Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for MAX_TFEB
Z-value: 0.29
Transcription factors associated with MAX_TFEB
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
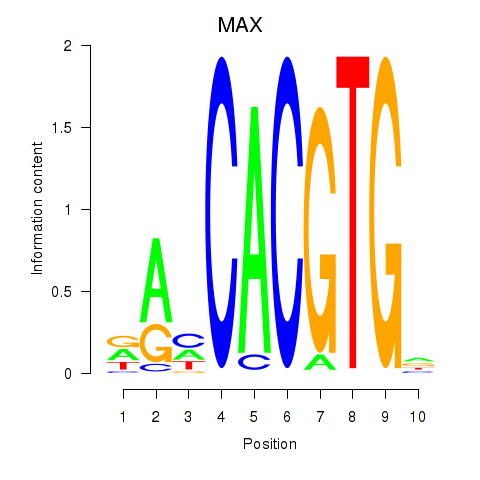
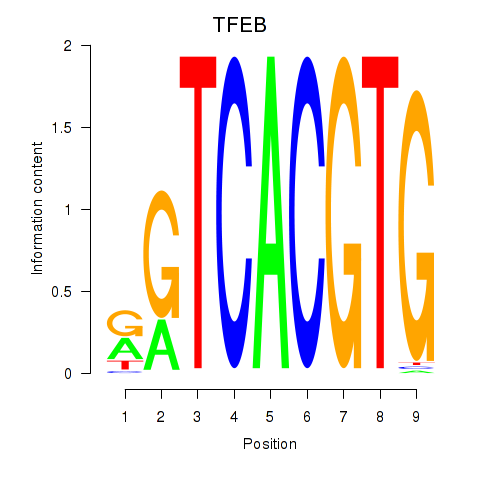
MAX
|
ENSG00000125952.14 | MAX |
|
TFEB
|
ENSG00000112561.13 | TFEB |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MAX | hg19_v2_chr14_-_65569057_65569119 | 0.95 | 3.8e-04 | Click! |
| TFEB | hg19_v2_chr6_-_41703296_41703367 | 0.70 | 5.5e-02 | Click! |
Activity profile of MAX_TFEB motif
Sorted Z-values of MAX_TFEB motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MAX_TFEB
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_+_49458107 | 1.03 |
ENST00000539787.1 ENST00000345358.7 ENST00000391871.3 ENST00000415969.2 ENST00000354470.3 ENST00000506183.1 ENST00000293288.8 |
BAX |
BCL2-associated X protein |
| chr10_-_93392811 | 0.80 |
ENST00000238994.5 |
PPP1R3C |
protein phosphatase 1, regulatory subunit 3C |
| chr1_+_183605200 | 0.58 |
ENST00000304685.4 |
RGL1 |
ral guanine nucleotide dissociation stimulator-like 1 |
| chr5_-_172198190 | 0.46 |
ENST00000239223.3 |
DUSP1 |
dual specificity phosphatase 1 |
| chr22_+_35776828 | 0.45 |
ENST00000216117.8 |
HMOX1 |
heme oxygenase (decycling) 1 |
| chr15_+_44084040 | 0.45 |
ENST00000249786.4 |
SERF2 |
small EDRK-rich factor 2 |
| chr15_+_44084503 | 0.39 |
ENST00000409960.2 ENST00000409646.1 ENST00000594896.1 ENST00000339624.5 ENST00000409291.1 ENST00000402131.1 ENST00000403425.1 ENST00000430901.1 |
SERF2 |
small EDRK-rich factor 2 |
| chr1_+_221051699 | 0.38 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr12_-_76953453 | 0.37 |
ENST00000549570.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr19_+_14544099 | 0.36 |
ENST00000242783.6 ENST00000586557.1 ENST00000590097.1 |
PKN1 |
protein kinase N1 |
| chr12_+_56110247 | 0.36 |
ENST00000551926.1 |
BLOC1S1 |
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr12_+_56109810 | 0.35 |
ENST00000550412.1 ENST00000257899.2 ENST00000548925.1 ENST00000549147.1 |
RP11-644F5.10 BLOC1S1 |
Uncharacterized protein biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr22_+_31003133 | 0.34 |
ENST00000405742.3 |
TCN2 |
transcobalamin II |
| chr9_-_79307096 | 0.31 |
ENST00000376717.2 ENST00000223609.6 ENST00000443509.2 |
PRUNE2 |
prune homolog 2 (Drosophila) |
| chr17_-_66951474 | 0.31 |
ENST00000269080.2 |
ABCA8 |
ATP-binding cassette, sub-family A (ABC1), member 8 |
| chr22_+_31002779 | 0.31 |
ENST00000215838.3 |
TCN2 |
transcobalamin II |
| chr14_-_65569057 | 0.31 |
ENST00000555419.1 ENST00000341653.2 |
MAX |
MYC associated factor X |
| chr14_-_65569244 | 0.30 |
ENST00000557277.1 ENST00000556892.1 |
MAX |
MYC associated factor X |
| chr22_-_36903069 | 0.30 |
ENST00000216187.6 ENST00000423980.1 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr12_+_56109926 | 0.30 |
ENST00000547076.1 |
BLOC1S1 |
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chr5_-_133706695 | 0.29 |
ENST00000521755.1 ENST00000523054.1 ENST00000435240.2 ENST00000609654.1 ENST00000536186.1 ENST00000609383.1 |
CDKL3 |
cyclin-dependent kinase-like 3 |
| chr19_-_48018203 | 0.28 |
ENST00000595227.1 ENST00000593761.1 ENST00000263354.3 |
NAPA |
N-ethylmaleimide-sensitive factor attachment protein, alpha |
| chr11_+_64781575 | 0.28 |
ENST00000246747.4 ENST00000529384.1 |
ARL2 |
ADP-ribosylation factor-like 2 |
| chr1_-_212873267 | 0.28 |
ENST00000243440.1 |
BATF3 |
basic leucine zipper transcription factor, ATF-like 3 |
| chr2_+_148778570 | 0.27 |
ENST00000407073.1 |
MBD5 |
methyl-CpG binding domain protein 5 |
| chr13_+_113951607 | 0.27 |
ENST00000397181.3 |
LAMP1 |
lysosomal-associated membrane protein 1 |
| chr22_-_36903101 | 0.27 |
ENST00000397224.4 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chrX_+_30671476 | 0.26 |
ENST00000378946.3 ENST00000378943.3 ENST00000378945.3 ENST00000427190.1 ENST00000378941.3 |
GK |
glycerol kinase |
| chr1_-_241520525 | 0.26 |
ENST00000366565.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr17_-_7137582 | 0.25 |
ENST00000575756.1 ENST00000575458.1 |
DVL2 |
dishevelled segment polarity protein 2 |
| chr7_+_99070527 | 0.25 |
ENST00000379724.3 |
ZNF789 |
zinc finger protein 789 |
| chr1_-_241520385 | 0.25 |
ENST00000366564.1 |
RGS7 |
regulator of G-protein signaling 7 |
| chr11_+_67776012 | 0.25 |
ENST00000539229.1 |
ALDH3B1 |
aldehyde dehydrogenase 3 family, member B1 |
| chr4_-_100242549 | 0.25 |
ENST00000305046.8 ENST00000394887.3 |
ADH1B |
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr12_-_76953513 | 0.25 |
ENST00000547540.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr3_-_178789220 | 0.24 |
ENST00000414084.1 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr16_-_4897266 | 0.23 |
ENST00000591451.1 ENST00000436648.5 ENST00000381983.3 ENST00000588297.1 ENST00000321919.9 |
GLYR1 |
glyoxylate reductase 1 homolog (Arabidopsis) |
| chrX_+_102883887 | 0.23 |
ENST00000372625.3 ENST00000372624.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chr11_+_64781657 | 0.22 |
ENST00000533729.1 |
ARL2 |
ADP-ribosylation factor-like 2 |
| chr1_+_151171012 | 0.22 |
ENST00000349792.5 ENST00000409426.1 ENST00000441902.2 ENST00000368890.4 ENST00000424999.1 ENST00000368888.4 |
PIP5K1A |
phosphatidylinositol-4-phosphate 5-kinase, type I, alpha |
| chr22_-_20850128 | 0.21 |
ENST00000328879.4 |
KLHL22 |
kelch-like family member 22 |
| chr8_+_104383728 | 0.21 |
ENST00000330295.5 |
CTHRC1 |
collagen triple helix repeat containing 1 |
| chr5_+_133706865 | 0.21 |
ENST00000265339.2 |
UBE2B |
ubiquitin-conjugating enzyme E2B |
| chr12_+_117176113 | 0.20 |
ENST00000319176.7 |
RNFT2 |
ring finger protein, transmembrane 2 |
| chr21_-_38639601 | 0.20 |
ENST00000539844.1 ENST00000476950.1 ENST00000399001.1 |
DSCR3 |
Down syndrome critical region gene 3 |
| chr19_-_12833361 | 0.20 |
ENST00000592287.1 |
TNPO2 |
transportin 2 |
| chr1_-_51425902 | 0.20 |
ENST00000396153.2 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
| chr7_+_150065879 | 0.20 |
ENST00000397281.2 ENST00000444957.1 ENST00000466559.1 ENST00000489432.2 ENST00000475514.1 ENST00000482680.1 ENST00000488943.1 ENST00000518514.1 ENST00000478789.1 |
REPIN1 ZNF775 |
replication initiator 1 zinc finger protein 775 |
| chr16_+_85061367 | 0.19 |
ENST00000538274.1 ENST00000258180.3 |
KIAA0513 |
KIAA0513 |
| chr12_-_58146048 | 0.19 |
ENST00000547281.1 ENST00000546489.1 ENST00000552388.1 ENST00000540325.1 ENST00000312990.6 |
CDK4 |
cyclin-dependent kinase 4 |
| chrX_-_102531717 | 0.19 |
ENST00000372680.1 |
TCEAL5 |
transcription elongation factor A (SII)-like 5 |
| chr22_-_36902522 | 0.18 |
ENST00000397223.4 |
FOXRED2 |
FAD-dependent oxidoreductase domain containing 2 |
| chr15_+_33022885 | 0.18 |
ENST00000322805.4 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chrX_+_102883620 | 0.18 |
ENST00000372626.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chr14_-_35183755 | 0.18 |
ENST00000555765.1 |
CFL2 |
cofilin 2 (muscle) |
| chr2_+_46926048 | 0.17 |
ENST00000306503.5 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr2_+_30370382 | 0.17 |
ENST00000402708.1 |
YPEL5 |
yippee-like 5 (Drosophila) |
| chr5_-_121413974 | 0.17 |
ENST00000231004.4 |
LOX |
lysyl oxidase |
| chr20_+_37590942 | 0.17 |
ENST00000373325.2 ENST00000252011.3 ENST00000373323.4 |
DHX35 |
DEAH (Asp-Glu-Ala-His) box polypeptide 35 |
| chr1_+_11866207 | 0.17 |
ENST00000312413.6 ENST00000346436.6 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chr19_+_39903185 | 0.17 |
ENST00000409794.3 |
PLEKHG2 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr22_-_20850070 | 0.17 |
ENST00000440659.2 ENST00000458248.1 ENST00000443285.1 ENST00000444967.1 ENST00000451553.1 ENST00000431430.1 |
KLHL22 |
kelch-like family member 22 |
| chr15_-_83316087 | 0.17 |
ENST00000568757.1 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr7_+_100464760 | 0.17 |
ENST00000200457.4 |
TRIP6 |
thyroid hormone receptor interactor 6 |
| chrX_-_102510045 | 0.16 |
ENST00000360000.4 |
TCEAL8 |
transcription elongation factor A (SII)-like 8 |
| chr7_+_99746514 | 0.16 |
ENST00000341942.5 ENST00000441173.1 |
LAMTOR4 |
late endosomal/lysosomal adaptor, MAPK and MTOR activator 4 |
| chrX_-_102510126 | 0.16 |
ENST00000372685.3 |
TCEAL8 |
transcription elongation factor A (SII)-like 8 |
| chr17_-_37886752 | 0.16 |
ENST00000577810.1 |
MIEN1 |
migration and invasion enhancer 1 |
| chr22_-_24622080 | 0.16 |
ENST00000425408.1 |
GGT5 |
gamma-glutamyltransferase 5 |
| chr5_-_176730676 | 0.16 |
ENST00000393611.2 ENST00000303251.6 ENST00000303270.6 |
RAB24 |
RAB24, member RAS oncogene family |
| chr1_-_31845914 | 0.16 |
ENST00000373713.2 |
FABP3 |
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chrX_+_102631248 | 0.16 |
ENST00000361298.4 ENST00000372645.3 ENST00000372635.1 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr14_-_20923195 | 0.16 |
ENST00000206542.4 |
OSGEP |
O-sialoglycoprotein endopeptidase |
| chr14_-_20929624 | 0.16 |
ENST00000398020.4 ENST00000250489.4 |
TMEM55B |
transmembrane protein 55B |
| chrX_+_55744166 | 0.15 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chr12_-_58146128 | 0.15 |
ENST00000551800.1 ENST00000549606.1 ENST00000257904.6 |
CDK4 |
cyclin-dependent kinase 4 |
| chr5_-_139944196 | 0.15 |
ENST00000357560.4 |
APBB3 |
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chrX_+_108780347 | 0.15 |
ENST00000372103.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr7_+_150065278 | 0.15 |
ENST00000519397.1 ENST00000479668.1 ENST00000540729.1 |
REPIN1 |
replication initiator 1 |
| chr6_+_127588020 | 0.15 |
ENST00000309649.3 ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146 |
ring finger protein 146 |
| chr17_-_66453562 | 0.15 |
ENST00000262139.5 ENST00000546360.1 |
WIPI1 |
WD repeat domain, phosphoinositide interacting 1 |
| chr1_+_159750776 | 0.14 |
ENST00000368107.1 |
DUSP23 |
dual specificity phosphatase 23 |
| chr14_+_96505659 | 0.14 |
ENST00000555004.1 |
C14orf132 |
chromosome 14 open reading frame 132 |
| chr1_-_28969517 | 0.14 |
ENST00000263974.4 ENST00000373824.4 |
TAF12 |
TAF12 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 20kDa |
| chr21_-_35987438 | 0.14 |
ENST00000313806.4 |
RCAN1 |
regulator of calcineurin 1 |
| chrX_+_102840408 | 0.14 |
ENST00000468024.1 ENST00000472484.1 ENST00000415568.2 ENST00000490644.1 ENST00000459722.1 ENST00000472745.1 ENST00000494801.1 ENST00000434216.2 ENST00000425011.1 |
TCEAL4 |
transcription elongation factor A (SII)-like 4 |
| chrX_+_55744228 | 0.14 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chrX_+_102631844 | 0.14 |
ENST00000372634.1 ENST00000299872.7 |
NGFRAP1 |
nerve growth factor receptor (TNFRSF16) associated protein 1 |
| chr20_-_2821271 | 0.14 |
ENST00000448755.1 ENST00000360652.2 |
PCED1A |
PC-esterase domain containing 1A |
| chr1_+_33116765 | 0.14 |
ENST00000544435.1 ENST00000373485.1 ENST00000458695.2 ENST00000490500.1 ENST00000445722.2 |
RBBP4 |
retinoblastoma binding protein 4 |
| chr6_+_127587755 | 0.14 |
ENST00000368314.1 ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146 |
ring finger protein 146 |
| chr2_+_27805880 | 0.14 |
ENST00000379717.1 ENST00000355467.4 ENST00000556601.1 ENST00000416005.2 |
ZNF512 |
zinc finger protein 512 |
| chr8_-_13134045 | 0.14 |
ENST00000512044.2 |
DLC1 |
deleted in liver cancer 1 |
| chr14_+_77564440 | 0.14 |
ENST00000361786.2 ENST00000555437.1 ENST00000555611.1 ENST00000554658.1 |
KIAA1737 |
CLOCK-interacting pacemaker |
| chr1_-_28559502 | 0.14 |
ENST00000263697.4 |
DNAJC8 |
DnaJ (Hsp40) homolog, subfamily C, member 8 |
| chrX_+_108779870 | 0.14 |
ENST00000372107.1 |
NXT2 |
nuclear transport factor 2-like export factor 2 |
| chr14_-_65569186 | 0.14 |
ENST00000555932.1 ENST00000358664.4 ENST00000284165.6 ENST00000358402.4 ENST00000246163.2 ENST00000556979.1 ENST00000555667.1 ENST00000557746.1 ENST00000556443.1 |
MAX |
MYC associated factor X |
| chr6_-_33385823 | 0.14 |
ENST00000494751.1 ENST00000374496.3 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr21_-_46237883 | 0.14 |
ENST00000397893.3 |
SUMO3 |
small ubiquitin-like modifier 3 |
| chr1_+_159750720 | 0.14 |
ENST00000368109.1 ENST00000368108.3 |
DUSP23 |
dual specificity phosphatase 23 |
| chr10_-_49860525 | 0.13 |
ENST00000435790.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr1_-_202896310 | 0.13 |
ENST00000367261.3 |
KLHL12 |
kelch-like family member 12 |
| chr12_+_6833237 | 0.13 |
ENST00000229251.3 ENST00000539735.1 ENST00000538410.1 |
COPS7A |
COP9 signalosome subunit 7A |
| chr2_-_211036051 | 0.13 |
ENST00000418791.1 ENST00000452086.1 ENST00000281772.9 |
KANSL1L |
KAT8 regulatory NSL complex subunit 1-like |
| chr17_-_57184260 | 0.13 |
ENST00000376149.3 ENST00000393066.3 |
TRIM37 |
tripartite motif containing 37 |
| chr12_-_76953573 | 0.13 |
ENST00000549646.1 ENST00000550628.1 ENST00000553139.1 ENST00000261183.3 ENST00000393250.4 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr3_-_49395705 | 0.13 |
ENST00000419349.1 |
GPX1 |
glutathione peroxidase 1 |
| chr5_+_156712372 | 0.13 |
ENST00000541131.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr14_-_74551096 | 0.13 |
ENST00000350259.4 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr12_-_63328817 | 0.13 |
ENST00000228705.6 |
PPM1H |
protein phosphatase, Mg2+/Mn2+ dependent, 1H |
| chr12_+_54332535 | 0.13 |
ENST00000243056.3 |
HOXC13 |
homeobox C13 |
| chr6_-_33385902 | 0.13 |
ENST00000374500.5 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr9_+_74526532 | 0.13 |
ENST00000486911.2 |
C9orf85 |
chromosome 9 open reading frame 85 |
| chr11_+_120894781 | 0.12 |
ENST00000529397.1 ENST00000528512.1 ENST00000422003.2 |
TBCEL |
tubulin folding cofactor E-like |
| chr2_+_201170703 | 0.12 |
ENST00000358677.5 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr12_+_90102729 | 0.12 |
ENST00000605386.1 |
LINC00936 |
long intergenic non-protein coding RNA 936 |
| chr20_+_48552908 | 0.12 |
ENST00000244061.2 |
RNF114 |
ring finger protein 114 |
| chr15_+_41913690 | 0.12 |
ENST00000563576.1 |
MGA |
MGA, MAX dimerization protein |
| chrX_-_34675391 | 0.12 |
ENST00000275954.3 |
TMEM47 |
transmembrane protein 47 |
| chr5_-_139943830 | 0.12 |
ENST00000412920.3 ENST00000511201.2 ENST00000356738.2 ENST00000354402.5 ENST00000358580.5 ENST00000508496.2 |
APBB3 |
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr17_+_42977122 | 0.12 |
ENST00000412523.2 ENST00000331733.4 ENST00000417826.2 |
FAM187A CCDC103 |
family with sequence similarity 187, member A coiled-coil domain containing 103 |
| chr19_+_50016610 | 0.12 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr20_-_443129 | 0.12 |
ENST00000354200.4 |
TBC1D20 |
TBC1 domain family, member 20 |
| chr2_+_46926326 | 0.12 |
ENST00000394861.2 |
SOCS5 |
suppressor of cytokine signaling 5 |
| chr1_-_222885770 | 0.12 |
ENST00000355727.2 ENST00000340020.6 |
AIDA |
axin interactor, dorsalization associated |
| chr7_+_100271446 | 0.12 |
ENST00000419828.1 ENST00000427895.1 |
GNB2 |
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr7_+_100271355 | 0.12 |
ENST00000436220.1 ENST00000424361.1 |
GNB2 |
guanine nucleotide binding protein (G protein), beta polypeptide 2 |
| chr19_+_50016411 | 0.12 |
ENST00000426395.3 ENST00000600273.1 ENST00000599988.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr6_+_42018614 | 0.12 |
ENST00000465926.1 ENST00000482432.1 |
TAF8 |
TAF8 RNA polymerase II, TATA box binding protein (TBP)-associated factor, 43kDa |
| chr17_+_57408994 | 0.12 |
ENST00000312655.4 |
YPEL2 |
yippee-like 2 (Drosophila) |
| chr8_+_42128812 | 0.12 |
ENST00000520810.1 ENST00000416505.2 ENST00000519735.1 ENST00000520835.1 ENST00000379708.3 |
IKBKB |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase beta |
| chr3_-_49395892 | 0.12 |
ENST00000419783.1 |
GPX1 |
glutathione peroxidase 1 |
| chr21_+_38445539 | 0.12 |
ENST00000418766.1 ENST00000450533.1 ENST00000438055.1 ENST00000355666.1 ENST00000540756.1 ENST00000399010.1 |
TTC3 |
tetratricopeptide repeat domain 3 |
| chr19_-_36545128 | 0.12 |
ENST00000538849.1 |
THAP8 |
THAP domain containing 8 |
| chr7_-_56118981 | 0.12 |
ENST00000419984.2 ENST00000413218.1 ENST00000424596.1 |
PSPH |
phosphoserine phosphatase |
| chr14_+_24630465 | 0.12 |
ENST00000557894.1 ENST00000559284.1 ENST00000560275.1 |
IRF9 |
interferon regulatory factor 9 |
| chr20_-_34638841 | 0.11 |
ENST00000565493.1 |
LINC00657 |
long intergenic non-protein coding RNA 657 |
| chr22_+_31003190 | 0.11 |
ENST00000407817.3 |
TCN2 |
transcobalamin II |
| chr6_-_43197189 | 0.11 |
ENST00000509253.1 ENST00000393987.2 ENST00000230431.6 |
DNPH1 |
2'-deoxynucleoside 5'-phosphate N-hydrolase 1 |
| chr12_+_51633061 | 0.11 |
ENST00000551313.1 |
DAZAP2 |
DAZ associated protein 2 |
| chr1_+_52521957 | 0.11 |
ENST00000472944.2 ENST00000484036.1 |
BTF3L4 |
basic transcription factor 3-like 4 |
| chr6_-_33385870 | 0.11 |
ENST00000488034.1 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr7_-_56119156 | 0.11 |
ENST00000421312.1 ENST00000416592.1 |
PSPH |
phosphoserine phosphatase |
| chr21_-_38639813 | 0.11 |
ENST00000309117.6 ENST00000398998.1 |
DSCR3 |
Down syndrome critical region gene 3 |
| chr4_+_76439665 | 0.11 |
ENST00000508105.1 ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6 |
THAP domain containing 6 |
| chr6_+_7590413 | 0.11 |
ENST00000342415.5 |
SNRNP48 |
small nuclear ribonucleoprotein 48kDa (U11/U12) |
| chr1_+_145611010 | 0.11 |
ENST00000369291.5 |
RNF115 |
ring finger protein 115 |
| chr6_-_33385854 | 0.11 |
ENST00000488478.1 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr19_-_45681482 | 0.11 |
ENST00000592647.1 ENST00000006275.4 ENST00000588062.1 ENST00000585934.1 |
TRAPPC6A |
trafficking protein particle complex 6A |
| chr22_-_50700140 | 0.11 |
ENST00000215659.8 |
MAPK12 |
mitogen-activated protein kinase 12 |
| chr20_-_2821756 | 0.11 |
ENST00000356872.3 ENST00000439542.1 |
PCED1A |
PC-esterase domain containing 1A |
| chr12_+_56110315 | 0.11 |
ENST00000548556.1 |
BLOC1S1 |
biogenesis of lysosomal organelles complex-1, subunit 1 |
| chrX_-_101397433 | 0.11 |
ENST00000372774.3 |
TCEAL6 |
transcription elongation factor A (SII)-like 6 |
| chr2_-_27545921 | 0.11 |
ENST00000402310.1 ENST00000405983.1 ENST00000403262.2 ENST00000428910.1 ENST00000402722.1 ENST00000399052.4 ENST00000380044.1 ENST00000405076.1 |
MPV17 |
MpV17 mitochondrial inner membrane protein |
| chr15_-_65281775 | 0.11 |
ENST00000433215.2 ENST00000558415.1 ENST00000557795.1 |
SPG21 |
spastic paraplegia 21 (autosomal recessive, Mast syndrome) |
| chr21_-_46238034 | 0.11 |
ENST00000332859.6 |
SUMO3 |
small ubiquitin-like modifier 3 |
| chr6_+_158589374 | 0.11 |
ENST00000607778.1 |
GTF2H5 |
general transcription factor IIH, polypeptide 5 |
| chr10_+_99258625 | 0.11 |
ENST00000370664.3 |
UBTD1 |
ubiquitin domain containing 1 |
| chrX_+_101380642 | 0.11 |
ENST00000372780.1 ENST00000329035.2 |
TCEAL2 |
transcription elongation factor A (SII)-like 2 |
| chr5_+_126112794 | 0.11 |
ENST00000261366.5 ENST00000395354.1 |
LMNB1 |
lamin B1 |
| chr1_+_52521797 | 0.11 |
ENST00000313334.8 |
BTF3L4 |
basic transcription factor 3-like 4 |
| chr19_-_8386238 | 0.11 |
ENST00000301457.2 |
NDUFA7 |
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 7, 14.5kDa |
| chr8_-_95961578 | 0.11 |
ENST00000448464.2 ENST00000342697.4 |
TP53INP1 |
tumor protein p53 inducible nuclear protein 1 |
| chr7_-_2354099 | 0.11 |
ENST00000222990.3 |
SNX8 |
sorting nexin 8 |
| chr2_-_131099897 | 0.11 |
ENST00000409127.1 ENST00000437688.2 ENST00000259229.2 |
CCDC115 |
coiled-coil domain containing 115 |
| chr16_-_20367584 | 0.10 |
ENST00000570689.1 |
UMOD |
uromodulin |
| chrX_-_15872914 | 0.10 |
ENST00000380291.1 ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2 |
adaptor-related protein complex 1, sigma 2 subunit |
| chr21_-_46237959 | 0.10 |
ENST00000397898.3 ENST00000411651.2 |
SUMO3 |
small ubiquitin-like modifier 3 |
| chr10_-_31320840 | 0.10 |
ENST00000375311.1 |
ZNF438 |
zinc finger protein 438 |
| chr5_+_43603229 | 0.10 |
ENST00000344920.4 ENST00000512996.2 |
NNT |
nicotinamide nucleotide transhydrogenase |
| chr6_-_41888843 | 0.10 |
ENST00000434077.1 ENST00000409312.1 |
MED20 |
mediator complex subunit 20 |
| chr6_-_33385655 | 0.10 |
ENST00000440279.3 ENST00000607266.1 |
CUTA |
cutA divalent cation tolerance homolog (E. coli) |
| chr15_-_40213080 | 0.10 |
ENST00000561100.1 |
GPR176 |
G protein-coupled receptor 176 |
| chr1_-_51425772 | 0.10 |
ENST00000371778.4 |
FAF1 |
Fas (TNFRSF6) associated factor 1 |
| chr3_-_127541679 | 0.10 |
ENST00000265052.5 |
MGLL |
monoglyceride lipase |
| chr12_+_117176090 | 0.10 |
ENST00000257575.4 ENST00000407967.3 ENST00000392549.2 |
RNFT2 |
ring finger protein, transmembrane 2 |
| chr1_-_26233423 | 0.10 |
ENST00000357865.2 |
STMN1 |
stathmin 1 |
| chr16_-_15736953 | 0.10 |
ENST00000548025.1 ENST00000551742.1 ENST00000602337.1 ENST00000344181.3 ENST00000396368.3 |
KIAA0430 |
KIAA0430 |
| chrX_-_100872911 | 0.10 |
ENST00000361910.4 ENST00000539247.1 ENST00000538627.1 |
ARMCX6 |
armadillo repeat containing, X-linked 6 |
| chr17_-_7137857 | 0.10 |
ENST00000005340.5 |
DVL2 |
dishevelled segment polarity protein 2 |
| chr14_-_54955721 | 0.10 |
ENST00000554908.1 |
GMFB |
glia maturation factor, beta |
| chr15_-_50978965 | 0.10 |
ENST00000560955.1 ENST00000313478.7 |
TRPM7 |
transient receptor potential cation channel, subfamily M, member 7 |
| chr1_+_52521928 | 0.10 |
ENST00000489308.2 |
BTF3L4 |
basic transcription factor 3-like 4 |
| chr9_+_133320301 | 0.10 |
ENST00000352480.5 |
ASS1 |
argininosuccinate synthase 1 |
| chr11_-_58611957 | 0.10 |
ENST00000532258.1 |
GLYATL2 |
glycine-N-acyltransferase-like 2 |
| chr3_+_19988885 | 0.10 |
ENST00000422242.1 |
RAB5A |
RAB5A, member RAS oncogene family |
| chr12_+_7023735 | 0.10 |
ENST00000538763.1 ENST00000544774.1 ENST00000545045.2 |
ENO2 |
enolase 2 (gamma, neuronal) |
| chr14_-_74551172 | 0.10 |
ENST00000553458.1 |
ALDH6A1 |
aldehyde dehydrogenase 6 family, member A1 |
| chr1_-_183604794 | 0.10 |
ENST00000367534.1 ENST00000359856.6 ENST00000294742.6 |
ARPC5 |
actin related protein 2/3 complex, subunit 5, 16kDa |
| chr7_-_27183263 | 0.10 |
ENST00000222726.3 |
HOXA5 |
homeobox A5 |
| chr20_+_43595115 | 0.10 |
ENST00000372806.3 ENST00000396731.4 ENST00000372801.1 ENST00000499879.2 |
STK4 |
serine/threonine kinase 4 |
| chr5_-_68628543 | 0.10 |
ENST00000396496.2 ENST00000511257.1 ENST00000383374.2 |
CCDC125 |
coiled-coil domain containing 125 |
| chr20_-_30458019 | 0.10 |
ENST00000486996.1 ENST00000398084.2 |
DUSP15 |
dual specificity phosphatase 15 |
| chr19_-_5340730 | 0.10 |
ENST00000372412.4 ENST00000357368.4 ENST00000262963.6 ENST00000348075.2 ENST00000353284.2 |
PTPRS |
protein tyrosine phosphatase, receptor type, S |
| chr12_+_72148614 | 0.10 |
ENST00000261263.3 |
RAB21 |
RAB21, member RAS oncogene family |
| chr1_-_154909329 | 0.10 |
ENST00000368467.3 |
PMVK |
phosphomevalonate kinase |
| chr11_+_65479702 | 0.09 |
ENST00000530446.1 ENST00000534104.1 ENST00000530605.1 ENST00000528198.1 ENST00000531880.1 ENST00000534650.1 |
KAT5 |
K(lysine) acetyltransferase 5 |
| chr1_-_3447967 | 0.09 |
ENST00000294599.4 |
MEGF6 |
multiple EGF-like-domains 6 |
| chr7_-_6312206 | 0.09 |
ENST00000350796.3 |
CYTH3 |
cytohesin 3 |
| chr12_+_71833756 | 0.09 |
ENST00000536515.1 ENST00000540815.2 |
LGR5 |
leucine-rich repeat containing G protein-coupled receptor 5 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:1903519 | B cell negative selection(GO:0002352) post-embryonic camera-type eye morphogenesis(GO:0048597) apoptotic process involved in mammary gland involution(GO:0060057) positive regulation of apoptotic process involved in mammary gland involution(GO:0060058) positive regulation of apoptotic process involved in morphogenesis(GO:1902339) positive regulation of apoptotic DNA fragmentation(GO:1902512) regulation of mammary gland involution(GO:1903519) positive regulation of mammary gland involution(GO:1903521) positive regulation of apoptotic process involved in development(GO:1904747) |
| 0.2 | 0.5 | GO:0006788 | heme oxidation(GO:0006788) smooth muscle hyperplasia(GO:0014806) negative regulation of mast cell cytokine production(GO:0032764) |
| 0.1 | 0.7 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.3 | GO:0010845 | positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.1 | 0.3 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.1 | 0.4 | GO:0014022 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
| 0.1 | 0.3 | GO:0046167 | glycerol-3-phosphate biosynthetic process(GO:0046167) |
| 0.1 | 0.8 | GO:0015889 | cobalamin transport(GO:0015889) |
| 0.1 | 1.1 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.1 | 0.9 | GO:0090204 | protein localization to nuclear pore(GO:0090204) |
| 0.1 | 0.1 | GO:0032456 | endocytic recycling(GO:0032456) |
| 0.1 | 0.2 | GO:0036353 | histone H2A-K119 monoubiquitination(GO:0036353) |
| 0.1 | 0.4 | GO:0035407 | histone H3-T11 phosphorylation(GO:0035407) |
| 0.1 | 0.2 | GO:0043321 | regulation of natural killer cell degranulation(GO:0043321) positive regulation of natural killer cell degranulation(GO:0043323) |
| 0.1 | 0.2 | GO:1900158 | negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.1 | 0.3 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.1 | 0.2 | GO:0006210 | pyrimidine nucleobase catabolic process(GO:0006208) thymine catabolic process(GO:0006210) thymine metabolic process(GO:0019859) |
| 0.1 | 0.2 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.2 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.2 | GO:0009609 | response to symbiont(GO:0009608) response to symbiotic bacterium(GO:0009609) |
| 0.0 | 0.2 | GO:1905098 | negative regulation of guanyl-nucleotide exchange factor activity(GO:1905098) |
| 0.0 | 0.1 | GO:0048203 | vesicle targeting, trans-Golgi to endosome(GO:0048203) |
| 0.0 | 0.3 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.2 | GO:0043473 | pigmentation(GO:0043473) |
| 0.0 | 0.1 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.0 | 0.1 | GO:1904760 | myofibroblast differentiation(GO:0036446) regulation of myofibroblast differentiation(GO:1904760) |
| 0.0 | 0.3 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.1 | GO:0046726 | positive regulation by virus of viral protein levels in host cell(GO:0046726) |
| 0.0 | 0.2 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0016340 | calcium-dependent cell-matrix adhesion(GO:0016340) |
| 0.0 | 0.1 | GO:0060435 | bronchiole development(GO:0060435) |
| 0.0 | 0.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:1904481 | response to tetrahydrofolate(GO:1904481) cellular response to tetrahydrofolate(GO:1904482) |
| 0.0 | 0.2 | GO:0043932 | ossification involved in bone remodeling(GO:0043932) |
| 0.0 | 0.1 | GO:1903347 | negative regulation of bicellular tight junction assembly(GO:1903347) |
| 0.0 | 0.4 | GO:0006564 | L-serine biosynthetic process(GO:0006564) |
| 0.0 | 0.3 | GO:0042908 | xenobiotic transport(GO:0042908) |
| 0.0 | 0.1 | GO:0006286 | base-excision repair, base-free sugar-phosphate removal(GO:0006286) |
| 0.0 | 0.0 | GO:0050787 | detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.1 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.0 | 0.1 | GO:0051031 | tRNA export from nucleus(GO:0006409) tRNA transport(GO:0051031) tRNA-containing ribonucleoprotein complex export from nucleus(GO:0071431) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.1 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.1 | GO:0097010 | eukaryotic translation initiation factor 4F complex assembly(GO:0097010) |
| 0.0 | 0.4 | GO:0030043 | actin filament fragmentation(GO:0030043) |
| 0.0 | 0.1 | GO:0070318 | positive regulation of G0 to G1 transition(GO:0070318) |
| 0.0 | 0.2 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:2000909 | regulation of cholesterol import(GO:0060620) regulation of sterol import(GO:2000909) |
| 0.0 | 0.4 | GO:0071850 | mitotic cell cycle arrest(GO:0071850) |
| 0.0 | 0.2 | GO:0031120 | snRNA pseudouridine synthesis(GO:0031120) |
| 0.0 | 0.1 | GO:0019243 | methylglyoxal catabolic process to D-lactate via S-lactoyl-glutathione(GO:0019243) methylglyoxal catabolic process(GO:0051596) methylglyoxal catabolic process to lactate(GO:0061727) |
| 0.0 | 0.1 | GO:1902728 | positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.0 | 0.1 | GO:0010133 | proline catabolic process to glutamate(GO:0010133) |
| 0.0 | 0.1 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.0 | GO:0071442 | regulation of histone H3-K14 acetylation(GO:0071440) positive regulation of histone H3-K14 acetylation(GO:0071442) |
| 0.0 | 0.1 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
| 0.0 | 0.1 | GO:0006290 | pyrimidine dimer repair(GO:0006290) |
| 0.0 | 0.1 | GO:1902544 | regulation of DNA N-glycosylase activity(GO:1902544) |
| 0.0 | 0.1 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.0 | 0.2 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 0.1 | GO:0035606 | peptidyl-cysteine S-trans-nitrosylation(GO:0035606) |
| 0.0 | 0.1 | GO:0060800 | regulation of cell differentiation involved in embryonic placenta development(GO:0060800) |
| 0.0 | 0.1 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.1 | GO:0061110 | histone H3-T6 phosphorylation(GO:0035408) dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.0 | 0.1 | GO:1902725 | negative regulation of satellite cell differentiation(GO:1902725) |
| 0.0 | 0.1 | GO:2000286 | receptor internalization involved in canonical Wnt signaling pathway(GO:2000286) |
| 0.0 | 0.1 | GO:0045643 | regulation of eosinophil differentiation(GO:0045643) positive regulation of eosinophil differentiation(GO:0045645) |
| 0.0 | 0.1 | GO:0000415 | negative regulation of histone H3-K36 methylation(GO:0000415) |
| 0.0 | 0.1 | GO:1904379 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.1 | GO:0042732 | D-xylose metabolic process(GO:0042732) |
| 0.0 | 0.1 | GO:0009236 | cobalamin biosynthetic process(GO:0009236) |
| 0.0 | 0.1 | GO:0098535 | de novo centriole assembly(GO:0098535) |
| 0.0 | 0.1 | GO:1900169 | regulation of glucocorticoid mediated signaling pathway(GO:1900169) |
| 0.0 | 0.1 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.3 | GO:0021894 | cerebral cortex GABAergic interneuron development(GO:0021894) |
| 0.0 | 0.0 | GO:0036146 | cellular response to mycotoxin(GO:0036146) |
| 0.0 | 0.3 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.1 | GO:1901857 | positive regulation of cellular respiration(GO:1901857) |
| 0.0 | 0.0 | GO:0002276 | basophil activation involved in immune response(GO:0002276) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.1 | GO:0000711 | meiotic DNA repair synthesis(GO:0000711) |
| 0.0 | 0.1 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.0 | GO:0014740 | negative regulation of muscle hyperplasia(GO:0014740) |
| 0.0 | 0.0 | GO:1903033 | regulation of microtubule plus-end binding(GO:1903031) positive regulation of microtubule plus-end binding(GO:1903033) |
| 0.0 | 0.1 | GO:0035513 | oxidative RNA demethylation(GO:0035513) oxidative single-stranded RNA demethylation(GO:0035553) |
| 0.0 | 0.0 | GO:1900075 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.0 | 0.2 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.1 | GO:0050917 | sensory perception of umami taste(GO:0050917) |
| 0.0 | 0.1 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.1 | GO:0007288 | sperm axoneme assembly(GO:0007288) |
| 0.0 | 0.1 | GO:0021769 | orbitofrontal cortex development(GO:0021769) |
| 0.0 | 0.1 | GO:1903566 | ciliary basal body organization(GO:0032053) positive regulation of protein localization to cilium(GO:1903566) |
| 0.0 | 0.1 | GO:0060265 | positive regulation of respiratory burst involved in inflammatory response(GO:0060265) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.1 | GO:2000828 | post-embryonic body morphogenesis(GO:0040032) regulation of parathyroid hormone secretion(GO:2000828) |
| 0.0 | 0.0 | GO:0015680 | intracellular copper ion transport(GO:0015680) |
| 0.0 | 0.1 | GO:0046604 | positive regulation of mitotic centrosome separation(GO:0046604) |
| 0.0 | 0.2 | GO:0001833 | inner cell mass cell proliferation(GO:0001833) |
| 0.0 | 0.2 | GO:0030050 | vesicle transport along actin filament(GO:0030050) |
| 0.0 | 0.3 | GO:0006068 | ethanol catabolic process(GO:0006068) |
| 0.0 | 0.2 | GO:0000338 | protein deneddylation(GO:0000338) |
| 0.0 | 0.0 | GO:0021860 | pyramidal neuron differentiation(GO:0021859) pyramidal neuron development(GO:0021860) |
| 0.0 | 0.1 | GO:0000079 | regulation of cyclin-dependent protein serine/threonine kinase activity(GO:0000079) |
| 0.0 | 0.1 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.0 | 0.1 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.1 | GO:1902475 | L-alpha-amino acid transmembrane transport(GO:1902475) |
| 0.0 | 0.1 | GO:0006104 | succinyl-CoA metabolic process(GO:0006104) |
| 0.0 | 0.0 | GO:1904617 | negative regulation of actin filament binding(GO:1904530) negative regulation of actin binding(GO:1904617) |
| 0.0 | 0.0 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.1 | GO:0006526 | arginine biosynthetic process(GO:0006526) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.0 | GO:0002904 | positive regulation of B cell apoptotic process(GO:0002904) |
| 0.0 | 0.5 | GO:0070911 | global genome nucleotide-excision repair(GO:0070911) |
| 0.0 | 0.8 | GO:0005978 | glycogen biosynthetic process(GO:0005978) glucan biosynthetic process(GO:0009250) |
| 0.0 | 0.1 | GO:1902459 | positive regulation of stem cell population maintenance(GO:1902459) |
| 0.0 | 0.0 | GO:0046452 | dihydrofolate metabolic process(GO:0046452) |
| 0.0 | 0.0 | GO:0061624 | fructose catabolic process(GO:0006001) fructose catabolic process to hydroxyacetone phosphate and glyceraldehyde-3-phosphate(GO:0061624) |
| 0.0 | 0.1 | GO:0046203 | spermidine catabolic process(GO:0046203) |
| 0.0 | 0.2 | GO:0070936 | protein K48-linked ubiquitination(GO:0070936) |
| 0.0 | 0.1 | GO:0051344 | regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051342) negative regulation of cyclic-nucleotide phosphodiesterase activity(GO:0051344) |
| 0.0 | 0.0 | GO:0000154 | rRNA modification(GO:0000154) |
| 0.0 | 0.1 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0043587 | tongue morphogenesis(GO:0043587) |
| 0.0 | 0.1 | GO:0019262 | N-acetylneuraminate catabolic process(GO:0019262) |
| 0.0 | 0.1 | GO:0006975 | DNA damage induced protein phosphorylation(GO:0006975) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | GO:0097136 | Bcl-2 family protein complex(GO:0097136) |
| 0.1 | 0.3 | GO:0070044 | synaptobrevin 2-SNAP-25-syntaxin-1a complex(GO:0070044) |
| 0.1 | 0.4 | GO:1990131 | Gtr1-Gtr2 GTPase complex(GO:1990131) |
| 0.1 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.2 | GO:0031084 | BLOC-2 complex(GO:0031084) |
| 0.0 | 1.2 | GO:0031083 | BLOC-1 complex(GO:0031083) |
| 0.0 | 0.4 | GO:0071986 | Ragulator complex(GO:0071986) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.4 | GO:0005827 | polar microtubule(GO:0005827) |
| 0.0 | 0.4 | GO:0010009 | cytoplasmic side of endosome membrane(GO:0010009) |
| 0.0 | 0.2 | GO:1990246 | uniplex complex(GO:1990246) |
| 0.0 | 0.2 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.2 | GO:0072589 | box H/ACA scaRNP complex(GO:0072589) box H/ACA telomerase RNP complex(GO:0090661) |
| 0.0 | 0.1 | GO:0031510 | SUMO activating enzyme complex(GO:0031510) |
| 0.0 | 0.2 | GO:0000015 | phosphopyruvate hydratase complex(GO:0000015) |
| 0.0 | 0.2 | GO:0033263 | CORVET complex(GO:0033263) |
| 0.0 | 0.2 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.2 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.2 | GO:0097413 | Lewy body(GO:0097413) |
| 0.0 | 0.1 | GO:0097452 | GAIT complex(GO:0097452) |
| 0.0 | 0.1 | GO:0000138 | Golgi trans cisterna(GO:0000138) |
| 0.0 | 0.8 | GO:0044665 | MLL1/2 complex(GO:0044665) MLL1 complex(GO:0071339) |
| 0.0 | 0.1 | GO:0097135 | cyclin E2-CDK2 complex(GO:0097135) |
| 0.0 | 0.1 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.0 | 0.1 | GO:0034274 | Atg12-Atg5-Atg16 complex(GO:0034274) |
| 0.0 | 0.1 | GO:0042406 | extrinsic component of endoplasmic reticulum membrane(GO:0042406) |
| 0.0 | 0.0 | GO:0009330 | DNA topoisomerase complex (ATP-hydrolyzing)(GO:0009330) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0030062 | mitochondrial tricarboxylic acid cycle enzyme complex(GO:0030062) |
| 0.0 | 0.2 | GO:0005664 | origin recognition complex(GO:0000808) nuclear origin of replication recognition complex(GO:0005664) |
| 0.0 | 0.1 | GO:0071149 | TEAD-2-YAP complex(GO:0071149) |
| 0.0 | 0.1 | GO:0002177 | manchette(GO:0002177) |
| 0.0 | 0.1 | GO:0033276 | transcription factor TFTC complex(GO:0033276) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0032044 | DSIF complex(GO:0032044) |
| 0.0 | 0.1 | GO:0033186 | CAF-1 complex(GO:0033186) |
| 0.0 | 0.1 | GO:0070381 | endosome to plasma membrane transport vesicle(GO:0070381) |
| 0.0 | 0.1 | GO:0000439 | core TFIIH complex(GO:0000439) |
| 0.0 | 0.3 | GO:0010369 | chromocenter(GO:0010369) |
| 0.0 | 0.2 | GO:0005847 | mRNA cleavage and polyadenylation specificity factor complex(GO:0005847) |
| 0.0 | 0.0 | GO:0034751 | aryl hydrocarbon receptor complex(GO:0034751) |
| 0.0 | 0.2 | GO:0000276 | mitochondrial proton-transporting ATP synthase complex, coupling factor F(o)(GO:0000276) |
| 0.0 | 0.6 | GO:0005834 | heterotrimeric G-protein complex(GO:0005834) |
| 0.0 | 0.1 | GO:0070552 | BRISC complex(GO:0070552) |
| 0.0 | 0.1 | GO:0035631 | CD40 receptor complex(GO:0035631) |
| 0.0 | 0.1 | GO:1990726 | Lsm1-7-Pat1 complex(GO:1990726) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0008330 | protein tyrosine/threonine phosphatase activity(GO:0008330) |
| 0.2 | 0.5 | GO:0004392 | heme oxygenase (decyclizing) activity(GO:0004392) |
| 0.1 | 0.6 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.1 | 1.0 | GO:0051434 | BH3 domain binding(GO:0051434) |
| 0.1 | 0.3 | GO:0005483 | soluble NSF attachment protein activity(GO:0005483) |
| 0.1 | 0.3 | GO:0004370 | glycerol kinase activity(GO:0004370) |
| 0.1 | 0.2 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.1 | 0.8 | GO:0031419 | cobalamin binding(GO:0031419) |
| 0.1 | 0.4 | GO:0035402 | histone kinase activity (H3-T11 specific)(GO:0035402) |
| 0.1 | 0.2 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.1 | 0.2 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.1 | 0.2 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.0 | 0.3 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.1 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.0 | 0.2 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.3 | GO:0008559 | xenobiotic-transporting ATPase activity(GO:0008559) |
| 0.0 | 0.5 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.3 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.0 | 0.4 | GO:0031386 | protein tag(GO:0031386) |
| 0.0 | 0.1 | GO:0008384 | IkappaB kinase activity(GO:0008384) |
| 0.0 | 0.2 | GO:0004634 | phosphopyruvate hydratase activity(GO:0004634) |
| 0.0 | 1.5 | GO:0050699 | WW domain binding(GO:0050699) |
| 0.0 | 0.1 | GO:0008309 | double-stranded DNA exodeoxyribonuclease activity(GO:0008309) |
| 0.0 | 0.1 | GO:0015439 | heme-transporting ATPase activity(GO:0015439) |
| 0.0 | 0.1 | GO:0043739 | G/U mismatch-specific uracil-DNA glycosylase activity(GO:0043739) |
| 0.0 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0004372 | glycine hydroxymethyltransferase activity(GO:0004372) threonine aldolase activity(GO:0004793) L-allo-threonine aldolase activity(GO:0008732) |
| 0.0 | 0.2 | GO:0019776 | Atg8 ligase activity(GO:0019776) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.0 | 0.1 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.0 | 0.1 | GO:0034057 | RNA strand-exchange activity(GO:0034057) |
| 0.0 | 0.1 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.9 | GO:0070273 | phosphatidylinositol-4-phosphate binding(GO:0070273) |
| 0.0 | 0.3 | GO:0004030 | aldehyde dehydrogenase [NAD(P)+] activity(GO:0004030) |
| 0.0 | 0.1 | GO:0004416 | hydroxyacylglutathione hydrolase activity(GO:0004416) |
| 0.0 | 0.1 | GO:0031751 | D2 dopamine receptor binding(GO:0031749) D4 dopamine receptor binding(GO:0031751) |
| 0.0 | 0.1 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.0 | GO:0048027 | mRNA 5'-UTR binding(GO:0048027) |
| 0.0 | 0.2 | GO:0034513 | box H/ACA snoRNA binding(GO:0034513) |
| 0.0 | 0.1 | GO:0047961 | glycine N-acyltransferase activity(GO:0047961) |
| 0.0 | 0.2 | GO:0034597 | phosphatidylinositol-4,5-bisphosphate 4-phosphatase activity(GO:0034597) |
| 0.0 | 0.1 | GO:0043891 | glyceraldehyde-3-phosphate dehydrogenase (NAD+) (phosphorylating) activity(GO:0004365) glyceraldehyde-3-phosphate dehydrogenase (NAD(P)+) (phosphorylating) activity(GO:0043891) |
| 0.0 | 0.1 | GO:0003826 | alpha-ketoacid dehydrogenase activity(GO:0003826) 3-methyl-2-oxobutanoate dehydrogenase (2-methylpropanoyl-transferring) activity(GO:0003863) |
| 0.0 | 0.1 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.0 | 0.1 | GO:0044388 | small protein activating enzyme binding(GO:0044388) |
| 0.0 | 0.1 | GO:0005137 | interleukin-5 receptor binding(GO:0005137) |
| 0.0 | 0.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.1 | GO:0034038 | deoxyhypusine synthase activity(GO:0034038) |
| 0.0 | 0.1 | GO:0019763 | immunoglobulin receptor activity(GO:0019763) |
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.1 | GO:1990050 | phosphatidic acid transporter activity(GO:1990050) |
| 0.0 | 0.0 | GO:0008948 | oxaloacetate decarboxylase activity(GO:0008948) |
| 0.0 | 0.1 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.7 | GO:0000983 | transcription factor activity, RNA polymerase II core promoter sequence-specific(GO:0000983) |
| 0.0 | 0.1 | GO:0035515 | oxidative RNA demethylase activity(GO:0035515) |
| 0.0 | 0.1 | GO:0032138 | single base insertion or deletion binding(GO:0032138) |
| 0.0 | 0.1 | GO:0016532 | superoxide dismutase copper chaperone activity(GO:0016532) |
| 0.0 | 0.1 | GO:0008479 | queuine tRNA-ribosyltransferase activity(GO:0008479) |
| 0.0 | 0.1 | GO:0004774 | succinate-CoA ligase activity(GO:0004774) |
| 0.0 | 0.0 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.3 | GO:0042609 | CD4 receptor binding(GO:0042609) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.6 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0035651 | AP-3 adaptor complex binding(GO:0035651) |
| 0.0 | 0.1 | GO:0008269 | JAK pathway signal transduction adaptor activity(GO:0008269) |
| 0.0 | 0.2 | GO:0000979 | RNA polymerase II core promoter sequence-specific DNA binding(GO:0000979) |
| 0.0 | 0.0 | GO:0010309 | acireductone dioxygenase [iron(II)-requiring] activity(GO:0010309) |
| 0.0 | 0.0 | GO:0001016 | RNA polymerase III regulatory region DNA binding(GO:0001016) |
| 0.0 | 0.0 | GO:0019203 | 6-phosphofructo-2-kinase activity(GO:0003873) fructose-2,6-bisphosphate 2-phosphatase activity(GO:0004331) phosphofructokinase activity(GO:0008443) carbohydrate phosphatase activity(GO:0019203) sugar-phosphatase activity(GO:0050308) |
| 0.0 | 0.1 | GO:0019864 | IgG binding(GO:0019864) |
| 0.0 | 0.1 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.0 | GO:0001588 | dopamine neurotransmitter receptor activity, coupled via Gs(GO:0001588) |
| 0.0 | 0.1 | GO:0003998 | acylphosphatase activity(GO:0003998) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.0 | SA PROGRAMMED CELL DEATH | Programmed cell death, or apoptosis, eliminates damaged or unneeded cells. |
| 0.0 | 0.7 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
| 0.0 | 0.4 | SA REG CASCADE OF CYCLIN EXPR | Expression of cyclins regulates progression through the cell cycle by activating cyclin-dependent kinases. |
| 0.0 | 1.1 | PID MYC PATHWAY | C-MYC pathway |
| 0.0 | 0.5 | PID WNT CANONICAL PATHWAY | Canonical Wnt signaling pathway |
| 0.0 | 0.4 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.2 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.1 | REACTOME TRANSPORT OF MATURE TRANSCRIPT TO CYTOPLASM | Genes involved in Transport of Mature Transcript to Cytoplasm |
| 0.0 | 1.1 | REACTOME INTRINSIC PATHWAY FOR APOPTOSIS | Genes involved in Intrinsic Pathway for Apoptosis |
| 0.0 | 0.3 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME METABOLISM OF PORPHYRINS | Genes involved in Metabolism of porphyrins |
| 0.0 | 0.5 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.2 | REACTOME PURINE CATABOLISM | Genes involved in Purine catabolism |
| 0.0 | 0.1 | REACTOME APOBEC3G MEDIATED RESISTANCE TO HIV1 INFECTION | Genes involved in APOBEC3G mediated resistance to HIV-1 infection |
| 0.0 | 0.1 | REACTOME RAF MAP KINASE CASCADE | Genes involved in RAF/MAP kinase cascade |
| 0.0 | 0.3 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.1 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.2 | REACTOME NFKB ACTIVATION THROUGH FADD RIP1 PATHWAY MEDIATED BY CASPASE 8 AND10 | Genes involved in NF-kB activation through FADD/RIP-1 pathway mediated by caspase-8 and -10 |
| 0.0 | 0.1 | REACTOME SOS MEDIATED SIGNALLING | Genes involved in SOS-mediated signalling |