Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
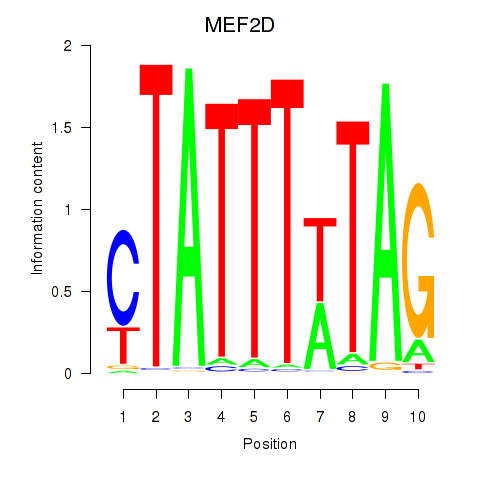
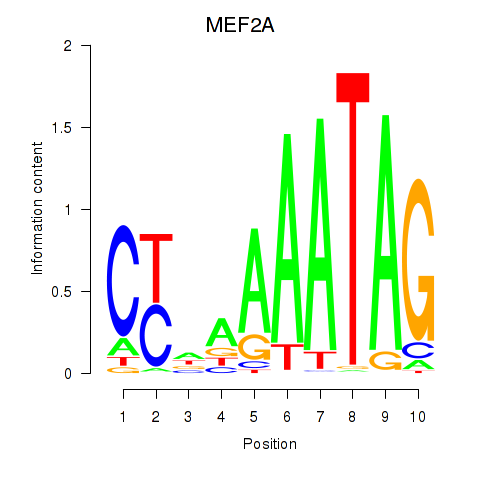
Results for MEF2D_MEF2A
Z-value: 0.50
Transcription factors associated with MEF2D_MEF2A
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEF2D
|
ENSG00000116604.13 | MEF2D |
|
MEF2A
|
ENSG00000068305.13 | MEF2A |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEF2A | hg19_v2_chr15_+_100106244_100106292 | 0.65 | 8.1e-02 | Click! |
| MEF2D | hg19_v2_chr1_-_156460391_156460417 | -0.41 | 3.1e-01 | Click! |
Activity profile of MEF2D_MEF2A motif
Sorted Z-values of MEF2D_MEF2A motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MEF2D_MEF2A
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr5_-_111091948 | 1.91 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr3_+_99357319 | 1.04 |
ENST00000452013.1 ENST00000261037.3 ENST00000273342.4 |
COL8A1 |
collagen, type VIII, alpha 1 |
| chr5_-_150521192 | 0.96 |
ENST00000523714.1 ENST00000521749.1 |
ANXA6 |
annexin A6 |
| chr2_-_190044480 | 0.82 |
ENST00000374866.3 |
COL5A2 |
collagen, type V, alpha 2 |
| chr12_-_91539918 | 0.73 |
ENST00000548218.1 |
DCN |
decorin |
| chr1_+_221051699 | 0.71 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr1_+_170633047 | 0.68 |
ENST00000239461.6 ENST00000497230.2 |
PRRX1 |
paired related homeobox 1 |
| chrX_-_142722897 | 0.63 |
ENST00000338017.4 |
SLITRK4 |
SLIT and NTRK-like family, member 4 |
| chr6_-_154751629 | 0.57 |
ENST00000424998.1 |
CNKSR3 |
CNKSR family member 3 |
| chr13_-_67802549 | 0.55 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr1_+_114522049 | 0.49 |
ENST00000369551.1 ENST00000320334.4 |
OLFML3 |
olfactomedin-like 3 |
| chr4_+_55095264 | 0.48 |
ENST00000257290.5 |
PDGFRA |
platelet-derived growth factor receptor, alpha polypeptide |
| chr2_+_233497931 | 0.45 |
ENST00000264059.3 |
EFHD1 |
EF-hand domain family, member D1 |
| chr1_+_145524891 | 0.43 |
ENST00000369304.3 |
ITGA10 |
integrin, alpha 10 |
| chr6_+_136172820 | 0.42 |
ENST00000308191.6 |
PDE7B |
phosphodiesterase 7B |
| chr5_+_173316341 | 0.37 |
ENST00000520867.1 ENST00000334035.5 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
| chr3_-_112360116 | 0.36 |
ENST00000206423.3 ENST00000439685.2 |
CCDC80 |
coiled-coil domain containing 80 |
| chr6_+_132129151 | 0.36 |
ENST00000360971.2 |
ENPP1 |
ectonucleotide pyrophosphatase/phosphodiesterase 1 |
| chr13_+_76378305 | 0.36 |
ENST00000526371.1 ENST00000526528.1 |
LMO7 |
LIM domain 7 |
| chr13_+_76210448 | 0.36 |
ENST00000377499.5 |
LMO7 |
LIM domain 7 |
| chr5_+_67584174 | 0.34 |
ENST00000320694.8 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr4_-_39640513 | 0.34 |
ENST00000511809.1 ENST00000505729.1 |
SMIM14 |
small integral membrane protein 14 |
| chr13_+_76378357 | 0.32 |
ENST00000489941.2 ENST00000525373.1 |
LMO7 |
LIM domain 7 |
| chr13_+_76334498 | 0.32 |
ENST00000534657.1 |
LMO7 |
LIM domain 7 |
| chr1_-_31845914 | 0.28 |
ENST00000373713.2 |
FABP3 |
fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) |
| chr4_-_57547454 | 0.28 |
ENST00000556376.2 |
HOPX |
HOP homeobox |
| chr4_-_57547870 | 0.28 |
ENST00000381260.3 ENST00000420433.1 ENST00000554144.1 ENST00000557328.1 |
HOPX |
HOP homeobox |
| chr3_+_69928256 | 0.28 |
ENST00000394355.2 |
MITF |
microphthalmia-associated transcription factor |
| chr1_-_153518270 | 0.27 |
ENST00000354332.4 ENST00000368716.4 |
S100A4 |
S100 calcium binding protein A4 |
| chr2_+_148778570 | 0.27 |
ENST00000407073.1 |
MBD5 |
methyl-CpG binding domain protein 5 |
| chr17_+_37821593 | 0.27 |
ENST00000578283.1 |
TCAP |
titin-cap |
| chr17_+_68165657 | 0.27 |
ENST00000243457.3 |
KCNJ2 |
potassium inwardly-rectifying channel, subfamily J, member 2 |
| chr10_-_62704005 | 0.27 |
ENST00000337910.5 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
| chrX_-_21776281 | 0.26 |
ENST00000379494.3 |
SMPX |
small muscle protein, X-linked |
| chr11_-_19223523 | 0.26 |
ENST00000265968.3 |
CSRP3 |
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr7_+_120629653 | 0.26 |
ENST00000450913.2 ENST00000340646.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chr3_+_69812877 | 0.25 |
ENST00000457080.1 ENST00000328528.6 |
MITF |
microphthalmia-associated transcription factor |
| chr1_+_156095951 | 0.24 |
ENST00000448611.2 ENST00000368297.1 |
LMNA |
lamin A/C |
| chr8_+_27183033 | 0.24 |
ENST00000420218.2 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr10_+_104178946 | 0.24 |
ENST00000432590.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chr8_+_27182862 | 0.24 |
ENST00000521164.1 ENST00000346049.5 |
PTK2B |
protein tyrosine kinase 2 beta |
| chr6_+_108977520 | 0.23 |
ENST00000540898.1 |
FOXO3 |
forkhead box O3 |
| chr2_-_211168332 | 0.22 |
ENST00000341685.4 |
MYL1 |
myosin, light chain 1, alkali; skeletal, fast |
| chr13_+_76334795 | 0.21 |
ENST00000526202.1 ENST00000465261.2 |
LMO7 |
LIM domain 7 |
| chr4_+_113970772 | 0.20 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr4_+_129730839 | 0.20 |
ENST00000511647.1 |
PHF17 |
jade family PHD finger 1 |
| chr17_-_1389228 | 0.19 |
ENST00000438665.2 |
MYO1C |
myosin IC |
| chr17_+_42248063 | 0.19 |
ENST00000293414.1 |
ASB16 |
ankyrin repeat and SOCS box containing 16 |
| chr17_-_1389419 | 0.19 |
ENST00000575158.1 |
MYO1C |
myosin IC |
| chr1_+_201617450 | 0.19 |
ENST00000295624.6 ENST00000367297.4 ENST00000367300.3 |
NAV1 |
neuron navigator 1 |
| chr1_+_171060018 | 0.19 |
ENST00000367755.4 ENST00000392085.2 ENST00000542847.1 ENST00000538429.1 ENST00000479749.1 |
FMO3 |
flavin containing monooxygenase 3 |
| chr3_+_8543393 | 0.19 |
ENST00000157600.3 ENST00000415597.1 ENST00000535732.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr7_-_134143841 | 0.19 |
ENST00000285930.4 |
AKR1B1 |
aldo-keto reductase family 1, member B1 (aldose reductase) |
| chr19_+_35630344 | 0.18 |
ENST00000455515.2 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr4_+_129730779 | 0.18 |
ENST00000226319.6 |
PHF17 |
jade family PHD finger 1 |
| chr13_+_76334567 | 0.18 |
ENST00000321797.8 |
LMO7 |
LIM domain 7 |
| chr10_-_49860525 | 0.18 |
ENST00000435790.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr1_+_201617264 | 0.18 |
ENST00000367296.4 |
NAV1 |
neuron navigator 1 |
| chr3_+_141106458 | 0.17 |
ENST00000509883.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr2_-_106054952 | 0.16 |
ENST00000336660.5 ENST00000393352.3 ENST00000607522.1 |
FHL2 |
four and a half LIM domains 2 |
| chr11_+_65266507 | 0.16 |
ENST00000544868.1 |
MALAT1 |
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr1_-_206945830 | 0.15 |
ENST00000423557.1 |
IL10 |
interleukin 10 |
| chr3_-_39234074 | 0.15 |
ENST00000340369.3 ENST00000421646.1 ENST00000396251.1 |
XIRP1 |
xin actin-binding repeat containing 1 |
| chr2_+_220143989 | 0.15 |
ENST00000336576.5 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr3_+_8543561 | 0.15 |
ENST00000397386.3 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr8_-_13372253 | 0.15 |
ENST00000316609.5 |
DLC1 |
deleted in liver cancer 1 |
| chr3_-_176914238 | 0.15 |
ENST00000430069.1 ENST00000428970.1 |
TBL1XR1 |
transducin (beta)-like 1 X-linked receptor 1 |
| chr4_-_186696425 | 0.14 |
ENST00000430503.1 ENST00000319454.6 ENST00000450341.1 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr15_-_83474806 | 0.14 |
ENST00000541889.1 ENST00000334574.8 ENST00000561368.1 |
FSD2 |
fibronectin type III and SPRY domain containing 2 |
| chr15_+_100106155 | 0.14 |
ENST00000557785.1 ENST00000558049.1 ENST00000449277.2 |
MEF2A |
myocyte enhancer factor 2A |
| chr3_+_141106643 | 0.14 |
ENST00000514251.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr6_-_41703952 | 0.14 |
ENST00000358871.2 ENST00000403298.4 |
TFEB |
transcription factor EB |
| chr2_+_220144052 | 0.14 |
ENST00000425450.1 ENST00000392086.4 ENST00000421532.1 |
DNAJB2 |
DnaJ (Hsp40) homolog, subfamily B, member 2 |
| chr1_+_156096336 | 0.14 |
ENST00000504687.1 ENST00000473598.2 |
LMNA |
lamin A/C |
| chr15_+_100106126 | 0.14 |
ENST00000558812.1 ENST00000338042.6 |
MEF2A |
myocyte enhancer factor 2A |
| chr15_+_100106244 | 0.14 |
ENST00000557942.1 |
MEF2A |
myocyte enhancer factor 2A |
| chr17_-_79900255 | 0.13 |
ENST00000330655.3 ENST00000582198.1 |
MYADML2 PYCR1 |
myeloid-associated differentiation marker-like 2 pyrroline-5-carboxylate reductase 1 |
| chr19_+_37096194 | 0.13 |
ENST00000460670.1 ENST00000292928.2 ENST00000439428.1 |
ZNF382 |
zinc finger protein 382 |
| chr2_+_161993465 | 0.13 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr3_+_8543533 | 0.12 |
ENST00000454244.1 |
LMCD1 |
LIM and cysteine-rich domains 1 |
| chr3_-_137834436 | 0.12 |
ENST00000327532.2 ENST00000467030.1 |
DZIP1L |
DAZ interacting zinc finger protein 1-like |
| chr7_-_150946015 | 0.12 |
ENST00000262188.8 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr1_+_150245177 | 0.12 |
ENST00000369098.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr1_-_46642154 | 0.12 |
ENST00000540385.1 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr21_+_17909594 | 0.12 |
ENST00000441820.1 ENST00000602280.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr3_+_179370517 | 0.12 |
ENST00000263966.3 |
USP13 |
ubiquitin specific peptidase 13 (isopeptidase T-3) |
| chrX_-_15332665 | 0.12 |
ENST00000537676.1 ENST00000344384.4 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr4_+_37892682 | 0.11 |
ENST00000508802.1 ENST00000261439.4 ENST00000402522.1 |
TBC1D1 |
TBC1 (tre-2/USP6, BUB2, cdc16) domain family, member 1 |
| chr3_-_114343039 | 0.11 |
ENST00000481632.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr12_+_12938541 | 0.11 |
ENST00000356591.4 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr8_-_27850141 | 0.11 |
ENST00000524352.1 |
SCARA5 |
scavenger receptor class A, member 5 (putative) |
| chr3_+_138067314 | 0.11 |
ENST00000423968.2 |
MRAS |
muscle RAS oncogene homolog |
| chr15_-_52944231 | 0.11 |
ENST00000546305.2 |
FAM214A |
family with sequence similarity 214, member A |
| chr9_+_127054217 | 0.11 |
ENST00000394199.2 ENST00000546191.1 |
NEK6 |
NIMA-related kinase 6 |
| chr8_-_13372395 | 0.11 |
ENST00000276297.4 ENST00000511869.1 |
DLC1 |
deleted in liver cancer 1 |
| chr4_+_120056939 | 0.11 |
ENST00000307128.5 |
MYOZ2 |
myozenin 2 |
| chr9_-_85882145 | 0.11 |
ENST00000328788.1 |
FRMD3 |
FERM domain containing 3 |
| chr5_-_66492562 | 0.11 |
ENST00000256447.4 |
CD180 |
CD180 molecule |
| chr11_+_12766583 | 0.11 |
ENST00000361985.2 |
TEAD1 |
TEA domain family member 1 (SV40 transcriptional enhancer factor) |
| chr4_-_114682224 | 0.11 |
ENST00000342666.5 ENST00000515496.1 ENST00000514328.1 ENST00000508738.1 ENST00000379773.2 |
CAMK2D |
calcium/calmodulin-dependent protein kinase II delta |
| chr1_-_59249732 | 0.11 |
ENST00000371222.2 |
JUN |
jun proto-oncogene |
| chr5_+_138609782 | 0.11 |
ENST00000361059.2 ENST00000514694.1 ENST00000504203.1 ENST00000502929.1 ENST00000394800.2 ENST00000509644.1 ENST00000505016.1 |
MATR3 |
matrin 3 |
| chr10_-_3827371 | 0.10 |
ENST00000469435.1 |
KLF6 |
Kruppel-like factor 6 |
| chr19_-_49926698 | 0.10 |
ENST00000270631.1 |
PTH2 |
parathyroid hormone 2 |
| chr1_+_150245099 | 0.10 |
ENST00000369099.3 |
C1orf54 |
chromosome 1 open reading frame 54 |
| chr2_-_211179883 | 0.10 |
ENST00000352451.3 |
MYL1 |
myosin, light chain 1, alkali; skeletal, fast |
| chr3_+_138067666 | 0.10 |
ENST00000475711.1 ENST00000464896.1 |
MRAS |
muscle RAS oncogene homolog |
| chr2_+_161993412 | 0.10 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr1_+_84630645 | 0.10 |
ENST00000394839.2 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr21_-_35016231 | 0.10 |
ENST00000438788.1 |
CRYZL1 |
crystallin, zeta (quinone reductase)-like 1 |
| chr2_+_44502597 | 0.10 |
ENST00000260649.6 ENST00000409387.1 |
SLC3A1 |
solute carrier family 3 (amino acid transporter heavy chain), member 1 |
| chr17_-_42200996 | 0.10 |
ENST00000587135.1 ENST00000225983.6 ENST00000393622.2 ENST00000588703.1 |
HDAC5 |
histone deacetylase 5 |
| chr11_+_57308979 | 0.10 |
ENST00000457912.1 |
SMTNL1 |
smoothelin-like 1 |
| chr2_-_166060571 | 0.10 |
ENST00000360093.3 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr3_+_50316458 | 0.10 |
ENST00000316436.3 |
LSMEM2 |
leucine-rich single-pass membrane protein 2 |
| chr16_-_15736881 | 0.10 |
ENST00000540441.2 |
KIAA0430 |
KIAA0430 |
| chr16_-_15736953 | 0.10 |
ENST00000548025.1 ENST00000551742.1 ENST00000602337.1 ENST00000344181.3 ENST00000396368.3 |
KIAA0430 |
KIAA0430 |
| chr2_+_42721689 | 0.10 |
ENST00000405592.1 |
MTA3 |
metastasis associated 1 family, member 3 |
| chr3_-_145878954 | 0.09 |
ENST00000282903.5 ENST00000360060.3 |
PLOD2 |
procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 |
| chr14_-_89021077 | 0.09 |
ENST00000556564.1 |
PTPN21 |
protein tyrosine phosphatase, non-receptor type 21 |
| chr12_+_15699286 | 0.09 |
ENST00000442921.2 ENST00000542557.1 ENST00000445537.2 ENST00000544244.1 |
PTPRO |
protein tyrosine phosphatase, receptor type, O |
| chr9_+_139871948 | 0.09 |
ENST00000224167.2 ENST00000457950.1 ENST00000371625.3 ENST00000371623.3 |
PTGDS |
prostaglandin D2 synthase 21kDa (brain) |
| chr8_-_62602327 | 0.09 |
ENST00000445642.3 ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH |
aspartate beta-hydroxylase |
| chr1_+_160051319 | 0.09 |
ENST00000368088.3 |
KCNJ9 |
potassium inwardly-rectifying channel, subfamily J, member 9 |
| chr11_+_45918092 | 0.09 |
ENST00000395629.2 |
MAPK8IP1 |
mitogen-activated protein kinase 8 interacting protein 1 |
| chr15_+_93443419 | 0.09 |
ENST00000557381.1 ENST00000420239.2 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
| chr22_+_32340481 | 0.09 |
ENST00000397492.1 |
YWHAH |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr19_+_35629702 | 0.09 |
ENST00000351325.4 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr4_-_186697044 | 0.09 |
ENST00000437304.2 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr2_-_166060552 | 0.09 |
ENST00000283254.7 ENST00000453007.1 |
SCN3A |
sodium channel, voltage-gated, type III, alpha subunit |
| chr8_-_72268889 | 0.09 |
ENST00000388742.4 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr2_+_11864458 | 0.09 |
ENST00000396098.1 ENST00000396099.1 ENST00000425416.2 |
LPIN1 |
lipin 1 |
| chr17_+_6918064 | 0.09 |
ENST00000546760.1 ENST00000552402.1 |
C17orf49 |
chromosome 17 open reading frame 49 |
| chr11_+_12399071 | 0.09 |
ENST00000539723.1 ENST00000550549.1 |
PARVA |
parvin, alpha |
| chr9_-_215744 | 0.08 |
ENST00000382387.2 |
C9orf66 |
chromosome 9 open reading frame 66 |
| chr17_+_7461613 | 0.08 |
ENST00000438470.1 ENST00000436057.1 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr1_+_151129135 | 0.08 |
ENST00000602841.1 |
SCNM1 |
sodium channel modifier 1 |
| chr14_-_70546897 | 0.08 |
ENST00000394330.2 ENST00000533541.1 ENST00000216568.7 |
SLC8A3 |
solute carrier family 8 (sodium/calcium exchanger), member 3 |
| chr10_-_3827417 | 0.08 |
ENST00000497571.1 ENST00000542957.1 |
KLF6 |
Kruppel-like factor 6 |
| chr17_-_42200958 | 0.08 |
ENST00000336057.5 |
HDAC5 |
histone deacetylase 5 |
| chr4_+_119809984 | 0.08 |
ENST00000307142.4 ENST00000448416.2 ENST00000429713.2 |
SYNPO2 |
synaptopodin 2 |
| chr9_-_130639997 | 0.08 |
ENST00000373176.1 |
AK1 |
adenylate kinase 1 |
| chr7_+_30951461 | 0.08 |
ENST00000311813.4 |
AQP1 |
aquaporin 1 (Colton blood group) |
| chr3_+_54157480 | 0.08 |
ENST00000490478.1 |
CACNA2D3 |
calcium channel, voltage-dependent, alpha 2/delta subunit 3 |
| chr2_+_162087577 | 0.08 |
ENST00000439442.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr1_+_90098606 | 0.08 |
ENST00000370454.4 |
LRRC8C |
leucine rich repeat containing 8 family, member C |
| chr8_-_72268968 | 0.08 |
ENST00000388740.3 |
EYA1 |
eyes absent homolog 1 (Drosophila) |
| chr19_+_35630022 | 0.08 |
ENST00000589209.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr17_+_6918093 | 0.07 |
ENST00000439424.2 |
C17orf49 |
chromosome 17 open reading frame 49 |
| chr18_-_3219847 | 0.07 |
ENST00000261606.7 |
MYOM1 |
myomesin 1 |
| chr22_-_36013368 | 0.07 |
ENST00000442617.1 ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB |
myoglobin |
| chr15_-_37391507 | 0.07 |
ENST00000557796.2 ENST00000397620.2 |
MEIS2 |
Meis homeobox 2 |
| chr2_+_33661382 | 0.07 |
ENST00000402538.3 |
RASGRP3 |
RAS guanyl releasing protein 3 (calcium and DAG-regulated) |
| chr22_+_25595817 | 0.07 |
ENST00000215855.2 ENST00000404334.1 |
CRYBB3 |
crystallin, beta B3 |
| chr5_+_161274685 | 0.07 |
ENST00000428797.2 |
GABRA1 |
gamma-aminobutyric acid (GABA) A receptor, alpha 1 |
| chr3_-_47934234 | 0.07 |
ENST00000420772.2 |
MAP4 |
microtubule-associated protein 4 |
| chr5_-_137475071 | 0.07 |
ENST00000265191.2 |
NME5 |
NME/NM23 family member 5 |
| chr11_-_18034701 | 0.07 |
ENST00000265965.5 |
SERGEF |
secretion regulating guanine nucleotide exchange factor |
| chr2_+_85766280 | 0.07 |
ENST00000306434.3 |
MAT2A |
methionine adenosyltransferase II, alpha |
| chrX_+_18725758 | 0.07 |
ENST00000472826.1 ENST00000544635.1 ENST00000496075.2 |
PPEF1 |
protein phosphatase, EF-hand calcium binding domain 1 |
| chr10_+_88428206 | 0.07 |
ENST00000429277.2 ENST00000458213.2 ENST00000352360.5 |
LDB3 |
LIM domain binding 3 |
| chr9_-_97356075 | 0.07 |
ENST00000375337.3 |
FBP2 |
fructose-1,6-bisphosphatase 2 |
| chr7_-_27169801 | 0.07 |
ENST00000511914.1 |
HOXA4 |
homeobox A4 |
| chr22_-_31742218 | 0.07 |
ENST00000266269.5 ENST00000405309.3 ENST00000351933.4 |
PATZ1 |
POZ (BTB) and AT hook containing zinc finger 1 |
| chr1_-_201391149 | 0.07 |
ENST00000555948.1 ENST00000556362.1 |
TNNI1 |
troponin I type 1 (skeletal, slow) |
| chr7_+_18535346 | 0.07 |
ENST00000405010.3 ENST00000406451.4 ENST00000428307.2 |
HDAC9 |
histone deacetylase 9 |
| chr8_-_70745575 | 0.07 |
ENST00000524945.1 |
SLCO5A1 |
solute carrier organic anion transporter family, member 5A1 |
| chr1_-_150693318 | 0.07 |
ENST00000442853.1 ENST00000368995.4 ENST00000368993.2 ENST00000361824.2 ENST00000322343.7 |
HORMAD1 |
HORMA domain containing 1 |
| chr17_+_80186908 | 0.06 |
ENST00000582743.1 ENST00000578684.1 ENST00000577650.1 ENST00000582715.1 |
SLC16A3 |
solute carrier family 16 (monocarboxylate transporter), member 3 |
| chr18_-_77276057 | 0.06 |
ENST00000597412.1 |
AC018445.1 |
Uncharacterized protein |
| chr19_+_50832943 | 0.06 |
ENST00000542413.1 |
NR1H2 |
nuclear receptor subfamily 1, group H, member 2 |
| chr15_-_37391614 | 0.06 |
ENST00000219869.9 |
MEIS2 |
Meis homeobox 2 |
| chr16_+_31225337 | 0.06 |
ENST00000322122.3 |
TRIM72 |
tripartite motif containing 72 |
| chr22_-_51016846 | 0.06 |
ENST00000312108.7 ENST00000395650.2 |
CPT1B |
carnitine palmitoyltransferase 1B (muscle) |
| chr10_+_88428370 | 0.06 |
ENST00000372066.3 ENST00000263066.6 ENST00000372056.4 ENST00000310944.6 ENST00000361373.4 ENST00000542786.1 |
LDB3 |
LIM domain binding 3 |
| chr9_+_129677123 | 0.06 |
ENST00000373436.1 |
RALGPS1 |
Ral GEF with PH domain and SH3 binding motif 1 |
| chr19_-_893200 | 0.06 |
ENST00000269814.4 ENST00000395808.3 ENST00000312090.6 ENST00000325464.1 |
MED16 |
mediator complex subunit 16 |
| chr1_+_156211753 | 0.06 |
ENST00000368272.4 |
BGLAP |
bone gamma-carboxyglutamate (gla) protein |
| chr8_-_7309887 | 0.06 |
ENST00000458665.1 ENST00000528168.1 |
SPAG11B |
sperm associated antigen 11B |
| chr2_+_168043793 | 0.06 |
ENST00000409273.1 ENST00000409605.1 |
XIRP2 |
xin actin-binding repeat containing 2 |
| chr11_-_88796803 | 0.06 |
ENST00000418177.2 ENST00000455756.2 |
GRM5 |
glutamate receptor, metabotropic 5 |
| chr13_+_76378407 | 0.06 |
ENST00000447038.1 |
LMO7 |
LIM domain 7 |
| chr1_-_159825137 | 0.06 |
ENST00000368102.1 |
C1orf204 |
chromosome 1 open reading frame 204 |
| chr16_+_30075463 | 0.06 |
ENST00000562168.1 ENST00000569545.1 |
ALDOA |
aldolase A, fructose-bisphosphate |
| chrX_-_30327495 | 0.05 |
ENST00000453287.1 |
NR0B1 |
nuclear receptor subfamily 0, group B, member 1 |
| chr19_-_40324255 | 0.05 |
ENST00000593685.1 ENST00000600611.1 |
DYRK1B |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 1B |
| chr1_+_182584314 | 0.05 |
ENST00000566297.1 |
RP11-317P15.4 |
RP11-317P15.4 |
| chr7_-_27702455 | 0.05 |
ENST00000265395.2 |
HIBADH |
3-hydroxyisobutyrate dehydrogenase |
| chr12_-_49393092 | 0.05 |
ENST00000421952.2 |
DDN |
dendrin |
| chr5_-_131347501 | 0.05 |
ENST00000543479.1 |
ACSL6 |
acyl-CoA synthetase long-chain family member 6 |
| chrX_-_70288234 | 0.05 |
ENST00000276105.3 ENST00000374274.3 |
SNX12 |
sorting nexin 12 |
| chr4_-_83765613 | 0.05 |
ENST00000503937.1 |
SEC31A |
SEC31 homolog A (S. cerevisiae) |
| chr22_+_32340447 | 0.05 |
ENST00000248975.5 |
YWHAH |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, eta |
| chr13_+_51483814 | 0.05 |
ENST00000336617.3 ENST00000422660.1 |
RNASEH2B |
ribonuclease H2, subunit B |
| chrX_-_71933888 | 0.05 |
ENST00000373542.4 ENST00000339490.3 ENST00000541944.1 ENST00000373539.3 ENST00000373545.3 |
PHKA1 |
phosphorylase kinase, alpha 1 (muscle) |
| chr10_-_104178857 | 0.05 |
ENST00000020673.5 |
PSD |
pleckstrin and Sec7 domain containing |
| chr3_+_138067521 | 0.05 |
ENST00000494949.1 |
MRAS |
muscle RAS oncogene homolog |
| chr5_-_141030943 | 0.05 |
ENST00000522783.1 ENST00000519800.1 ENST00000435817.2 |
FCHSD1 |
FCH and double SH3 domains 1 |
| chr16_+_15737124 | 0.05 |
ENST00000396355.1 ENST00000396353.2 |
NDE1 |
nudE neurodevelopment protein 1 |
| chr6_-_169654139 | 0.05 |
ENST00000366787.3 |
THBS2 |
thrombospondin 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:1903225 | negative regulation of endodermal cell differentiation(GO:1903225) |
| 0.1 | 0.5 | GO:2000537 | regulation of cGMP-mediated signaling(GO:0010752) regulation of B cell chemotaxis(GO:2000537) positive regulation of B cell chemotaxis(GO:2000538) |
| 0.1 | 0.4 | GO:0030505 | inorganic diphosphate transport(GO:0030505) |
| 0.1 | 0.4 | GO:0072277 | metanephric glomerulus morphogenesis(GO:0072275) metanephric glomerulus vasculature morphogenesis(GO:0072276) metanephric glomerular capillary formation(GO:0072277) |
| 0.1 | 0.3 | GO:2001245 | regulation of phosphatidylcholine biosynthetic process(GO:2001245) |
| 0.1 | 0.3 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.7 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.5 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.1 | 0.2 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.4 | GO:0035105 | sterol regulatory element binding protein import into nucleus(GO:0035105) |
| 0.1 | 0.3 | GO:1903644 | regulation of chaperone-mediated protein folding(GO:1903644) |
| 0.1 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.4 | GO:1900748 | positive regulation of vascular endothelial growth factor signaling pathway(GO:1900748) |
| 0.1 | 0.2 | GO:0006059 | hexitol metabolic process(GO:0006059) inner medullary collecting duct development(GO:0072061) |
| 0.1 | 0.4 | GO:0055005 | ventricular cardiac myofibril assembly(GO:0055005) ERK5 cascade(GO:0070375) |
| 0.1 | 0.3 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.6 | GO:0048664 | neuron fate determination(GO:0048664) |
| 0.1 | 0.3 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.1 | 0.2 | GO:0002874 | regulation of chronic inflammatory response to antigenic stimulus(GO:0002874) |
| 0.0 | 0.3 | GO:0060399 | positive regulation of growth hormone receptor signaling pathway(GO:0060399) |
| 0.0 | 0.2 | GO:0055011 | atrial cardiac muscle cell differentiation(GO:0055011) atrial cardiac muscle cell development(GO:0055014) |
| 0.0 | 0.1 | GO:0044313 | protein K6-linked deubiquitination(GO:0044313) |
| 0.0 | 0.7 | GO:0051901 | positive regulation of mitochondrial depolarization(GO:0051901) |
| 0.0 | 1.0 | GO:0051560 | mitochondrial calcium ion homeostasis(GO:0051560) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.1 | GO:0036060 | filtration diaphragm assembly(GO:0036058) slit diaphragm assembly(GO:0036060) |
| 0.0 | 0.4 | GO:2000766 | negative regulation of cytoplasmic translation(GO:2000766) |
| 0.0 | 0.1 | GO:0035026 | leading edge cell differentiation(GO:0035026) |
| 0.0 | 0.1 | GO:0072229 | carbon dioxide transmembrane transport(GO:0035378) proximal convoluted tubule development(GO:0072019) metanephric proximal convoluted tubule development(GO:0072229) |
| 0.0 | 0.1 | GO:0032485 | Ral protein signal transduction(GO:0032484) regulation of Ral protein signal transduction(GO:0032485) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:0042138 | meiotic DNA double-strand break formation(GO:0042138) meiotic recombination checkpoint(GO:0051598) |
| 0.0 | 0.6 | GO:0001829 | trophectodermal cell differentiation(GO:0001829) |
| 0.0 | 0.1 | GO:0090187 | positive regulation of pancreatic juice secretion(GO:0090187) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.0 | 0.1 | GO:0055129 | L-proline biosynthetic process(GO:0055129) |
| 0.0 | 0.5 | GO:0070886 | positive regulation of calcineurin-NFAT signaling cascade(GO:0070886) |
| 0.0 | 0.3 | GO:0051531 | NFAT protein import into nucleus(GO:0051531) |
| 0.0 | 0.1 | GO:0002322 | B cell proliferation involved in immune response(GO:0002322) |
| 0.0 | 0.2 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.0 | 0.1 | GO:0006556 | S-adenosylmethionine biosynthetic process(GO:0006556) |
| 0.0 | 0.1 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.6 | GO:2000651 | positive regulation of sodium ion transmembrane transporter activity(GO:2000651) |
| 0.0 | 0.1 | GO:1990822 | L-cystine transport(GO:0015811) basic amino acid transmembrane transport(GO:1990822) |
| 0.0 | 0.3 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.0 | 0.1 | GO:0060613 | fat pad development(GO:0060613) |
| 0.0 | 0.0 | GO:0010737 | protein kinase A signaling(GO:0010737) |
| 0.0 | 0.0 | GO:0036309 | protein localization to M-band(GO:0036309) |
| 0.0 | 0.0 | GO:0071947 | protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) |
| 0.0 | 0.1 | GO:0006642 | triglyceride mobilization(GO:0006642) |
| 0.0 | 0.1 | GO:0034983 | peptidyl-lysine deacetylation(GO:0034983) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0032571 | response to vitamin K(GO:0032571) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.0 | GO:1903279 | regulation of calcium:sodium antiporter activity(GO:1903279) |
| 0.0 | 0.0 | GO:0014053 | negative regulation of gamma-aminobutyric acid secretion(GO:0014053) aspartate secretion(GO:0061528) regulation of aspartate secretion(GO:1904448) positive regulation of aspartate secretion(GO:1904450) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.1 | GO:1903336 | negative regulation of vacuolar transport(GO:1903336) |
| 0.0 | 0.6 | GO:0035987 | endodermal cell differentiation(GO:0035987) |
| 0.0 | 0.0 | GO:0033387 | putrescine biosynthetic process from ornithine(GO:0033387) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.1 | 0.4 | GO:0034680 | integrin alpha10-beta1 complex(GO:0034680) |
| 0.1 | 1.0 | GO:0098642 | network-forming collagen trimer(GO:0098642) collagen network(GO:0098645) |
| 0.1 | 0.4 | GO:0045160 | myosin I complex(GO:0045160) |
| 0.1 | 0.3 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 0.7 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.3 | GO:0005927 | muscle tendon junction(GO:0005927) |
| 0.0 | 0.4 | GO:0005638 | lamin filament(GO:0005638) |
| 0.0 | 0.2 | GO:0097454 | Schwann cell microvillus(GO:0097454) |
| 0.0 | 0.5 | GO:0017146 | NMDA selective glutamate receptor complex(GO:0017146) |
| 0.0 | 0.1 | GO:0020005 | symbiont-containing vacuole(GO:0020003) symbiont-containing vacuole membrane(GO:0020005) |
| 0.0 | 0.3 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0048269 | methionine adenosyltransferase complex(GO:0048269) |
| 0.0 | 0.1 | GO:0071148 | TEAD-1-YAP complex(GO:0071148) |
| 0.0 | 0.1 | GO:0032299 | ribonuclease H2 complex(GO:0032299) |
| 0.0 | 0.4 | GO:0043194 | axon initial segment(GO:0043194) |
| 0.0 | 0.4 | GO:0005890 | sodium:potassium-exchanging ATPase complex(GO:0005890) |
| 0.0 | 0.4 | GO:1990124 | messenger ribonucleoprotein complex(GO:1990124) |
| 0.0 | 0.4 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 0.2 | GO:0001518 | voltage-gated sodium channel complex(GO:0001518) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.1 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.0 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 1.0 | GO:0044291 | cell-cell contact zone(GO:0044291) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0043423 | 3-phosphoinositide-dependent protein kinase binding(GO:0043423) |
| 0.1 | 0.4 | GO:0005017 | platelet-derived growth factor-activated receptor activity(GO:0005017) |
| 0.1 | 0.3 | GO:0070538 | oleic acid binding(GO:0070538) |
| 0.1 | 0.4 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.1 | 0.4 | GO:0035529 | NADH pyrophosphatase activity(GO:0035529) |
| 0.1 | 0.4 | GO:0051373 | FATZ binding(GO:0051373) |
| 0.1 | 0.3 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.3 | GO:0043559 | insulin binding(GO:0043559) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.2 | GO:0001025 | RNA polymerase III transcription factor binding(GO:0001025) |
| 0.0 | 0.1 | GO:0008475 | procollagen-lysine 5-dioxygenase activity(GO:0008475) procollagen glucosyltransferase activity(GO:0033823) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.7 | GO:0071837 | HMG box domain binding(GO:0071837) |
| 0.0 | 0.3 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.1 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.0 | 0.2 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0032038 | myosin II heavy chain binding(GO:0032038) |
| 0.0 | 0.1 | GO:0004667 | prostaglandin-D synthase activity(GO:0004667) |
| 0.0 | 0.1 | GO:0004735 | pyrroline-5-carboxylate reductase activity(GO:0004735) |
| 0.0 | 0.1 | GO:0008195 | phosphatidate phosphatase activity(GO:0008195) |
| 0.0 | 0.4 | GO:0015467 | G-protein activated inward rectifier potassium channel activity(GO:0015467) |
| 0.0 | 0.1 | GO:0010861 | thyroid hormone receptor activator activity(GO:0010861) thyroid hormone receptor coactivator activity(GO:0030375) |
| 0.0 | 0.1 | GO:0046848 | hydroxyapatite binding(GO:0046848) |
| 0.0 | 0.1 | GO:0004095 | carnitine O-palmitoyltransferase activity(GO:0004095) |
| 0.0 | 0.1 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.2 | GO:0004032 | alditol:NADP+ 1-oxidoreductase activity(GO:0004032) |
| 0.0 | 0.1 | GO:0015184 | L-cystine transmembrane transporter activity(GO:0015184) |
| 0.0 | 0.1 | GO:0008321 | Ral guanyl-nucleotide exchange factor activity(GO:0008321) |
| 0.0 | 0.4 | GO:0030898 | actin-dependent ATPase activity(GO:0030898) |
| 0.0 | 0.1 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0003960 | NADPH:quinone reductase activity(GO:0003960) |
| 0.0 | 1.0 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0004616 | phosphogluconate dehydrogenase (decarboxylating) activity(GO:0004616) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.1 | GO:0004332 | fructose-bisphosphate aldolase activity(GO:0004332) |
| 0.0 | 0.1 | GO:0034191 | transcription factor activity, sequence-specific DNA binding transcription factor recruiting(GO:0001010) apolipoprotein A-I receptor binding(GO:0034191) |
| 0.0 | 0.1 | GO:0004972 | NMDA glutamate receptor activity(GO:0004972) |
| 0.0 | 0.1 | GO:0099529 | neurotransmitter receptor activity involved in regulation of postsynaptic membrane potential(GO:0099529) transmitter-gated ion channel activity involved in regulation of postsynaptic membrane potential(GO:1904315) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0098960 | postsynaptic neurotransmitter receptor activity(GO:0098960) neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.0 | GO:0032145 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.7 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.4 | GO:0001968 | fibronectin binding(GO:0001968) |
| 0.0 | 0.1 | GO:0004523 | RNA-DNA hybrid ribonuclease activity(GO:0004523) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 1.3 | NABA COLLAGENS | Genes encoding collagen proteins |
| 0.0 | 0.9 | PID PDGFRA PATHWAY | PDGFR-alpha signaling pathway |
| 0.0 | 0.5 | SA PTEN PATHWAY | PTEN is a tumor suppressor that dephosphorylates the lipid messenger phosphatidylinositol triphosphate. |
| 0.0 | 0.7 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.8 | PID P38 ALPHA BETA DOWNSTREAM PATHWAY | Signaling mediated by p38-alpha and p38-beta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.5 | REACTOME IL 7 SIGNALING | Genes involved in Interleukin-7 signaling |
| 0.0 | 1.4 | REACTOME COLLAGEN FORMATION | Genes involved in Collagen formation |
| 0.0 | 0.4 | REACTOME ERK MAPK TARGETS | Genes involved in ERK/MAPK targets |