyltransferase complex(GO:0043527) |
| 0.0 | 0.3 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.0 | 0.1 | GO:0005736 | DNA-directed RNA polymerase I complex(GO:0005736) |
| 0.0 | 0.3 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.0 | GO:0032937 | SREBP-SCAP-Insig complex(GO:0032937) |
| 0.0 | 0.2 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.1 | 0.3 | GO:0004939 | beta-adrenergic receptor activity(GO:0004939) |
| 0.1 | 0.2 | GO:0017095 | heparan sulfate 6-O-sulfotransferase activity(GO:0017095) |
| 0.1 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.8 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.5 | GO:0001087 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.3 | GO:0086075 | gap junction channel activity involved in cardiac conduction electrical coupling(GO:0086075) |
| 0.0 | 0.2 | GO:0010465 | nerve growth factor receptor activity(GO:0010465) |
| 0.0 | 0.2 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.7 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.1 | GO:0016992 | lipoate synthase activity(GO:0016992) radical SAM enzyme activity(GO:0070283) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0015349 | thyroid hormone transmembrane transporter activity(GO:0015349) |
| 0.0 | 0.1 | GO:0005151 | interleukin-1, Type II receptor binding(GO:0005151) |
| 0.0 | 0.6 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.1 | GO:0004169 | dolichyl-phosphate-mannose-protein mannosyltransferase activity(GO:0004169) |
| 0.0 | 0.2 | GO:1903136 | cuprous ion binding(GO:1903136) |
| 0.0 | 0.2 | GO:0004687 | myosin light chain kinase activity(GO:0004687) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.2 | GO:0035381 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.0 | GO:0042954 | lipoprotein transporter activity(GO:0042954) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.1 | GO:0004591 | oxoglutarate dehydrogenase (succinyl-transferring) activity(GO:0004591) |
| 0.0 | 0.8 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.1 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0008422 | beta-glucosidase activity(GO:0008422) |
| 0.0 | 0.0 | GO:0050528 | acyloxyacyl hydrolase activity(GO:0050528) |
| 0.0 | 0.1 | GO:0015333 | peptide:proton symporter activity(GO:0015333) proton-dependent peptide secondary active transmembrane transporter activity(GO:0022897) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.0 | GO:0005136 | interleukin-4 receptor binding(GO:0005136) |
| 0.0 | 0.2 | GO:0052650 | NADP-retinol dehydrogenase activity(GO:0052650) |
| 0.0 | 0.2 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0004716 | receptor signaling protein tyrosine kinase activity(GO:0004716) |
| 0.0 | 0.0 | GO:0030375 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.2 | GO:0097027 | ubiquitin-protein transferase activator activity(GO:0097027) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.3 | GO:0034237 | protein kinase A regulatory subunit binding(GO:0034237) |
| 0.0 | 1.7 | GO:0031490 | chromatin DNA binding(GO:0031490) |
| 0.0 | 0.0 | GO:0005223 | intracellular cGMP activated cation channel activity(GO:0005223) |
| 0.0 | 0.1 | GO:0004117 | calmodulin-dependent cyclic-nucleotide phosphodiesterase activity(GO:0004117) |
| 0.0 | 0.2 | GO:0030280 | structural constituent of epidermis(GO:0030280) |
| 0.0 | 0.0 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0016724 | ferroxidase activity(GO:0004322) oxidoreductase activity, oxidizing metal ions, oxygen as acceptor(GO:0016724) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.0 | GO:0000035 | acyl binding(GO:0000035) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.0 | GO:0016749 | 5-aminolevulinate synthase activity(GO:0003870) N-succinyltransferase activity(GO:0016749) |
| 0.0 | 0.1 | GO:0004719 | protein-L-isoaspartate (D-aspartate) O-methyltransferase activity(GO:0004719) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID ANGIOPOIETIN RECEPTOR PATHWAY | Angiopoietin receptor Tie2-mediated signaling |
| 0.0 | 0.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.4 | PID INSULIN GLUCOSE PATHWAY | Insulin-mediated glucose transport |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.6 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.4 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.6 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.3 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.0 | 0.3 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |