Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
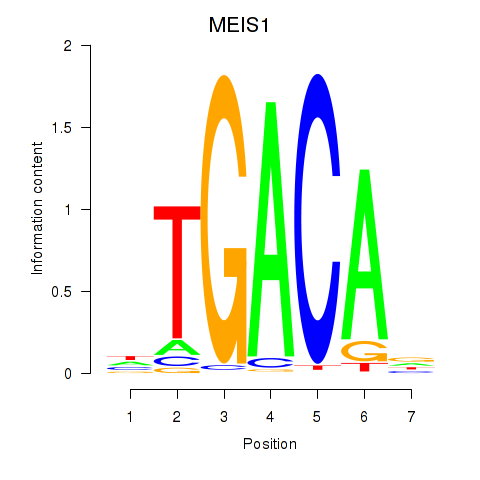
Results for MEIS1
Z-value: 1.27
Transcription factors associated with MEIS1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MEIS1
|
ENSG00000143995.15 | MEIS1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MEIS1 | hg19_v2_chr2_+_66666432_66666462 | -0.92 | 1.4e-03 | Click! |
Activity profile of MEIS1 motif
Sorted Z-values of MEIS1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MEIS1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_14113592 | 4.51 |
ENST00000502759.1 ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085 |
long intergenic non-protein coding RNA 1085 |
| chr14_-_92413353 | 2.91 |
ENST00000556154.1 |
FBLN5 |
fibulin 5 |
| chr5_+_92919043 | 2.28 |
ENST00000327111.3 |
NR2F1 |
nuclear receptor subfamily 2, group F, member 1 |
| chr2_-_202562774 | 2.16 |
ENST00000396886.3 ENST00000409143.1 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr13_+_102104980 | 2.12 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr8_-_120651020 | 2.08 |
ENST00000522826.1 ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr2_+_33359646 | 2.02 |
ENST00000390003.4 ENST00000418533.2 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr13_-_67802549 | 2.00 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr2_+_33359687 | 1.99 |
ENST00000402934.1 ENST00000404525.1 ENST00000407925.1 |
LTBP1 |
latent transforming growth factor beta binding protein 1 |
| chr1_+_78956651 | 1.93 |
ENST00000370757.3 ENST00000370756.3 |
PTGFR |
prostaglandin F receptor (FP) |
| chr13_+_102104952 | 1.90 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr9_-_79307096 | 1.85 |
ENST00000376717.2 ENST00000223609.6 ENST00000443509.2 |
PRUNE2 |
prune homolog 2 (Drosophila) |
| chr2_-_202562716 | 1.77 |
ENST00000428900.2 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr4_-_156787425 | 1.72 |
ENST00000537611.2 |
ASIC5 |
acid-sensing (proton-gated) ion channel family member 5 |
| chr2_-_68547061 | 1.62 |
ENST00000263655.3 |
CNRIP1 |
cannabinoid receptor interacting protein 1 |
| chr2_-_202563414 | 1.59 |
ENST00000409474.3 ENST00000315506.7 ENST00000359962.5 |
MPP4 |
membrane protein, palmitoylated 4 (MAGUK p55 subfamily member 4) |
| chr8_-_13372395 | 1.51 |
ENST00000276297.4 ENST00000511869.1 |
DLC1 |
deleted in liver cancer 1 |
| chr16_-_3350614 | 1.40 |
ENST00000268674.2 |
TIGD7 |
tigger transposable element derived 7 |
| chr15_+_33010175 | 1.36 |
ENST00000300177.4 ENST00000560677.1 ENST00000560830.1 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr8_+_97597148 | 1.35 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr8_-_120685608 | 1.24 |
ENST00000427067.2 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr8_-_13372253 | 1.21 |
ENST00000316609.5 |
DLC1 |
deleted in liver cancer 1 |
| chr6_-_46293378 | 1.20 |
ENST00000330430.6 |
RCAN2 |
regulator of calcineurin 2 |
| chr5_-_20575959 | 1.12 |
ENST00000507958.1 |
CDH18 |
cadherin 18, type 2 |
| chr17_-_29648761 | 1.11 |
ENST00000247270.3 ENST00000462804.2 |
EVI2A |
ecotropic viral integration site 2A |
| chr5_+_156693091 | 1.10 |
ENST00000318218.6 ENST00000442283.2 ENST00000522463.1 ENST00000521420.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr5_+_156693159 | 1.09 |
ENST00000347377.6 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr4_-_175443943 | 1.04 |
ENST00000296522.6 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr20_-_39946237 | 1.03 |
ENST00000441102.2 ENST00000559234.1 |
ZHX3 |
zinc fingers and homeoboxes 3 |
| chr6_+_155537771 | 1.01 |
ENST00000275246.7 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
| chr3_-_100712352 | 1.00 |
ENST00000471714.1 ENST00000284322.5 |
ABI3BP |
ABI family, member 3 (NESH) binding protein |
| chr3_+_141103634 | 0.98 |
ENST00000507722.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr1_+_114522049 | 0.93 |
ENST00000369551.1 ENST00000320334.4 |
OLFML3 |
olfactomedin-like 3 |
| chr15_-_82338460 | 0.93 |
ENST00000558133.1 ENST00000329713.4 |
MEX3B |
mex-3 RNA binding family member B |
| chr17_-_53809473 | 0.92 |
ENST00000575734.1 |
TMEM100 |
transmembrane protein 100 |
| chr2_+_24714729 | 0.88 |
ENST00000406961.1 ENST00000405141.1 |
NCOA1 |
nuclear receptor coactivator 1 |
| chr13_-_33780133 | 0.88 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr10_+_54074033 | 0.87 |
ENST00000373970.3 |
DKK1 |
dickkopf WNT signaling pathway inhibitor 1 |
| chr5_-_172756506 | 0.83 |
ENST00000265087.4 |
STC2 |
stanniocalcin 2 |
| chr17_-_67138015 | 0.83 |
ENST00000284425.2 ENST00000590645.1 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr5_+_82767487 | 0.82 |
ENST00000343200.5 ENST00000342785.4 |
VCAN |
versican |
| chr5_+_82767284 | 0.82 |
ENST00000265077.3 |
VCAN |
versican |
| chr6_-_87804815 | 0.82 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr3_+_12392971 | 0.80 |
ENST00000287820.6 |
PPARG |
peroxisome proliferator-activated receptor gamma |
| chr7_+_149571045 | 0.80 |
ENST00000479613.1 ENST00000606024.1 ENST00000464662.1 ENST00000425642.2 |
ATP6V0E2 |
ATPase, H+ transporting V0 subunit e2 |
| chr9_-_16727978 | 0.79 |
ENST00000418777.1 ENST00000468187.2 |
BNC2 |
basonuclin 2 |
| chr13_+_76378305 | 0.78 |
ENST00000526371.1 ENST00000526528.1 |
LMO7 |
LIM domain 7 |
| chr14_+_52164820 | 0.76 |
ENST00000554167.1 |
FRMD6 |
FERM domain containing 6 |
| chr8_-_108510224 | 0.72 |
ENST00000517746.1 ENST00000297450.3 |
ANGPT1 |
angiopoietin 1 |
| chr2_-_79315112 | 0.72 |
ENST00000305089.3 |
REG1B |
regenerating islet-derived 1 beta |
| chr1_+_210406121 | 0.72 |
ENST00000367012.3 |
SERTAD4 |
SERTA domain containing 4 |
| chr10_-_29084886 | 0.71 |
ENST00000608061.1 ENST00000443246.2 ENST00000446012.1 |
LINC00837 |
long intergenic non-protein coding RNA 837 |
| chr13_+_76378357 | 0.70 |
ENST00000489941.2 ENST00000525373.1 |
LMO7 |
LIM domain 7 |
| chr11_-_118550346 | 0.69 |
ENST00000530256.1 |
TREH |
trehalase (brush-border membrane glycoprotein) |
| chr3_+_151986709 | 0.69 |
ENST00000495875.2 ENST00000493459.1 ENST00000324210.5 ENST00000459747.1 |
MBNL1 |
muscleblind-like splicing regulator 1 |
| chr9_-_16728161 | 0.69 |
ENST00000603713.1 ENST00000603313.1 |
BNC2 |
basonuclin 2 |
| chr2_+_201170703 | 0.68 |
ENST00000358677.5 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr10_-_33623564 | 0.68 |
ENST00000374875.1 ENST00000374822.4 |
NRP1 |
neuropilin 1 |
| chr12_-_76953453 | 0.68 |
ENST00000549570.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr2_-_145275228 | 0.67 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr22_-_44708731 | 0.66 |
ENST00000381176.4 |
KIAA1644 |
KIAA1644 |
| chr5_+_82767583 | 0.66 |
ENST00000512590.2 ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN |
versican |
| chrX_-_62974941 | 0.65 |
ENST00000374872.1 ENST00000253401.6 ENST00000374870.4 |
ARHGEF9 |
Cdc42 guanine nucleotide exchange factor (GEF) 9 |
| chr7_+_95401851 | 0.64 |
ENST00000447467.2 |
DYNC1I1 |
dynein, cytoplasmic 1, intermediate chain 1 |
| chr12_+_1738363 | 0.64 |
ENST00000397196.2 |
WNT5B |
wingless-type MMTV integration site family, member 5B |
| chr11_-_27723158 | 0.63 |
ENST00000395980.2 |
BDNF |
brain-derived neurotrophic factor |
| chr1_-_85870177 | 0.62 |
ENST00000542148.1 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr5_-_168727713 | 0.62 |
ENST00000404867.3 |
SLIT3 |
slit homolog 3 (Drosophila) |
| chr7_+_120628731 | 0.62 |
ENST00000310396.5 |
CPED1 |
cadherin-like and PC-esterase domain containing 1 |
| chrX_+_152082969 | 0.61 |
ENST00000535861.1 ENST00000539731.1 ENST00000449285.2 ENST00000318504.7 ENST00000324823.6 ENST00000370268.4 ENST00000370270.2 |
ZNF185 |
zinc finger protein 185 (LIM domain) |
| chrX_-_142722897 | 0.60 |
ENST00000338017.4 |
SLITRK4 |
SLIT and NTRK-like family, member 4 |
| chr2_-_179343226 | 0.59 |
ENST00000434643.2 |
FKBP7 |
FK506 binding protein 7 |
| chr2_-_179343268 | 0.59 |
ENST00000424785.2 |
FKBP7 |
FK506 binding protein 7 |
| chrX_+_100333709 | 0.59 |
ENST00000372930.4 |
TMEM35 |
transmembrane protein 35 |
| chr20_-_3996036 | 0.58 |
ENST00000336095.6 |
RNF24 |
ring finger protein 24 |
| chr2_+_210444142 | 0.58 |
ENST00000360351.4 ENST00000361559.4 |
MAP2 |
microtubule-associated protein 2 |
| chr3_-_178789220 | 0.57 |
ENST00000414084.1 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr1_+_221051699 | 0.57 |
ENST00000366903.6 |
HLX |
H2.0-like homeobox |
| chr2_+_109237717 | 0.57 |
ENST00000409441.1 |
LIMS1 |
LIM and senescent cell antigen-like domains 1 |
| chr14_+_22615942 | 0.57 |
ENST00000390457.2 |
TRAV27 |
T cell receptor alpha variable 27 |
| chr7_+_129932974 | 0.56 |
ENST00000445470.2 ENST00000222482.4 ENST00000492072.1 ENST00000473956.1 ENST00000493259.1 ENST00000486598.1 |
CPA4 |
carboxypeptidase A4 |
| chr6_+_39760783 | 0.55 |
ENST00000398904.2 ENST00000538976.1 |
DAAM2 |
dishevelled associated activator of morphogenesis 2 |
| chr3_-_178789993 | 0.55 |
ENST00000432729.1 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr12_+_55248289 | 0.54 |
ENST00000308796.6 |
MUCL1 |
mucin-like 1 |
| chrX_-_92928557 | 0.53 |
ENST00000373079.3 ENST00000475430.2 |
NAP1L3 |
nucleosome assembly protein 1-like 3 |
| chr9_-_22009297 | 0.53 |
ENST00000276925.6 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr16_+_2880157 | 0.53 |
ENST00000382280.3 |
ZG16B |
zymogen granule protein 16B |
| chr9_+_2157655 | 0.53 |
ENST00000452193.1 |
SMARCA2 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| chr12_+_21679220 | 0.53 |
ENST00000256969.2 |
C12orf39 |
chromosome 12 open reading frame 39 |
| chrX_-_107018969 | 0.52 |
ENST00000372383.4 |
TSC22D3 |
TSC22 domain family, member 3 |
| chr12_+_51785057 | 0.52 |
ENST00000535225.2 |
SLC4A8 |
solute carrier family 4, sodium bicarbonate cotransporter, member 8 |
| chr10_+_85954377 | 0.52 |
ENST00000332904.3 ENST00000372117.3 |
CDHR1 |
cadherin-related family member 1 |
| chr11_+_71938925 | 0.51 |
ENST00000538751.1 |
INPPL1 |
inositol polyphosphate phosphatase-like 1 |
| chr12_+_65672702 | 0.51 |
ENST00000538045.1 ENST00000535239.1 |
MSRB3 |
methionine sulfoxide reductase B3 |
| chr10_-_33623310 | 0.51 |
ENST00000395995.1 ENST00000374823.5 ENST00000374821.5 ENST00000374816.3 |
NRP1 |
neuropilin 1 |
| chr3_+_49449636 | 0.51 |
ENST00000273590.3 |
TCTA |
T-cell leukemia translocation altered |
| chr10_-_49813090 | 0.51 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr5_-_139943830 | 0.51 |
ENST00000412920.3 ENST00000511201.2 ENST00000356738.2 ENST00000354402.5 ENST00000358580.5 ENST00000508496.2 |
APBB3 |
amyloid beta (A4) precursor protein-binding, family B, member 3 |
| chr10_+_124768482 | 0.50 |
ENST00000368869.4 ENST00000358776.4 |
ACADSB |
acyl-CoA dehydrogenase, short/branched chain |
| chr7_-_100881041 | 0.50 |
ENST00000412417.1 ENST00000414035.1 |
CLDN15 |
claudin 15 |
| chrX_+_9431324 | 0.50 |
ENST00000407597.2 ENST00000424279.1 ENST00000536365.1 ENST00000441088.1 ENST00000380961.1 ENST00000415293.1 |
TBL1X |
transducin (beta)-like 1X-linked |
| chr16_-_30006922 | 0.50 |
ENST00000564026.1 |
HIRIP3 |
HIRA interacting protein 3 |
| chr9_-_113341985 | 0.50 |
ENST00000374469.1 |
SVEP1 |
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr20_-_45035223 | 0.50 |
ENST00000450812.1 ENST00000290246.6 ENST00000439931.2 ENST00000396391.1 |
ELMO2 |
engulfment and cell motility 2 |
| chr3_+_141121164 | 0.50 |
ENST00000510338.1 ENST00000504673.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr5_-_139930713 | 0.50 |
ENST00000602657.1 |
SRA1 |
steroid receptor RNA activator 1 |
| chr5_+_139505520 | 0.49 |
ENST00000333305.3 |
IGIP |
IgA-inducing protein |
| chr18_+_32455201 | 0.49 |
ENST00000590831.2 |
DTNA |
dystrobrevin, alpha |
| chrX_+_102883887 | 0.49 |
ENST00000372625.3 ENST00000372624.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chrX_+_66764375 | 0.49 |
ENST00000374690.3 |
AR |
androgen receptor |
| chr16_+_2588012 | 0.48 |
ENST00000354836.5 ENST00000389224.3 |
PDPK1 |
3-phosphoinositide dependent protein kinase-1 |
| chr21_-_31744557 | 0.48 |
ENST00000399889.2 |
KRTAP13-2 |
keratin associated protein 13-2 |
| chr5_+_49962772 | 0.48 |
ENST00000281631.5 ENST00000513738.1 ENST00000503665.1 ENST00000514067.2 ENST00000503046.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr1_-_46598371 | 0.48 |
ENST00000372006.1 ENST00000425892.1 ENST00000420542.1 ENST00000354242.4 ENST00000340332.6 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr22_-_36013368 | 0.47 |
ENST00000442617.1 ENST00000397326.2 ENST00000397328.1 ENST00000451685.1 |
MB |
myoglobin |
| chr6_-_11382478 | 0.47 |
ENST00000397378.3 ENST00000513989.1 ENST00000508546.1 ENST00000504387.1 |
NEDD9 |
neural precursor cell expressed, developmentally down-regulated 9 |
| chr8_+_81397876 | 0.47 |
ENST00000430430.1 |
ZBTB10 |
zinc finger and BTB domain containing 10 |
| chr6_-_154751629 | 0.47 |
ENST00000424998.1 |
CNKSR3 |
CNKSR family member 3 |
| chr16_+_53133070 | 0.47 |
ENST00000565832.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chr10_-_49860525 | 0.46 |
ENST00000435790.2 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr1_+_26605618 | 0.46 |
ENST00000270792.5 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
| chr15_+_43809797 | 0.46 |
ENST00000399453.1 ENST00000300231.5 |
MAP1A |
microtubule-associated protein 1A |
| chr6_+_72922590 | 0.46 |
ENST00000523963.1 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr20_-_45061695 | 0.46 |
ENST00000445496.2 |
ELMO2 |
engulfment and cell motility 2 |
| chr6_+_72922505 | 0.45 |
ENST00000401910.3 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr3_+_148457585 | 0.45 |
ENST00000402260.1 |
AGTR1 |
angiotensin II receptor, type 1 |
| chr4_+_77870960 | 0.45 |
ENST00000505788.1 ENST00000510515.1 ENST00000504637.1 |
SEPT11 |
septin 11 |
| chr1_-_155211017 | 0.45 |
ENST00000536770.1 ENST00000368373.3 |
GBA |
glucosidase, beta, acid |
| chr8_-_27462822 | 0.45 |
ENST00000522098.1 |
CLU |
clusterin |
| chr5_+_67586465 | 0.44 |
ENST00000336483.5 |
PIK3R1 |
phosphoinositide-3-kinase, regulatory subunit 1 (alpha) |
| chr8_-_93029520 | 0.44 |
ENST00000521553.1 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr7_+_90338712 | 0.44 |
ENST00000265741.3 ENST00000406263.1 |
CDK14 |
cyclin-dependent kinase 14 |
| chr10_+_45495898 | 0.44 |
ENST00000298299.3 |
ZNF22 |
zinc finger protein 22 |
| chr1_+_109102652 | 0.44 |
ENST00000370035.3 ENST00000405454.1 |
FAM102B |
family with sequence similarity 102, member B |
| chr20_-_35580104 | 0.43 |
ENST00000373694.5 |
SAMHD1 |
SAM domain and HD domain 1 |
| chr17_-_7145475 | 0.43 |
ENST00000571129.1 ENST00000571253.1 ENST00000573928.1 |
GABARAP |
GABA(A) receptor-associated protein |
| chr2_-_99279928 | 0.43 |
ENST00000414521.2 |
MGAT4A |
mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A |
| chr9_-_113342160 | 0.43 |
ENST00000401783.2 ENST00000374461.1 |
SVEP1 |
sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 |
| chr14_-_80677815 | 0.42 |
ENST00000557125.1 ENST00000555750.1 |
DIO2 |
deiodinase, iodothyronine, type II |
| chr3_-_127541679 | 0.42 |
ENST00000265052.5 |
MGLL |
monoglyceride lipase |
| chr11_-_64052111 | 0.42 |
ENST00000394532.3 ENST00000394531.3 ENST00000309032.3 |
BAD |
BCL2-associated agonist of cell death |
| chr8_+_81398444 | 0.42 |
ENST00000455036.3 ENST00000426744.2 |
ZBTB10 |
zinc finger and BTB domain containing 10 |
| chr20_+_33464407 | 0.42 |
ENST00000253382.5 |
ACSS2 |
acyl-CoA synthetase short-chain family member 2 |
| chr19_-_12595586 | 0.42 |
ENST00000397732.3 |
ZNF709 |
zinc finger protein 709 |
| chr9_-_123239632 | 0.41 |
ENST00000416449.1 |
CDK5RAP2 |
CDK5 regulatory subunit associated protein 2 |
| chr19_+_36266433 | 0.41 |
ENST00000314737.5 |
ARHGAP33 |
Rho GTPase activating protein 33 |
| chr11_+_12115543 | 0.41 |
ENST00000537344.1 ENST00000532179.1 ENST00000526065.1 |
MICAL2 |
microtubule associated monooxygenase, calponin and LIM domain containing 2 |
| chr5_-_127674883 | 0.41 |
ENST00000507835.1 |
FBN2 |
fibrillin 2 |
| chr19_+_35630628 | 0.41 |
ENST00000588715.1 ENST00000588607.1 |
FXYD1 |
FXYD domain containing ion transport regulator 1 |
| chr9_+_4985016 | 0.41 |
ENST00000539801.1 |
JAK2 |
Janus kinase 2 |
| chrX_+_51636629 | 0.40 |
ENST00000375722.1 ENST00000326587.7 ENST00000375695.2 |
MAGED1 |
melanoma antigen family D, 1 |
| chr2_-_200322723 | 0.40 |
ENST00000417098.1 |
SATB2 |
SATB homeobox 2 |
| chr17_-_53800217 | 0.40 |
ENST00000424486.2 |
TMEM100 |
transmembrane protein 100 |
| chr9_-_22009241 | 0.40 |
ENST00000380142.4 |
CDKN2B |
cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4) |
| chr1_+_61330931 | 0.39 |
ENST00000371191.1 |
NFIA |
nuclear factor I/A |
| chr20_-_45984401 | 0.39 |
ENST00000311275.7 |
ZMYND8 |
zinc finger, MYND-type containing 8 |
| chr12_+_100041527 | 0.39 |
ENST00000324341.1 |
FAM71C |
family with sequence similarity 71, member C |
| chr1_-_156828810 | 0.39 |
ENST00000368195.3 |
INSRR |
insulin receptor-related receptor |
| chr16_+_30006615 | 0.38 |
ENST00000563197.1 |
INO80E |
INO80 complex subunit E |
| chr16_+_30907927 | 0.38 |
ENST00000279804.2 ENST00000395019.3 |
CTF1 |
cardiotrophin 1 |
| chr5_+_140810132 | 0.38 |
ENST00000252085.3 |
PCDHGA12 |
protocadherin gamma subfamily A, 12 |
| chr1_-_241799232 | 0.38 |
ENST00000366553.1 |
CHML |
choroideremia-like (Rab escort protein 2) |
| chr3_-_127541194 | 0.38 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr4_-_175443484 | 0.38 |
ENST00000514584.1 ENST00000542498.1 ENST00000296521.7 ENST00000422112.2 ENST00000504433.1 |
HPGD |
hydroxyprostaglandin dehydrogenase 15-(NAD) |
| chr2_-_50574856 | 0.38 |
ENST00000342183.5 |
NRXN1 |
neurexin 1 |
| chrX_+_55478538 | 0.38 |
ENST00000342972.1 |
MAGEH1 |
melanoma antigen family H, 1 |
| chr3_-_47934234 | 0.37 |
ENST00000420772.2 |
MAP4 |
microtubule-associated protein 4 |
| chr12_+_65672423 | 0.37 |
ENST00000355192.3 ENST00000308259.5 ENST00000540804.1 ENST00000535664.1 ENST00000541189.1 |
MSRB3 |
methionine sulfoxide reductase B3 |
| chr20_+_33464238 | 0.37 |
ENST00000360596.2 |
ACSS2 |
acyl-CoA synthetase short-chain family member 2 |
| chr9_+_4985228 | 0.37 |
ENST00000381652.3 |
JAK2 |
Janus kinase 2 |
| chr3_-_120170052 | 0.37 |
ENST00000295633.3 |
FSTL1 |
follistatin-like 1 |
| chr20_-_35580240 | 0.36 |
ENST00000262878.4 |
SAMHD1 |
SAM domain and HD domain 1 |
| chr9_+_34652164 | 0.36 |
ENST00000441545.2 ENST00000553620.1 |
IL11RA |
interleukin 11 receptor, alpha |
| chr21_-_34100341 | 0.36 |
ENST00000382499.2 ENST00000433931.2 |
SYNJ1 |
synaptojanin 1 |
| chr1_-_154909329 | 0.36 |
ENST00000368467.3 |
PMVK |
phosphomevalonate kinase |
| chr5_+_118965244 | 0.36 |
ENST00000515256.1 ENST00000509264.1 |
FAM170A |
family with sequence similarity 170, member A |
| chrX_-_83757399 | 0.36 |
ENST00000373177.2 ENST00000297977.5 ENST00000506585.2 ENST00000449553.2 |
HDX |
highly divergent homeobox |
| chr12_-_76953513 | 0.36 |
ENST00000547540.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr8_-_72987810 | 0.35 |
ENST00000262209.4 |
TRPA1 |
transient receptor potential cation channel, subfamily A, member 1 |
| chrX_-_153775426 | 0.35 |
ENST00000393562.2 |
G6PD |
glucose-6-phosphate dehydrogenase |
| chr6_+_108881012 | 0.35 |
ENST00000343882.6 |
FOXO3 |
forkhead box O3 |
| chr1_+_110210644 | 0.35 |
ENST00000369831.2 ENST00000442650.1 ENST00000369827.3 ENST00000460717.3 ENST00000241337.4 ENST00000467579.3 ENST00000414179.2 ENST00000369829.2 |
GSTM2 |
glutathione S-transferase mu 2 (muscle) |
| chr12_+_12938541 | 0.35 |
ENST00000356591.4 |
APOLD1 |
apolipoprotein L domain containing 1 |
| chr17_+_41158742 | 0.35 |
ENST00000415816.2 ENST00000438323.2 |
IFI35 |
interferon-induced protein 35 |
| chr5_+_49962495 | 0.35 |
ENST00000515175.1 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr6_+_30524663 | 0.34 |
ENST00000376560.3 |
PRR3 |
proline rich 3 |
| chr5_+_140864649 | 0.34 |
ENST00000306593.1 |
PCDHGC4 |
protocadherin gamma subfamily C, 4 |
| chr18_+_2655849 | 0.34 |
ENST00000261598.8 |
SMCHD1 |
structural maintenance of chromosomes flexible hinge domain containing 1 |
| chr19_-_38714847 | 0.34 |
ENST00000420980.2 ENST00000355526.4 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
| chr1_+_45478568 | 0.34 |
ENST00000428106.1 |
UROD |
uroporphyrinogen decarboxylase |
| chr7_+_8008418 | 0.33 |
ENST00000223145.5 |
GLCCI1 |
glucocorticoid induced transcript 1 |
| chr14_+_53019822 | 0.33 |
ENST00000321662.6 |
GPR137C |
G protein-coupled receptor 137C |
| chrX_+_54947229 | 0.33 |
ENST00000442098.1 ENST00000430420.1 ENST00000453081.1 ENST00000173898.7 ENST00000319167.8 ENST00000375022.4 ENST00000399736.1 ENST00000440072.1 ENST00000420798.2 ENST00000431115.1 ENST00000440759.1 ENST00000375041.2 |
TRO |
trophinin |
| chr11_-_2323290 | 0.33 |
ENST00000381153.3 |
C11orf21 |
chromosome 11 open reading frame 21 |
| chr3_-_52719810 | 0.33 |
ENST00000424867.1 ENST00000394830.3 ENST00000431678.1 ENST00000450271.1 |
PBRM1 |
polybromo 1 |
| chr9_-_73029540 | 0.33 |
ENST00000377126.2 |
KLF9 |
Kruppel-like factor 9 |
| chr11_+_131240373 | 0.33 |
ENST00000374791.3 ENST00000436745.1 |
NTM |
neurotrimin |
| chr19_+_36266417 | 0.33 |
ENST00000378944.5 ENST00000007510.4 |
ARHGAP33 |
Rho GTPase activating protein 33 |
| chr6_-_41168920 | 0.32 |
ENST00000483722.1 |
TREML2 |
triggering receptor expressed on myeloid cells-like 2 |
| chr10_-_92681033 | 0.32 |
ENST00000371697.3 |
ANKRD1 |
ankyrin repeat domain 1 (cardiac muscle) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.5 | 1.4 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.4 | 4.4 | GO:0035583 | sequestering of TGFbeta in extracellular matrix(GO:0035583) |
| 0.3 | 0.9 | GO:0045925 | positive regulation of female receptivity(GO:0045925) |
| 0.3 | 0.9 | GO:1904956 | regulation of endodermal cell fate specification(GO:0042663) regulation of midbrain dopaminergic neuron differentiation(GO:1904956) |
| 0.2 | 0.7 | GO:0019417 | sulfur oxidation(GO:0019417) |
| 0.2 | 1.2 | GO:0021649 | vestibulocochlear nerve structural organization(GO:0021649) positive regulation of cytokine activity(GO:0060301) ganglion morphogenesis(GO:0061552) VEGF-activated neuropilin signaling pathway involved in axon guidance(GO:1902378) dorsal root ganglion morphogenesis(GO:1904835) otic placode development(GO:1905040) |
| 0.2 | 1.3 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.2 | 3.4 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.2 | 0.6 | GO:0031548 | regulation of brain-derived neurotrophic factor receptor signaling pathway(GO:0031548) |
| 0.2 | 2.1 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.2 | 2.7 | GO:0048251 | elastic fiber assembly(GO:0048251) |
| 0.2 | 1.8 | GO:2001300 | lipoxin metabolic process(GO:2001300) |
| 0.2 | 0.8 | GO:2000230 | negative regulation of pancreatic stellate cell proliferation(GO:2000230) |
| 0.2 | 0.8 | GO:0019541 | acetate biosynthetic process(GO:0019413) acetyl-CoA biosynthetic process from acetate(GO:0019427) propionate metabolic process(GO:0019541) propionate biosynthetic process(GO:0019542) |
| 0.2 | 0.8 | GO:0086097 | phospholipase C-activating angiotensin-activated signaling pathway(GO:0086097) |
| 0.2 | 0.8 | GO:1904457 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.2 | 1.7 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.2 | 1.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.2 | 2.8 | GO:1900119 | positive regulation of execution phase of apoptosis(GO:1900119) |
| 0.2 | 0.2 | GO:0014724 | regulation of twitch skeletal muscle contraction(GO:0014724) |
| 0.2 | 0.7 | GO:1904978 | regulation of endosome organization(GO:1904978) |
| 0.2 | 0.5 | GO:1900073 | regulation of neuromuscular synaptic transmission(GO:1900073) positive regulation of neuromuscular synaptic transmission(GO:1900075) |
| 0.2 | 0.5 | GO:0060599 | lateral sprouting involved in mammary gland duct morphogenesis(GO:0060599) |
| 0.2 | 1.4 | GO:0060842 | arterial endothelial cell differentiation(GO:0060842) |
| 0.2 | 0.8 | GO:1902724 | mineralocorticoid receptor signaling pathway(GO:0031959) positive regulation of skeletal muscle satellite cell proliferation(GO:1902724) positive regulation of growth factor dependent skeletal muscle satellite cell proliferation(GO:1902728) |
| 0.2 | 0.2 | GO:0006097 | glyoxylate cycle(GO:0006097) |
| 0.2 | 0.5 | GO:1990108 | protein linear deubiquitination(GO:1990108) |
| 0.1 | 0.7 | GO:0006740 | NADPH regeneration(GO:0006740) |
| 0.1 | 1.5 | GO:0010891 | negative regulation of sequestering of triglyceride(GO:0010891) |
| 0.1 | 0.4 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.1 | 0.9 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.1 | 0.8 | GO:0046061 | dATP catabolic process(GO:0046061) |
| 0.1 | 0.4 | GO:0019287 | isopentenyl diphosphate biosynthetic process, mevalonate pathway(GO:0019287) |
| 0.1 | 0.4 | GO:0001544 | initiation of primordial ovarian follicle growth(GO:0001544) |
| 0.1 | 0.8 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.1 | 0.7 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.1 | 0.8 | GO:0003383 | apical constriction(GO:0003383) |
| 0.1 | 1.0 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.1 | 1.3 | GO:2000576 | positive regulation of microtubule motor activity(GO:2000576) regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000580) positive regulation of ATP-dependent microtubule motor activity, plus-end-directed(GO:2000582) |
| 0.1 | 0.3 | GO:0090076 | relaxation of skeletal muscle(GO:0090076) |
| 0.1 | 0.3 | GO:0014876 | response to injury involved in regulation of muscle adaptation(GO:0014876) |
| 0.1 | 0.2 | GO:0003162 | atrioventricular node development(GO:0003162) |
| 0.1 | 0.7 | GO:0048014 | Tie signaling pathway(GO:0048014) |
| 0.1 | 0.3 | GO:1904395 | positive regulation of skeletal muscle acetylcholine-gated channel clustering(GO:1904395) negative regulation of neuromuscular junction development(GO:1904397) |
| 0.1 | 0.3 | GO:0098746 | fast, calcium ion-dependent exocytosis of neurotransmitter(GO:0098746) |
| 0.1 | 0.4 | GO:0071469 | cellular response to alkaline pH(GO:0071469) |
| 0.1 | 0.3 | GO:1902568 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.1 | 0.3 | GO:2000705 | dense core granule biogenesis(GO:0061110) regulation of dense core granule biogenesis(GO:2000705) |
| 0.1 | 0.8 | GO:2001256 | regulation of store-operated calcium entry(GO:2001256) |
| 0.1 | 0.3 | GO:0010701 | positive regulation of norepinephrine secretion(GO:0010701) |
| 0.1 | 0.7 | GO:0043353 | enucleate erythrocyte differentiation(GO:0043353) |
| 0.1 | 0.3 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.1 | 0.3 | GO:0001971 | negative regulation of activation of membrane attack complex(GO:0001971) |
| 0.1 | 1.6 | GO:0098828 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.1 | 0.3 | GO:0036333 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.1 | 0.3 | GO:1903248 | regulation of cellular amine catabolic process(GO:0033241) negative regulation of cellular amine catabolic process(GO:0033242) negative regulation of the force of heart contraction(GO:0098736) regulation of arginine catabolic process(GO:1900081) negative regulation of arginine catabolic process(GO:1900082) regulation of citrulline biosynthetic process(GO:1903248) negative regulation of citrulline biosynthetic process(GO:1903249) negative regulation of cellular amino acid biosynthetic process(GO:2000283) |
| 0.1 | 0.5 | GO:0072674 | multinuclear osteoclast differentiation(GO:0072674) osteoclast fusion(GO:0072675) |
| 0.1 | 2.3 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.1 | 0.3 | GO:0045362 | regulation of interleukin-1 biosynthetic process(GO:0045360) positive regulation of interleukin-1 biosynthetic process(GO:0045362) |
| 0.1 | 0.2 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.1 | 0.4 | GO:0010732 | protein glutathionylation(GO:0010731) regulation of protein glutathionylation(GO:0010732) negative regulation of protein glutathionylation(GO:0010734) |
| 0.1 | 0.3 | GO:1904647 | response to rotenone(GO:1904647) |
| 0.1 | 0.6 | GO:0045629 | negative regulation of T-helper 2 cell differentiation(GO:0045629) |
| 0.1 | 0.4 | GO:0048861 | leukemia inhibitory factor signaling pathway(GO:0048861) |
| 0.1 | 0.4 | GO:0061517 | macrophage proliferation(GO:0061517) microglial cell proliferation(GO:0061518) regulation of neuronal signal transduction(GO:1902847) positive regulation of neurofibrillary tangle assembly(GO:1902998) |
| 0.1 | 0.2 | GO:0006657 | CDP-choline pathway(GO:0006657) |
| 0.1 | 0.8 | GO:0030091 | protein repair(GO:0030091) |
| 0.1 | 0.3 | GO:0006850 | mitochondrial pyruvate transport(GO:0006850) mitochondrial pyruvate transmembrane transport(GO:1902361) |
| 0.1 | 0.4 | GO:0050968 | thermoception(GO:0050955) detection of chemical stimulus involved in sensory perception of pain(GO:0050968) |
| 0.1 | 0.6 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.1 | 0.3 | GO:1990502 | dense core granule maturation(GO:1990502) |
| 0.1 | 0.3 | GO:1902766 | skeletal muscle satellite cell migration(GO:1902766) |
| 0.1 | 0.2 | GO:0050760 | negative regulation of thymidylate synthase biosynthetic process(GO:0050760) |
| 0.1 | 0.3 | GO:0071484 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.1 | 0.4 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.1 | 0.2 | GO:0003363 | lamellipodium assembly involved in ameboidal cell migration(GO:0003363) extension of a leading process involved in cell motility in cerebral cortex radial glia guided migration(GO:0021816) |
| 0.1 | 0.4 | GO:1903758 | regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903756) negative regulation of transcription from RNA polymerase II promoter by histone modification(GO:1903758) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.4 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.1 | 0.6 | GO:1904781 | positive regulation of protein localization to centrosome(GO:1904781) |
| 0.1 | 0.2 | GO:0033364 | mast cell secretory granule organization(GO:0033364) |
| 0.1 | 0.7 | GO:0043951 | negative regulation of cAMP-mediated signaling(GO:0043951) |
| 0.1 | 0.4 | GO:0070458 | cellular detoxification of nitrogen compound(GO:0070458) |
| 0.1 | 0.2 | GO:1905244 | regulation of modification of synaptic structure(GO:1905244) |
| 0.1 | 0.4 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.1 | 0.1 | GO:0035162 | embryonic hemopoiesis(GO:0035162) |
| 0.1 | 0.5 | GO:0072513 | positive regulation of secondary heart field cardioblast proliferation(GO:0072513) |
| 0.1 | 0.2 | GO:1904694 | negative regulation of vascular smooth muscle contraction(GO:1904694) |
| 0.1 | 0.3 | GO:0090131 | mesenchyme migration(GO:0090131) |
| 0.0 | 0.3 | GO:0000720 | pyrimidine dimer repair by nucleotide-excision repair(GO:0000720) |
| 0.0 | 0.3 | GO:0048733 | sebaceous gland development(GO:0048733) |
| 0.0 | 0.2 | GO:0051835 | positive regulation of synapse structural plasticity(GO:0051835) |
| 0.0 | 0.4 | GO:0051013 | microtubule severing(GO:0051013) |
| 0.0 | 0.4 | GO:0097116 | gephyrin clustering involved in postsynaptic density assembly(GO:0097116) |
| 0.0 | 0.7 | GO:0005984 | disaccharide metabolic process(GO:0005984) |
| 0.0 | 0.2 | GO:1903377 | negative regulation of oxidative stress-induced neuron intrinsic apoptotic signaling pathway(GO:1903377) |
| 0.0 | 0.3 | GO:0044339 | canonical Wnt signaling pathway involved in osteoblast differentiation(GO:0044339) |
| 0.0 | 0.3 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0060406 | positive regulation of penile erection(GO:0060406) |
| 0.0 | 0.2 | GO:0046462 | monoacylglycerol metabolic process(GO:0046462) monoacylglycerol catabolic process(GO:0052651) |
| 0.0 | 0.6 | GO:0006527 | arginine catabolic process(GO:0006527) negative regulation of cellular response to hypoxia(GO:1900038) |
| 0.0 | 0.1 | GO:0006581 | acetylcholine catabolic process in synaptic cleft(GO:0001507) acetylcholine catabolic process(GO:0006581) |
| 0.0 | 0.3 | GO:0042866 | pyruvate biosynthetic process(GO:0042866) |
| 0.0 | 0.4 | GO:0021902 | commitment of neuronal cell to specific neuron type in forebrain(GO:0021902) |
| 0.0 | 0.9 | GO:2001275 | positive regulation of glucose import in response to insulin stimulus(GO:2001275) |
| 0.0 | 0.4 | GO:0070236 | negative regulation of activation-induced cell death of T cells(GO:0070236) |
| 0.0 | 0.2 | GO:0046013 | T cell homeostatic proliferation(GO:0001777) regulation of T cell homeostatic proliferation(GO:0046013) |
| 0.0 | 0.1 | GO:1902523 | positive regulation of protein K63-linked ubiquitination(GO:1902523) |
| 0.0 | 0.2 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.0 | 0.1 | GO:0006106 | fumarate metabolic process(GO:0006106) |
| 0.0 | 0.1 | GO:0003050 | regulation of systemic arterial blood pressure by atrial natriuretic peptide(GO:0003050) |
| 0.0 | 0.3 | GO:0009048 | dosage compensation by inactivation of X chromosome(GO:0009048) |
| 0.0 | 0.3 | GO:0031274 | positive regulation of pseudopodium assembly(GO:0031274) |
| 0.0 | 0.3 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.7 | GO:0006995 | cellular response to nitrogen starvation(GO:0006995) cellular response to nitrogen levels(GO:0043562) |
| 0.0 | 0.6 | GO:0070307 | lens fiber cell development(GO:0070307) |
| 0.0 | 0.1 | GO:0045578 | negative regulation of B cell differentiation(GO:0045578) |
| 0.0 | 0.2 | GO:0045204 | MAPK export from nucleus(GO:0045204) |
| 0.0 | 0.1 | GO:0002384 | hepatic immune response(GO:0002384) |
| 0.0 | 0.1 | GO:0007231 | osmosensory signaling pathway(GO:0007231) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.5 | GO:0001514 | selenocysteine incorporation(GO:0001514) translational readthrough(GO:0006451) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.2 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.2 | GO:0051490 | negative regulation of filopodium assembly(GO:0051490) |
| 0.0 | 0.1 | GO:1903595 | positive regulation of histamine secretion by mast cell(GO:1903595) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0043376 | regulation of CD8-positive, alpha-beta T cell differentiation(GO:0043376) |
| 0.0 | 0.2 | GO:0060023 | soft palate development(GO:0060023) |
| 0.0 | 0.4 | GO:0014722 | regulation of skeletal muscle contraction by calcium ion signaling(GO:0014722) |
| 0.0 | 0.4 | GO:0046600 | negative regulation of centriole replication(GO:0046600) |
| 0.0 | 0.1 | GO:0016240 | autophagosome docking(GO:0016240) |
| 0.0 | 0.2 | GO:0010825 | positive regulation of centrosome duplication(GO:0010825) |
| 0.0 | 0.2 | GO:1903435 | positive regulation of constitutive secretory pathway(GO:1903435) |
| 0.0 | 0.6 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0018343 | protein farnesylation(GO:0018343) |
| 0.0 | 0.1 | GO:0031064 | negative regulation of histone deacetylation(GO:0031064) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.1 | GO:0021913 | regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) |
| 0.0 | 0.2 | GO:0070294 | renal sodium ion absorption(GO:0070294) |
| 0.0 | 0.1 | GO:1902896 | terminal web assembly(GO:1902896) |
| 0.0 | 0.3 | GO:0048102 | autophagic cell death(GO:0048102) |
| 0.0 | 0.1 | GO:0032877 | positive regulation of DNA endoreduplication(GO:0032877) |
| 0.0 | 0.1 | GO:0019046 | release from viral latency(GO:0019046) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 1.4 | GO:0000413 | protein peptidyl-prolyl isomerization(GO:0000413) |
| 0.0 | 0.5 | GO:0071578 | zinc II ion transmembrane import(GO:0071578) |
| 0.0 | 0.1 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.1 | GO:0035359 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) |
| 0.0 | 0.9 | GO:0030511 | positive regulation of transforming growth factor beta receptor signaling pathway(GO:0030511) positive regulation of cellular response to transforming growth factor beta stimulus(GO:1903846) |
| 0.0 | 0.3 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0001555 | oocyte growth(GO:0001555) |
| 0.0 | 1.1 | GO:0015991 | ATP hydrolysis coupled proton transport(GO:0015991) |
| 0.0 | 0.1 | GO:0070253 | somatostatin secretion(GO:0070253) |
| 0.0 | 0.1 | GO:0014835 | myoblast differentiation involved in skeletal muscle regeneration(GO:0014835) |
| 0.0 | 0.1 | GO:0048250 | mitochondrial iron ion transport(GO:0048250) |
| 0.0 | 0.3 | GO:0006782 | protoporphyrinogen IX biosynthetic process(GO:0006782) |
| 0.0 | 0.2 | GO:0050957 | equilibrioception(GO:0050957) |
| 0.0 | 0.2 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.7 | GO:0043586 | tongue development(GO:0043586) |
| 0.0 | 0.1 | GO:0033615 | mitochondrial proton-transporting ATP synthase complex assembly(GO:0033615) |
| 0.0 | 0.1 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.3 | GO:0042118 | endothelial cell activation(GO:0042118) |
| 0.0 | 0.2 | GO:0017062 | respiratory chain complex III assembly(GO:0017062) mitochondrial respiratory chain complex III assembly(GO:0034551) mitochondrial respiratory chain complex III biogenesis(GO:0097033) |
| 0.0 | 0.4 | GO:0006020 | inositol metabolic process(GO:0006020) |
| 0.0 | 0.4 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.2 | GO:0040038 | polar body extrusion after meiotic divisions(GO:0040038) formin-nucleated actin cable assembly(GO:0070649) |
| 0.0 | 0.1 | GO:0015798 | myo-inositol transport(GO:0015798) |
| 0.0 | 0.3 | GO:0045721 | negative regulation of gluconeogenesis(GO:0045721) |
| 0.0 | 0.1 | GO:0009447 | putrescine catabolic process(GO:0009447) |
| 0.0 | 0.1 | GO:0006499 | N-terminal protein myristoylation(GO:0006499) |
| 0.0 | 3.9 | GO:0007156 | homophilic cell adhesion via plasma membrane adhesion molecules(GO:0007156) |
| 0.0 | 0.3 | GO:0015074 | DNA integration(GO:0015074) |
| 0.0 | 0.5 | GO:0016048 | detection of temperature stimulus(GO:0016048) |
| 0.0 | 0.8 | GO:0016486 | peptide hormone processing(GO:0016486) |
| 0.0 | 0.1 | GO:0070213 | protein auto-ADP-ribosylation(GO:0070213) |
| 0.0 | 0.2 | GO:1901725 | regulation of histone deacetylase activity(GO:1901725) |
| 0.0 | 0.1 | GO:0035965 | cardiolipin acyl-chain remodeling(GO:0035965) |
| 0.0 | 0.1 | GO:0051661 | maintenance of centrosome location(GO:0051661) |
| 0.0 | 0.1 | GO:1903715 | regulation of aerobic respiration(GO:1903715) |
| 0.0 | 0.1 | GO:0086092 | regulation of the force of heart contraction by cardiac conduction(GO:0086092) regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.0 | 0.1 | GO:0052027 | modulation of signal transduction in other organism(GO:0044501) modulation by symbiont of host signal transduction pathway(GO:0052027) modulation of signal transduction in other organism involved in symbiotic interaction(GO:0052250) modulation by symbiont of host I-kappaB kinase/NF-kappaB cascade(GO:0085032) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.8 | GO:0007274 | neuromuscular synaptic transmission(GO:0007274) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0010735 | positive regulation of transcription via serum response element binding(GO:0010735) |
| 0.0 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.0 | 0.1 | GO:0002725 | negative regulation of T cell cytokine production(GO:0002725) |
| 0.0 | 0.1 | GO:0061470 | T follicular helper cell differentiation(GO:0061470) |
| 0.0 | 0.2 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0051970 | negative regulation of transmission of nerve impulse(GO:0051970) |
| 0.0 | 0.2 | GO:0070914 | UV-damage excision repair(GO:0070914) |
| 0.0 | 0.1 | GO:0045356 | positive regulation of interferon-alpha biosynthetic process(GO:0045356) |
| 0.0 | 0.3 | GO:0034393 | positive regulation of smooth muscle cell apoptotic process(GO:0034393) |
| 0.0 | 0.3 | GO:0051291 | protein heterooligomerization(GO:0051291) |
| 0.0 | 0.2 | GO:0042905 | 9-cis-retinoic acid biosynthetic process(GO:0042904) 9-cis-retinoic acid metabolic process(GO:0042905) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.1 | GO:1904073 | trophectodermal cell proliferation(GO:0001834) regulation of trophectodermal cell proliferation(GO:1904073) positive regulation of trophectodermal cell proliferation(GO:1904075) |
| 0.0 | 0.2 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.3 | GO:0007253 | cytoplasmic sequestering of NF-kappaB(GO:0007253) |
| 0.0 | 0.0 | GO:0035349 | coenzyme A transport(GO:0015880) coenzyme A transmembrane transport(GO:0035349) adenosine 3',5'-bisphosphate transmembrane transport(GO:0071106) AMP transport(GO:0080121) |
| 0.0 | 0.1 | GO:0035973 | aggrephagy(GO:0035973) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0072734 | response to staurosporine(GO:0072733) cellular response to staurosporine(GO:0072734) |
| 0.0 | 0.1 | GO:0000707 | meiotic DNA recombinase assembly(GO:0000707) |
| 0.0 | 0.1 | GO:0010847 | regulation of chromatin assembly(GO:0010847) |
| 0.0 | 0.4 | GO:0006646 | phosphatidylethanolamine biosynthetic process(GO:0006646) |
| 0.0 | 0.1 | GO:0070836 | caveola assembly(GO:0070836) |
| 0.0 | 0.6 | GO:0051894 | positive regulation of focal adhesion assembly(GO:0051894) |
| 0.0 | 0.0 | GO:0070781 | arginine biosynthetic process via ornithine(GO:0042450) response to biotin(GO:0070781) |
| 0.0 | 0.5 | GO:0009083 | branched-chain amino acid catabolic process(GO:0009083) |
| 0.0 | 0.2 | GO:0034058 | endosomal vesicle fusion(GO:0034058) |
| 0.0 | 0.3 | GO:0043508 | negative regulation of JUN kinase activity(GO:0043508) |
| 0.0 | 0.1 | GO:0016554 | cytidine to uridine editing(GO:0016554) |
| 0.0 | 0.3 | GO:0035428 | hexose transmembrane transport(GO:0035428) glucose transmembrane transport(GO:1904659) |
| 0.0 | 0.1 | GO:0033211 | adiponectin-activated signaling pathway(GO:0033211) |
| 0.0 | 0.2 | GO:0034498 | early endosome to Golgi transport(GO:0034498) |
| 0.0 | 0.1 | GO:0050860 | negative regulation of T cell receptor signaling pathway(GO:0050860) |
| 0.0 | 0.1 | GO:0034201 | response to oleic acid(GO:0034201) |
| 0.0 | 0.3 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.0 | GO:0016128 | phytosteroid metabolic process(GO:0016128) phytosteroid biosynthetic process(GO:0016129) |
| 0.0 | 0.1 | GO:0018094 | protein polyglycylation(GO:0018094) |
| 0.0 | 0.0 | GO:0060266 | negative regulation of respiratory burst involved in inflammatory response(GO:0060266) |
| 0.0 | 0.0 | GO:0016260 | selenocysteine biosynthetic process(GO:0016260) |
| 0.0 | 0.1 | GO:0045792 | negative regulation of cell size(GO:0045792) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:0070862 | negative regulation of protein exit from endoplasmic reticulum(GO:0070862) negative regulation of retrograde protein transport, ER to cytosol(GO:1904153) |
| 0.0 | 0.0 | GO:0032536 | regulation of cell projection size(GO:0032536) |
| 0.0 | 0.2 | GO:2000353 | positive regulation of endothelial cell apoptotic process(GO:2000353) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.1 | GO:1900121 | negative regulation of receptor binding(GO:1900121) |
| 0.0 | 0.5 | GO:0006607 | NLS-bearing protein import into nucleus(GO:0006607) |
| 0.0 | 0.3 | GO:0043517 | positive regulation of DNA damage response, signal transduction by p53 class mediator(GO:0043517) |
| 0.0 | 0.1 | GO:0006177 | GMP biosynthetic process(GO:0006177) |
| 0.0 | 0.5 | GO:0046326 | positive regulation of glucose import(GO:0046326) |
| 0.0 | 0.1 | GO:0015015 | heparan sulfate proteoglycan biosynthetic process, enzymatic modification(GO:0015015) |
| 0.0 | 0.2 | GO:0051775 | response to redox state(GO:0051775) |
| 0.0 | 0.2 | GO:0060004 | reflex(GO:0060004) |
| 0.0 | 0.1 | GO:0045654 | positive regulation of megakaryocyte differentiation(GO:0045654) |
| 0.0 | 0.4 | GO:0033137 | negative regulation of peptidyl-serine phosphorylation(GO:0033137) |
| 0.0 | 0.3 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0043569 | negative regulation of insulin-like growth factor receptor signaling pathway(GO:0043569) |
| 0.0 | 0.2 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:0035549 | positive regulation of interferon-beta secretion(GO:0035549) |
| 0.0 | 0.1 | GO:0007158 | neuron cell-cell adhesion(GO:0007158) |
| 0.0 | 0.2 | GO:2000480 | negative regulation of cAMP-dependent protein kinase activity(GO:2000480) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.1 | GO:0007525 | somatic muscle development(GO:0007525) |
| 0.0 | 0.2 | GO:0045116 | protein neddylation(GO:0045116) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 3.0 | GO:0071953 | elastic fiber(GO:0071953) |
| 0.2 | 4.3 | GO:0001527 | microfibril(GO:0001527) fibril(GO:0043205) |
| 0.2 | 0.7 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.1 | 0.6 | GO:0097442 | CA3 pyramidal cell dendrite(GO:0097442) |
| 0.1 | 0.2 | GO:0097208 | alveolar lamellar body(GO:0097208) lamellar body membrane(GO:0097232) alveolar lamellar body membrane(GO:0097233) |
| 0.1 | 0.4 | GO:1990578 | perinuclear endoplasmic reticulum membrane(GO:1990578) |
| 0.1 | 1.1 | GO:0097443 | sorting endosome(GO:0097443) |
| 0.1 | 0.4 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.1 | 0.3 | GO:0034657 | GID complex(GO:0034657) |
| 0.1 | 0.4 | GO:0009328 | phenylalanine-tRNA ligase complex(GO:0009328) |
| 0.1 | 1.1 | GO:0033179 | proton-transporting V-type ATPase, V0 domain(GO:0033179) |
| 0.1 | 0.5 | GO:0000110 | nucleotide-excision repair factor 1 complex(GO:0000110) |
| 0.1 | 0.4 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
| 0.1 | 0.3 | GO:0060201 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.4 | GO:0042622 | photoreceptor outer segment membrane(GO:0042622) |
| 0.1 | 0.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.2 | GO:0030934 | anchoring collagen complex(GO:0030934) |
| 0.0 | 0.2 | GO:0005971 | ribonucleoside-diphosphate reductase complex(GO:0005971) |
| 0.0 | 0.0 | GO:0044609 | DBIRD complex(GO:0044609) |
| 0.0 | 0.3 | GO:0070545 | PeBoW complex(GO:0070545) |
| 0.0 | 0.3 | GO:0001740 | Barr body(GO:0001740) |
| 0.0 | 0.2 | GO:1990761 | growth cone lamellipodium(GO:1990761) growth cone filopodium(GO:1990812) |
| 0.0 | 0.5 | GO:1990909 | Wnt signalosome(GO:1990909) |
| 0.0 | 1.4 | GO:0005868 | cytoplasmic dynein complex(GO:0005868) |
| 0.0 | 0.2 | GO:0032144 | 4-aminobutyrate transaminase complex(GO:0032144) |
| 0.0 | 0.2 | GO:0098837 | postsynaptic recycling endosome(GO:0098837) |
| 0.0 | 0.2 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 0.3 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.9 | GO:0005614 | interstitial matrix(GO:0005614) |
| 0.0 | 0.1 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.0 | 5.2 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 1.9 | GO:0048786 | presynaptic active zone(GO:0048786) |
| 0.0 | 0.1 | GO:0035189 | Rb-E2F complex(GO:0035189) |
| 0.0 | 0.2 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.9 | GO:0071565 | nBAF complex(GO:0071565) |
| 0.0 | 0.7 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.5 | GO:0099634 | postsynaptic specialization membrane(GO:0099634) |
| 0.0 | 3.9 | GO:0005901 | caveola(GO:0005901) |
| 0.0 | 0.2 | GO:0030485 | smooth muscle contractile fiber(GO:0030485) |
| 0.0 | 0.4 | GO:0042583 | chromaffin granule(GO:0042583) |
| 0.0 | 0.1 | GO:0005965 | protein farnesyltransferase complex(GO:0005965) |
| 0.0 | 0.1 | GO:0005594 | collagen type IX trimer(GO:0005594) |
| 0.0 | 0.2 | GO:0030905 | retromer, tubulation complex(GO:0030905) |
| 0.0 | 0.1 | GO:0005896 | interleukin-6 receptor complex(GO:0005896) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.3 | GO:0034098 | VCP-NPL4-UFD1 AAA ATPase complex(GO:0034098) |
| 0.0 | 0.3 | GO:0070775 | H3 histone acetyltransferase complex(GO:0070775) MOZ/MORF histone acetyltransferase complex(GO:0070776) |
| 0.0 | 0.5 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.3 | GO:0090544 | BAF-type complex(GO:0090544) |
| 0.0 | 0.1 | GO:0020018 | ciliary pocket(GO:0020016) ciliary pocket membrane(GO:0020018) |
| 0.0 | 0.3 | GO:0035749 | myelin sheath adaxonal region(GO:0035749) lateral loop(GO:0043219) |
| 0.0 | 0.6 | GO:0043218 | compact myelin(GO:0043218) |
| 0.0 | 0.1 | GO:0002193 | MAML1-RBP-Jkappa- ICN1 complex(GO:0002193) |
| 0.0 | 0.1 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.0 | 0.2 | GO:0042382 | paraspeckles(GO:0042382) |
| 0.0 | 0.2 | GO:0044327 | dendritic spine head(GO:0044327) |
| 0.0 | 1.8 | GO:0044291 | cell-cell contact zone(GO:0044291) |
| 0.0 | 0.5 | GO:0005942 | phosphatidylinositol 3-kinase complex(GO:0005942) |
| 0.0 | 0.1 | GO:0072357 | PTW/PP1 phosphatase complex(GO:0072357) |
| 0.0 | 0.3 | GO:0043083 | synaptic cleft(GO:0043083) |
| 0.0 | 0.2 | GO:0071203 | WASH complex(GO:0071203) |
| 0.0 | 0.1 | GO:1990745 | EARP complex(GO:1990745) |
| 0.0 | 0.1 | GO:0045272 | plasma membrane respiratory chain complex I(GO:0045272) |
| 0.0 | 0.4 | GO:0097431 | mitotic spindle pole(GO:0097431) |
| 0.0 | 0.2 | GO:0031314 | extrinsic component of mitochondrial inner membrane(GO:0031314) |
| 0.0 | 0.1 | GO:0032777 | Piccolo NuA4 histone acetyltransferase complex(GO:0032777) |
| 0.0 | 0.8 | GO:0032809 | neuronal cell body membrane(GO:0032809) cell body membrane(GO:0044298) |
| 0.0 | 0.1 | GO:0033063 | Rad51B-Rad51C-Rad51D-XRCC2 complex(GO:0033063) |
| 0.0 | 0.3 | GO:0090543 | Flemming body(GO:0090543) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:0045120 | pronucleus(GO:0045120) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
| 0.0 | 0.2 | GO:0032584 | growth cone membrane(GO:0032584) |
| 0.0 | 0.0 | GO:0097124 | cyclin A2-CDK2 complex(GO:0097124) |
| 0.0 | 0.5 | GO:0031011 | Ino80 complex(GO:0031011) DNA helicase complex(GO:0033202) |
| 0.0 | 0.3 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.2 | GO:0001931 | uropod(GO:0001931) cell trailing edge(GO:0031254) |
| 0.0 | 0.4 | GO:0000145 | exocyst(GO:0000145) |
| 0.0 | 0.3 | GO:0030127 | COPII vesicle coat(GO:0030127) |
| 0.0 | 0.1 | GO:0097425 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.1 | GO:0005885 | Arp2/3 protein complex(GO:0005885) |
| 0.0 | 0.3 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.5 | GO:0031304 | intrinsic component of mitochondrial inner membrane(GO:0031304) integral component of mitochondrial inner membrane(GO:0031305) |
| 0.0 | 0.0 | GO:0005955 | calcineurin complex(GO:0005955) |
| 0.0 | 0.3 | GO:0005892 | acetylcholine-gated channel complex(GO:0005892) |
| 0.0 | 0.2 | GO:0033093 | Weibel-Palade body(GO:0033093) |
| 0.0 | 0.1 | GO:0005726 | perichromatin fibrils(GO:0005726) |
| 0.0 | 0.3 | GO:0032982 | myosin filament(GO:0032982) |
| 0.0 | 0.1 | GO:0042587 | glycogen granule(GO:0042587) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0031616 | spindle pole centrosome(GO:0031616) |
| 0.0 | 0.4 | GO:0031307 | integral component of mitochondrial outer membrane(GO:0031307) |
| 0.0 | 1.1 | GO:0097610 | cleavage furrow(GO:0032154) cell surface furrow(GO:0097610) |
| 0.0 | 0.1 | GO:0035867 | alphav-beta3 integrin-IGF-1-IGF1R complex(GO:0035867) |
| 0.0 | 0.1 | GO:0030897 | HOPS complex(GO:0030897) |
| 0.0 | 0.1 | GO:0031931 | TORC1 complex(GO:0031931) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
| 0.0 | 0.2 | GO:0005859 | muscle myosin complex(GO:0005859) |
| 0.0 | 0.1 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 1.2 | GO:0005902 | microvillus(GO:0005902) |
| 0.0 | 0.2 | GO:0031143 | pseudopodium(GO:0031143) |
| 0.0 | 1.0 | GO:0005811 | lipid particle(GO:0005811) |
| 0.0 | 0.1 | GO:0005602 | complement component C1 complex(GO:0005602) |
| 0.0 | 0.1 | GO:0000109 | nucleotide-excision repair complex(GO:0000109) |
| 0.0 | 0.1 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0005751 | mitochondrial respiratory chain complex IV(GO:0005751) |
| 0.0 | 0.8 | GO:0010494 | cytoplasmic stress granule(GO:0010494) |
| 0.0 | 1.2 | GO:0005924 | cell-substrate adherens junction(GO:0005924) focal adhesion(GO:0005925) cell-substrate junction(GO:0030055) |
| 0.0 | 0.4 | GO:0030286 | dynein complex(GO:0030286) |
| 0.0 | 0.6 | GO:0045171 | intercellular bridge(GO:0045171) |
| 0.0 | 0.4 | GO:0005720 | nuclear heterochromatin(GO:0005720) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.8 | 3.3 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.8 | 4.0 | GO:0050436 | microfibril binding(GO:0050436) |
| 0.6 | 1.9 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.4 | 1.7 | GO:0016404 | 15-hydroxyprostaglandin dehydrogenase (NAD+) activity(GO:0016404) |
| 0.3 | 1.0 | GO:0004676 | 3-phosphoinositide-dependent protein kinase activity(GO:0004676) |
| 0.3 | 2.2 | GO:0044323 | retinoic acid-responsive element binding(GO:0044323) |
| 0.3 | 0.8 | GO:0001596 | angiotensin type I receptor activity(GO:0001596) |
| 0.2 | 0.9 | GO:0001069 | regulatory region RNA binding(GO:0001069) |
| 0.2 | 1.3 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.2 | 0.6 | GO:0005169 | neurotrophin TRKB receptor binding(GO:0005169) |
| 0.2 | 1.2 | GO:0008597 | calcium-dependent protein serine/threonine phosphatase regulator activity(GO:0008597) |
| 0.2 | 0.8 | GO:0003987 | acetate-CoA ligase activity(GO:0003987) |
| 0.2 | 0.6 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.2 | 0.8 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.2 | 1.5 | GO:0038085 | vascular endothelial growth factor binding(GO:0038085) |
| 0.1 | 0.9 | GO:0033743 | peptide-methionine (R)-S-oxide reductase activity(GO:0033743) |
| 0.1 | 0.4 | GO:0016652 | NAD(P)+ transhydrogenase activity(GO:0008746) oxidoreductase activity, acting on NAD(P)H, NAD(P) as acceptor(GO:0016652) |
| 0.1 | 1.2 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.1 | 0.4 | GO:0004345 | glucose-6-phosphate dehydrogenase activity(GO:0004345) |
| 0.1 | 0.1 | GO:0046934 | phosphatidylinositol-4,5-bisphosphate 3-kinase activity(GO:0046934) phosphatidylinositol bisphosphate kinase activity(GO:0052813) |
| 0.1 | 0.8 | GO:0050692 | DBD domain binding(GO:0050692) |
| 0.1 | 0.3 | GO:0004853 | uroporphyrinogen decarboxylase activity(GO:0004853) |
| 0.1 | 0.3 | GO:0061629 | RNA polymerase II sequence-specific DNA binding transcription factor binding(GO:0061629) |
| 0.1 | 0.8 | GO:0032554 | purine deoxyribonucleotide binding(GO:0032554) |
| 0.1 | 0.5 | GO:0008454 | alpha-1,3-mannosylglycoprotein 4-beta-N-acetylglucosaminyltransferase activity(GO:0008454) |
| 0.1 | 0.4 | GO:0097604 | temperature-gated cation channel activity(GO:0097604) |
| 0.1 | 0.6 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.1 | 0.3 | GO:0036487 | nitric-oxide synthase inhibitor activity(GO:0036487) |
| 0.1 | 0.3 | GO:0031731 | CCR6 chemokine receptor binding(GO:0031731) |
| 0.1 | 0.5 | GO:0004882 | androgen receptor activity(GO:0004882) |
| 0.1 | 0.3 | GO:0004743 | pyruvate kinase activity(GO:0004743) |
| 0.1 | 1.7 | GO:0043560 | insulin receptor substrate binding(GO:0043560) |
| 0.1 | 0.2 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 0.4 | GO:0004306 | ethanolamine-phosphate cytidylyltransferase activity(GO:0004306) |
| 0.1 | 0.1 | GO:0008142 | oxysterol binding(GO:0008142) |
| 0.1 | 0.4 | GO:0033857 | diphosphoinositol-pentakisphosphate kinase activity(GO:0033857) |
| 0.1 | 0.3 | GO:0016230 | sphingomyelin phosphodiesterase activator activity(GO:0016230) |
| 0.1 | 0.4 | GO:0004800 | thyroxine 5'-deiodinase activity(GO:0004800) |
| 0.1 | 0.6 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.1 | 0.3 | GO:0003990 | acetylcholinesterase activity(GO:0003990) |
| 0.1 | 0.3 | GO:0050309 | glucose-6-phosphatase activity(GO:0004346) sugar-terminal-phosphatase activity(GO:0050309) |
| 0.1 | 0.9 | GO:0017162 | aryl hydrocarbon receptor binding(GO:0017162) |
| 0.1 | 0.2 | GO:0005174 | CD40 receptor binding(GO:0005174) |
| 0.1 | 0.1 | GO:0004660 | protein farnesyltransferase activity(GO:0004660) |
| 0.1 | 0.5 | GO:0046935 | 1-phosphatidylinositol-3-kinase regulator activity(GO:0046935) |
| 0.1 | 0.3 | GO:0004663 | Rab geranylgeranyltransferase activity(GO:0004663) |
| 0.1 | 0.5 | GO:0043426 | MRF binding(GO:0043426) |
| 0.1 | 1.3 | GO:0004861 | cyclin-dependent protein serine/threonine kinase inhibitor activity(GO:0004861) |
| 0.1 | 0.4 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.1 | 0.2 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.1 | 1.2 | GO:0045504 | dynein heavy chain binding(GO:0045504) |
| 0.1 | 1.8 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.1 | 1.9 | GO:0050750 | low-density lipoprotein particle receptor binding(GO:0050750) |
| 0.1 | 1.0 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.1 | 0.2 | GO:0016649 | electron-transferring-flavoprotein dehydrogenase activity(GO:0004174) oxidoreductase activity, acting on the CH-NH group of donors, quinone or similar compound as acceptor(GO:0016649) |
| 0.1 | 0.2 | GO:0004450 | isocitrate dehydrogenase (NADP+) activity(GO:0004450) |
| 0.1 | 0.5 | GO:0030346 | protein phosphatase 2B binding(GO:0030346) |
| 0.1 | 0.2 | GO:0008798 | beta-aspartyl-peptidase activity(GO:0008798) |
| 0.1 | 1.6 | GO:0005528 | macrolide binding(GO:0005527) FK506 binding(GO:0005528) |
| 0.0 | 0.2 | GO:0016728 | ribonucleoside-diphosphate reductase activity, thioredoxin disulfide as acceptor(GO:0004748) oxidoreductase activity, acting on CH or CH2 groups, disulfide as acceptor(GO:0016728) ribonucleoside-diphosphate reductase activity(GO:0061731) |
| 0.0 | 0.4 | GO:0005412 | glucose:sodium symporter activity(GO:0005412) |
| 0.0 | 0.3 | GO:0030197 | extracellular matrix constituent, lubricant activity(GO:0030197) |
| 0.0 | 0.3 | GO:0052811 | 1-phosphatidylinositol-3-phosphate 4-kinase activity(GO:0052811) |
| 0.0 | 0.1 | GO:0030366 | molybdopterin synthase activity(GO:0030366) |
| 0.0 | 0.4 | GO:0030023 | extracellular matrix constituent conferring elasticity(GO:0030023) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.3 | GO:0004396 | glucokinase activity(GO:0004340) hexokinase activity(GO:0004396) fructokinase activity(GO:0008865) mannokinase activity(GO:0019158) |
| 0.0 | 0.2 | GO:1990460 | leptin receptor binding(GO:1990460) |
| 0.0 | 0.3 | GO:1990599 | 3' overhang single-stranded DNA endodeoxyribonuclease activity(GO:1990599) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.5 | GO:1990380 | Lys48-specific deubiquitinase activity(GO:1990380) |
| 0.0 | 2.4 | GO:0042169 | SH2 domain binding(GO:0042169) |
| 0.0 | 0.9 | GO:0033130 | acetylcholine receptor binding(GO:0033130) |
| 0.0 | 0.2 | GO:0051021 | GDP-dissociation inhibitor binding(GO:0051021) Rho GDP-dissociation inhibitor binding(GO:0051022) |
| 0.0 | 0.1 | GO:0045518 | interleukin-22 receptor binding(GO:0045518) |
| 0.0 | 0.2 | GO:0003867 | 4-aminobutyrate transaminase activity(GO:0003867) succinate-semialdehyde dehydrogenase binding(GO:0032145) (S)-3-amino-2-methylpropionate transaminase activity(GO:0047298) |
| 0.0 | 0.3 | GO:0050833 | pyruvate transmembrane transporter activity(GO:0050833) |
| 0.0 | 0.1 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.1 | GO:0003985 | acetyl-CoA C-acetyltransferase activity(GO:0003985) |
| 0.0 | 0.1 | GO:0019981 | interleukin-6 receptor activity(GO:0004915) interleukin-6 binding(GO:0019981) |
| 0.0 | 1.1 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.1 | GO:0000253 | 3-keto sterol reductase activity(GO:0000253) |
| 0.0 | 0.2 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.2 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.2 | GO:0042030 | ATPase inhibitor activity(GO:0042030) |
| 0.0 | 0.2 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.2 | GO:0061676 | importin-alpha family protein binding(GO:0061676) |
| 0.0 | 0.4 | GO:0008432 | JUN kinase binding(GO:0008432) |
| 0.0 | 0.6 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.8 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.3 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.3 | GO:0046976 | histone methyltransferase activity (H3-K27 specific)(GO:0046976) |
| 0.0 | 0.3 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.3 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.7 | GO:0071949 | FAD binding(GO:0071949) |
| 0.0 | 0.5 | GO:0003995 | acyl-CoA dehydrogenase activity(GO:0003995) |
| 0.0 | 0.1 | GO:0035473 | lipase binding(GO:0035473) |
| 0.0 | 0.2 | GO:0008568 | microtubule-severing ATPase activity(GO:0008568) |
| 0.0 | 0.1 | GO:0048273 | mitogen-activated protein kinase p38 binding(GO:0048273) |
| 0.0 | 0.2 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.0 | GO:0003956 | NAD(P)+-protein-arginine ADP-ribosyltransferase activity(GO:0003956) |
| 0.0 | 0.2 | GO:0001190 | transcriptional activator activity, RNA polymerase II transcription factor binding(GO:0001190) transcriptional repressor activity, RNA polymerase II activating transcription factor binding(GO:0098811) |
| 0.0 | 0.1 | GO:0033858 | N-acetylgalactosamine kinase activity(GO:0033858) |
| 0.0 | 0.1 | GO:0035727 | lysophosphatidic acid binding(GO:0035727) |
| 0.0 | 0.6 | GO:0015035 | protein disulfide oxidoreductase activity(GO:0015035) |
| 0.0 | 0.8 | GO:0004181 | metallocarboxypeptidase activity(GO:0004181) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.2 | GO:0047035 | testosterone dehydrogenase (NAD+) activity(GO:0047035) |
| 0.0 | 0.8 | GO:0008327 | methyl-CpG binding(GO:0008327) |
| 0.0 | 0.4 | GO:1900750 | glutathione binding(GO:0043295) oligopeptide binding(GO:1900750) |
| 0.0 | 0.2 | GO:0008081 | phosphoric diester hydrolase activity(GO:0008081) |
| 0.0 | 0.1 | GO:0015207 | ATP:ADP antiporter activity(GO:0005471) adenine transmembrane transporter activity(GO:0015207) |
| 0.0 | 0.1 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.0 | 0.3 | GO:0000150 | recombinase activity(GO:0000150) |
| 0.0 | 0.1 | GO:0044378 | non-sequence-specific DNA binding, bending(GO:0044378) |
| 0.0 | 0.3 | GO:0031681 | G-protein beta-subunit binding(GO:0031681) |
| 0.0 | 0.2 | GO:0035877 | death effector domain binding(GO:0035877) |
| 0.0 | 0.8 | GO:0003950 | NAD+ ADP-ribosyltransferase activity(GO:0003950) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.6 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
| 0.0 | 1.3 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.5 | GO:0070410 | co-SMAD binding(GO:0070410) |
| 0.0 | 0.3 | GO:0086008 | voltage-gated potassium channel activity involved in cardiac muscle cell action potential repolarization(GO:0086008) |
| 0.0 | 0.1 | GO:0030020 | extracellular matrix structural constituent conferring tensile strength(GO:0030020) |
| 0.0 | 2.0 | GO:0030165 | PDZ domain binding(GO:0030165) |
| 0.0 | 0.0 | GO:0015228 | coenzyme A transmembrane transporter activity(GO:0015228) adenosine 3',5'-bisphosphate transmembrane transporter activity(GO:0071077) AMP transmembrane transporter activity(GO:0080122) |
| 0.0 | 0.2 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0004471 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 2.2 | GO:0005179 | hormone activity(GO:0005179) |
| 0.0 | 0.1 | GO:0070740 | tubulin-glutamic acid ligase activity(GO:0070740) |
| 0.0 | 0.2 | GO:0003777 | microtubule motor activity(GO:0003777) |
| 0.0 | 0.1 | GO:0004126 | cytidine deaminase activity(GO:0004126) |
| 0.0 | 0.1 | GO:0045125 | bioactive lipid receptor activity(GO:0045125) |
| 0.0 | 0.3 | GO:0005344 | oxygen transporter activity(GO:0005344) |
| 0.0 | 0.1 | GO:0003875 | ADP-ribosylarginine hydrolase activity(GO:0003875) |
| 0.0 | 0.1 | GO:0070735 | protein-glycine ligase activity(GO:0070735) |
| 0.0 | 0.0 | GO:0019976 | interleukin-2 receptor activity(GO:0004911) interleukin-2 binding(GO:0019976) |
| 0.0 | 0.0 | GO:0016781 | selenide, water dikinase activity(GO:0004756) phosphotransferase activity, paired acceptors(GO:0016781) |
| 0.0 | 0.1 | GO:0001609 | G-protein coupled adenosine receptor activity(GO:0001609) |
| 0.0 | 0.1 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.3 | GO:0002162 | dystroglycan binding(GO:0002162) |
| 0.0 | 0.2 | GO:0102391 | decanoate--CoA ligase activity(GO:0102391) |
| 0.0 | 0.3 | GO:0051010 | microtubule plus-end binding(GO:0051010) |
| 0.0 | 0.2 | GO:0048156 | tau protein binding(GO:0048156) |
| 0.0 | 0.3 | GO:0048531 | beta-1,3-galactosyltransferase activity(GO:0048531) |
| 0.0 | 0.1 | GO:0030169 | low-density lipoprotein particle binding(GO:0030169) |
| 0.0 | 0.4 | GO:0005092 | GDP-dissociation inhibitor activity(GO:0005092) |
| 0.0 | 0.5 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0000400 | four-way junction DNA binding(GO:0000400) |
| 0.0 | 0.3 | GO:0017160 | Ral GTPase binding(GO:0017160) |
| 0.0 | 1.4 | GO:0017137 | Rab GTPase binding(GO:0017137) |
| 0.0 | 0.4 | GO:0070003 | threonine-type endopeptidase activity(GO:0004298) threonine-type peptidase activity(GO:0070003) |
| 0.0 | 0.1 | GO:0046975 | histone methyltransferase activity (H3-K36 specific)(GO:0046975) |
| 0.0 | 0.5 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.0 | GO:0004980 | melanocyte-stimulating hormone receptor activity(GO:0004980) |
| 0.0 | 0.2 | GO:0004697 | protein kinase C activity(GO:0004697) |
| 0.0 | 0.5 | GO:1990939 | ATP-dependent microtubule motor activity(GO:1990939) |
| 0.0 | 0.0 | GO:0015173 | aromatic amino acid transmembrane transporter activity(GO:0015173) |
| 0.0 | 0.3 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.0 | GO:0001632 | leukotriene B4 receptor activity(GO:0001632) |
| 0.0 | 0.1 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.1 | GO:0008172 | S-methyltransferase activity(GO:0008172) |
| 0.0 | 0.1 | GO:0008429 | phosphatidylethanolamine binding(GO:0008429) |
| 0.0 | 0.1 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | ST IL 13 PATHWAY | Interleukin 13 (IL-13) Pathway |
| 0.1 | 1.2 | PID VEGF VEGFR PATHWAY | VEGF and VEGFR signaling network |
| 0.1 | 1.9 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
| 0.0 | 1.1 | PID CDC42 REG PATHWAY | Regulation of CDC42 activity |
| 0.0 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 2.1 | SIG IL4RECEPTOR IN B LYPHOCYTES | Genes related to IL4 rceptor signaling in B lymphocytes |
| 0.0 | 0.3 | PID ERBB4 PATHWAY | ErbB4 signaling events |
| 0.0 | 2.0 | PID ECADHERIN NASCENT AJ PATHWAY | E-cadherin signaling in the nascent adherens junction |
| 0.0 | 7.8 | NABA ECM GLYCOPROTEINS | Genes encoding structural ECM glycoproteins |
| 0.0 | 2.0 | PID SHP2 PATHWAY | SHP2 signaling |
| 0.0 | 2.4 | PID RHOA REG PATHWAY | Regulation of RhoA activity |
| 0.0 | 1.1 | PID RAC1 REG PATHWAY | Regulation of RAC1 activity |
| 0.0 | 1.0 | PID INTEGRIN2 PATHWAY | Beta2 integrin cell surface interactions |
| 0.0 | 1.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.9 | PID HDAC CLASSIII PATHWAY | Signaling events mediated by HDAC Class III |
| 0.0 | 1.3 | PID ERBB1 INTERNALIZATION PATHWAY | Internalization of ErbB1 |
| 0.0 | 1.3 | PID ARF6 TRAFFICKING PATHWAY | Arf6 trafficking events |
| 0.0 | 0.6 | PID PRL SIGNALING EVENTS PATHWAY | Signaling events mediated by PRL |
| 0.0 | 0.8 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 2.5 | PID MYC REPRESS PATHWAY | Validated targets of C-MYC transcriptional repression |
| 0.0 | 0.1 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 0.7 | PID RET PATHWAY | Signaling events regulated by Ret tyrosine kinase |
| 0.0 | 1.3 | PID NFAT 3PATHWAY | Role of Calcineurin-dependent NFAT signaling in lymphocytes |
| 0.0 | 0.1 | ST TYPE I INTERFERON PATHWAY | Type I Interferon (alpha/beta IFN) Pathway |
| 0.0 | 0.2 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 1.1 | PID AR TF PATHWAY | Regulation of Androgen receptor activity |
| 0.0 | 0.1 | PID VEGFR1 PATHWAY | VEGFR1 specific signals |
| 0.0 | 0.3 | PID SYNDECAN 3 PATHWAY | Syndecan-3-mediated signaling events |
| 0.0 | 0.3 | SIG CD40PATHWAYMAP | Genes related to CD40 signaling |
| 0.0 | 0.4 | PID TRKR PATHWAY | Neurotrophic factor-mediated Trk receptor signaling |
| 0.0 | 2.5 | NABA ECM AFFILIATED | Genes encoding proteins affiliated structurally or functionally to extracellular matrix proteins |
| 0.0 | 0.9 | PID ERA GENOMIC PATHWAY | Validated nuclear estrogen receptor alpha network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 1.9 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.1 | 1.8 | REACTOME NOREPINEPHRINE NEUROTRANSMITTER RELEASE CYCLE | Genes involved in Norepinephrine Neurotransmitter Release Cycle |
| 0.1 | 3.7 | REACTOME A TETRASACCHARIDE LINKER SEQUENCE IS REQUIRED FOR GAG SYNTHESIS | Genes involved in A tetrasaccharide linker sequence is required for GAG synthesis |
| 0.1 | 1.4 | REACTOME SEMA3A PLEXIN REPULSION SIGNALING BY INHIBITING INTEGRIN ADHESION | Genes involved in SEMA3A-Plexin repulsion signaling by inhibiting Integrin adhesion |
| 0.1 | 0.9 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.1 | 0.7 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.1 | 0.8 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 1.9 | REACTOME CD28 DEPENDENT PI3K AKT SIGNALING | Genes involved in CD28 dependent PI3K/Akt signaling |
| 0.0 | 0.9 | REACTOME IL 6 SIGNALING | Genes involved in Interleukin-6 signaling |
| 0.0 | 1.0 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |
| 0.0 | 1.1 | REACTOME CIRCADIAN REPRESSION OF EXPRESSION BY REV ERBA | Genes involved in Circadian Repression of Expression by REV-ERBA |
| 0.0 | 1.2 | REACTOME SYNTHESIS OF PIPS AT THE PLASMA MEMBRANE | Genes involved in Synthesis of PIPs at the plasma membrane |
| 0.0 | 0.4 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 0.5 | REACTOME VITAMIN B5 PANTOTHENATE METABOLISM | Genes involved in Vitamin B5 (pantothenate) metabolism |
| 0.0 | 0.4 | REACTOME ROLE OF DCC IN REGULATING APOPTOSIS | Genes involved in Role of DCC in regulating apoptosis |
| 0.0 | 0.1 | REACTOME TRAF3 DEPENDENT IRF ACTIVATION PATHWAY | Genes involved in TRAF3-dependent IRF activation pathway |
| 0.0 | 0.8 | REACTOME INSULIN SYNTHESIS AND PROCESSING | Genes involved in Insulin Synthesis and Processing |
| 0.0 | 0.7 | REACTOME CITRIC ACID CYCLE TCA CYCLE | Genes involved in Citric acid cycle (TCA cycle) |
| 0.0 | 0.3 | REACTOME REGULATION OF INSULIN SECRETION BY ACETYLCHOLINE | Genes involved in Regulation of Insulin Secretion by Acetylcholine |
| 0.0 | 0.6 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME SIGNAL ATTENUATION | Genes involved in Signal attenuation |
| 0.0 | 0.5 | REACTOME N GLYCAN ANTENNAE ELONGATION | Genes involved in N-Glycan antennae elongation |
| 0.0 | 0.5 | REACTOME PI3K AKT ACTIVATION | Genes involved in PI3K/AKT activation |
| 0.0 | 2.7 | REACTOME PPARA ACTIVATES GENE EXPRESSION | Genes involved in PPARA Activates Gene Expression |
| 0.0 | 0.5 | REACTOME TERMINATION OF O GLYCAN BIOSYNTHESIS | Genes involved in Termination of O-glycan biosynthesis |
| 0.0 | 0.6 | REACTOME CELL EXTRACELLULAR MATRIX INTERACTIONS | Genes involved in Cell-extracellular matrix interactions |
| 0.0 | 0.5 | REACTOME CHOLESTEROL BIOSYNTHESIS | Genes involved in Cholesterol biosynthesis |
| 0.0 | 1.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.3 | REACTOME CTNNB1 PHOSPHORYLATION CASCADE | Genes involved in Beta-catenin phosphorylation cascade |
| 0.0 | 0.4 | REACTOME FORMATION OF INCISION COMPLEX IN GG NER | Genes involved in Formation of incision complex in GG-NER |
| 0.0 | 0.5 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |
| 0.0 | 0.3 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.5 | REACTOME GABA A RECEPTOR ACTIVATION | Genes involved in GABA A receptor activation |
| 0.0 | 0.5 | REACTOME HORMONE SENSITIVE LIPASE HSL MEDIATED TRIACYLGLYCEROL HYDROLYSIS | Genes involved in Hormone-sensitive lipase (HSL)-mediated triacylglycerol hydrolysis |
| 0.0 | 0.4 | REACTOME SYNTHESIS OF PE | Genes involved in Synthesis of PE |
| 0.0 | 0.2 | REACTOME REPAIR SYNTHESIS FOR GAP FILLING BY DNA POL IN TC NER | Genes involved in Repair synthesis for gap-filling by DNA polymerase in TC-NER |
| 0.0 | 0.5 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.9 | REACTOME GLUCOSE TRANSPORT | Genes involved in Glucose transport |
| 0.0 | 0.3 | REACTOME G0 AND EARLY G1 | Genes involved in G0 and Early G1 |
| 0.0 | 0.1 | REACTOME SHC MEDIATED SIGNALLING | Genes involved in SHC-mediated signalling |
| 0.0 | 0.2 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |
| 0.0 | 0.2 | REACTOME PRESYNAPTIC NICOTINIC ACETYLCHOLINE RECEPTORS | Genes involved in Presynaptic nicotinic acetylcholine receptors |
| 0.0 | 0.3 | REACTOME AMINE DERIVED HORMONES | Genes involved in Amine-derived hormones |
| 0.0 | 2.6 | REACTOME SIGNALING BY RHO GTPASES | Genes involved in Signaling by Rho GTPases |
| 0.0 | 0.3 | REACTOME PKA MEDIATED PHOSPHORYLATION OF CREB | Genes involved in PKA-mediated phosphorylation of CREB |
| 0.0 | 0.7 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.1 | REACTOME CREATION OF C4 AND C2 ACTIVATORS | Genes involved in Creation of C4 and C2 activators |
| 0.0 | 0.3 | REACTOME LYSOSOME VESICLE BIOGENESIS | Genes involved in Lysosome Vesicle Biogenesis |
| 0.0 | 0.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |
| 0.0 | 0.2 | REACTOME PLATELET CALCIUM HOMEOSTASIS | Genes involved in Platelet calcium homeostasis |
| 0.0 | 0.2 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.1 | REACTOME COSTIMULATION BY THE CD28 FAMILY | Genes involved in Costimulation by the CD28 family |