Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
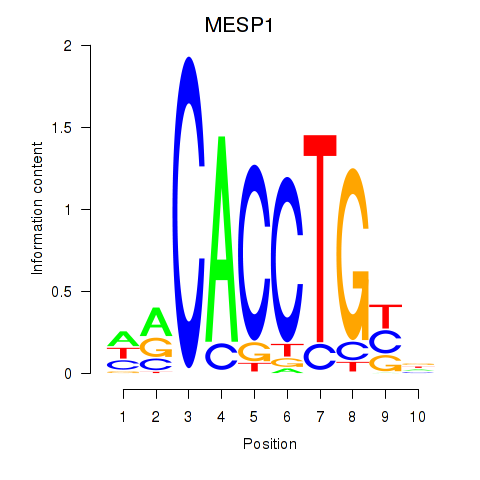
Results for MESP1
Z-value: 0.73
Transcription factors associated with MESP1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
MESP1
|
ENSG00000166823.5 | MESP1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MESP1 | hg19_v2_chr15_-_90294523_90294541 | -0.92 | 1.0e-03 | Click! |
Activity profile of MESP1 motif
Sorted Z-values of MESP1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MESP1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_60280458 | 1.31 |
ENST00000455990.1 ENST00000371208.3 |
HOOK1 |
hook microtubule-tethering protein 1 |
| chr17_-_39274606 | 1.15 |
ENST00000391413.2 |
KRTAP4-11 |
keratin associated protein 4-11 |
| chr1_-_209979465 | 0.92 |
ENST00000542854.1 |
IRF6 |
interferon regulatory factor 6 |
| chr2_+_47596287 | 0.92 |
ENST00000263735.4 |
EPCAM |
epithelial cell adhesion molecule |
| chr13_-_20806440 | 0.80 |
ENST00000400066.3 ENST00000400065.3 ENST00000356192.6 |
GJB6 |
gap junction protein, beta 6, 30kDa |
| chr16_+_68679193 | 0.80 |
ENST00000581171.1 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr17_+_39382900 | 0.75 |
ENST00000377721.3 ENST00000455970.2 |
KRTAP9-2 |
keratin associated protein 9-2 |
| chr1_-_160990886 | 0.71 |
ENST00000537746.1 |
F11R |
F11 receptor |
| chr1_-_209979375 | 0.66 |
ENST00000367021.3 |
IRF6 |
interferon regulatory factor 6 |
| chr4_+_40198527 | 0.63 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr11_-_108464465 | 0.63 |
ENST00000525344.1 |
EXPH5 |
exophilin 5 |
| chr19_+_35739597 | 0.61 |
ENST00000361790.3 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr19_+_35739631 | 0.61 |
ENST00000602003.1 ENST00000360798.3 ENST00000354900.3 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr19_+_35739782 | 0.61 |
ENST00000347609.4 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr8_+_120220561 | 0.61 |
ENST00000276681.6 |
MAL2 |
mal, T-cell differentiation protein 2 (gene/pseudogene) |
| chr16_+_68771128 | 0.60 |
ENST00000261769.5 ENST00000422392.2 |
CDH1 |
cadherin 1, type 1, E-cadherin (epithelial) |
| chr16_+_68678739 | 0.59 |
ENST00000264012.4 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr11_-_108464321 | 0.58 |
ENST00000265843.4 |
EXPH5 |
exophilin 5 |
| chr18_+_47088401 | 0.58 |
ENST00000261292.4 ENST00000427224.2 ENST00000580036.1 |
LIPG |
lipase, endothelial |
| chr17_-_39674668 | 0.57 |
ENST00000393981.3 |
KRT15 |
keratin 15 |
| chr1_+_183155373 | 0.56 |
ENST00000493293.1 ENST00000264144.4 |
LAMC2 |
laminin, gamma 2 |
| chr1_+_44399466 | 0.56 |
ENST00000498139.2 ENST00000491846.1 |
ARTN |
artemin |
| chr16_+_68678892 | 0.55 |
ENST00000429102.2 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr7_+_16793160 | 0.53 |
ENST00000262067.4 |
TSPAN13 |
tetraspanin 13 |
| chr1_+_2005425 | 0.53 |
ENST00000461106.2 |
PRKCZ |
protein kinase C, zeta |
| chr19_+_35739280 | 0.53 |
ENST00000602122.1 |
LSR |
lipolysis stimulated lipoprotein receptor |
| chr12_+_41086297 | 0.51 |
ENST00000551295.2 |
CNTN1 |
contactin 1 |
| chr17_-_7165662 | 0.51 |
ENST00000571881.2 ENST00000360325.7 |
CLDN7 |
claudin 7 |
| chr13_-_20767037 | 0.51 |
ENST00000382848.4 |
GJB2 |
gap junction protein, beta 2, 26kDa |
| chr17_+_9548845 | 0.49 |
ENST00000570475.1 ENST00000285199.7 |
USP43 |
ubiquitin specific peptidase 43 |
| chr15_+_41136586 | 0.49 |
ENST00000431806.1 |
SPINT1 |
serine peptidase inhibitor, Kunitz type 1 |
| chr1_+_109792641 | 0.49 |
ENST00000271332.3 |
CELSR2 |
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr6_+_150464155 | 0.48 |
ENST00000361131.4 |
PPP1R14C |
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr19_-_51456198 | 0.47 |
ENST00000594846.1 |
KLK5 |
kallikrein-related peptidase 5 |
| chr19_+_38755203 | 0.47 |
ENST00000587090.1 ENST00000454580.3 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
| chr3_-_111314230 | 0.47 |
ENST00000317012.4 |
ZBED2 |
zinc finger, BED-type containing 2 |
| chr8_+_102504651 | 0.46 |
ENST00000251808.3 ENST00000521085.1 |
GRHL2 |
grainyhead-like 2 (Drosophila) |
| chr1_+_24646002 | 0.45 |
ENST00000356046.2 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr1_+_24645807 | 0.44 |
ENST00000361548.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr19_-_35992780 | 0.44 |
ENST00000593342.1 ENST00000601650.1 ENST00000408915.2 |
DMKN |
dermokine |
| chr1_+_24645865 | 0.44 |
ENST00000342072.4 |
GRHL3 |
grainyhead-like 3 (Drosophila) |
| chr17_-_7493390 | 0.44 |
ENST00000538513.2 ENST00000570788.1 ENST00000250055.2 |
SOX15 |
SRY (sex determining region Y)-box 15 |
| chr6_+_7541808 | 0.43 |
ENST00000379802.3 |
DSP |
desmoplakin |
| chr19_+_38755042 | 0.42 |
ENST00000301244.7 |
SPINT2 |
serine peptidase inhibitor, Kunitz type, 2 |
| chr6_+_7541845 | 0.42 |
ENST00000418664.2 |
DSP |
desmoplakin |
| chr5_+_68788594 | 0.41 |
ENST00000396442.2 ENST00000380766.2 |
OCLN |
occludin |
| chr8_+_32405785 | 0.41 |
ENST00000287842.3 |
NRG1 |
neuregulin 1 |
| chr11_+_1860200 | 0.41 |
ENST00000381911.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr2_-_238499303 | 0.41 |
ENST00000409576.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr7_+_26331541 | 0.40 |
ENST00000416246.1 ENST00000338523.4 ENST00000412416.1 |
SNX10 |
sorting nexin 10 |
| chr8_+_32405728 | 0.40 |
ENST00000523079.1 ENST00000338921.4 ENST00000356819.4 ENST00000287845.5 ENST00000341377.5 |
NRG1 |
neuregulin 1 |
| chr3_-_190040223 | 0.39 |
ENST00000295522.3 |
CLDN1 |
claudin 1 |
| chr12_+_53342625 | 0.39 |
ENST00000388837.2 ENST00000550600.1 ENST00000388835.3 |
KRT18 |
keratin 18 |
| chr4_-_6202247 | 0.38 |
ENST00000409021.3 ENST00000409371.3 |
JAKMIP1 |
janus kinase and microtubule interacting protein 1 |
| chr1_+_1981890 | 0.37 |
ENST00000378567.3 ENST00000468310.1 |
PRKCZ |
protein kinase C, zeta |
| chr1_+_95582881 | 0.37 |
ENST00000370203.4 ENST00000456991.1 |
TMEM56 |
transmembrane protein 56 |
| chr2_+_120770686 | 0.37 |
ENST00000331393.4 ENST00000443124.1 |
EPB41L5 |
erythrocyte membrane protein band 4.1 like 5 |
| chr6_+_36098262 | 0.37 |
ENST00000373761.6 ENST00000373766.5 |
MAPK13 |
mitogen-activated protein kinase 13 |
| chr8_-_127570603 | 0.36 |
ENST00000304916.3 |
FAM84B |
family with sequence similarity 84, member B |
| chr19_-_51471362 | 0.35 |
ENST00000376853.4 ENST00000424910.2 |
KLK6 |
kallikrein-related peptidase 6 |
| chr1_+_95286151 | 0.35 |
ENST00000467909.1 ENST00000422520.2 ENST00000532427.1 |
SLC44A3 |
solute carrier family 44, member 3 |
| chrX_-_102565932 | 0.35 |
ENST00000372674.1 ENST00000372677.3 |
BEX2 |
brain expressed X-linked 2 |
| chr17_-_39324424 | 0.35 |
ENST00000391356.2 |
KRTAP4-3 |
keratin associated protein 4-3 |
| chr19_+_6464243 | 0.35 |
ENST00000600229.1 ENST00000356762.3 |
CRB3 |
crumbs homolog 3 (Drosophila) |
| chr18_+_29077990 | 0.35 |
ENST00000261590.8 |
DSG2 |
desmoglein 2 |
| chr3_-_133748913 | 0.34 |
ENST00000310926.4 |
SLCO2A1 |
solute carrier organic anion transporter family, member 2A1 |
| chr1_+_186798073 | 0.34 |
ENST00000367466.3 ENST00000442353.2 |
PLA2G4A |
phospholipase A2, group IVA (cytosolic, calcium-dependent) |
| chr11_+_1855645 | 0.34 |
ENST00000381968.3 ENST00000381978.3 |
SYT8 |
synaptotagmin VIII |
| chr19_-_51471381 | 0.34 |
ENST00000594641.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chr17_+_80693427 | 0.33 |
ENST00000300784.7 |
FN3K |
fructosamine 3 kinase |
| chr19_-_49565254 | 0.33 |
ENST00000593537.1 |
NTF4 |
neurotrophin 4 |
| chr18_-_47721447 | 0.32 |
ENST00000285039.7 |
MYO5B |
myosin VB |
| chr1_+_95285896 | 0.32 |
ENST00000446120.2 ENST00000271227.6 ENST00000527077.1 ENST00000529450.1 |
SLC44A3 |
solute carrier family 44, member 3 |
| chr1_+_13910479 | 0.32 |
ENST00000509009.1 |
PDPN |
podoplanin |
| chr19_+_6464502 | 0.32 |
ENST00000308243.7 |
CRB3 |
crumbs homolog 3 (Drosophila) |
| chr16_-_4987065 | 0.31 |
ENST00000590782.2 ENST00000345988.2 |
PPL |
periplakin |
| chr17_+_73521763 | 0.31 |
ENST00000167462.5 ENST00000375227.4 ENST00000392550.3 ENST00000578363.1 ENST00000579392.1 |
LLGL2 |
lethal giant larvae homolog 2 (Drosophila) |
| chr11_+_1856034 | 0.31 |
ENST00000341958.3 |
SYT8 |
synaptotagmin VIII |
| chr4_+_48018781 | 0.31 |
ENST00000295461.5 |
NIPAL1 |
NIPA-like domain containing 1 |
| chr12_-_53343602 | 0.30 |
ENST00000546897.1 ENST00000552551.1 |
KRT8 |
keratin 8 |
| chr14_+_75746340 | 0.30 |
ENST00000555686.1 ENST00000555672.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr15_+_74833518 | 0.30 |
ENST00000346246.5 |
ARID3B |
AT rich interactive domain 3B (BRIGHT-like) |
| chr15_-_63674034 | 0.29 |
ENST00000344366.3 ENST00000422263.2 |
CA12 |
carbonic anhydrase XII |
| chr11_-_88070920 | 0.29 |
ENST00000524463.1 ENST00000227266.5 |
CTSC |
cathepsin C |
| chr7_+_18535786 | 0.28 |
ENST00000406072.1 |
HDAC9 |
histone deacetylase 9 |
| chr11_+_1860682 | 0.28 |
ENST00000381906.1 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr22_+_29876197 | 0.28 |
ENST00000310624.6 |
NEFH |
neurofilament, heavy polypeptide |
| chr7_-_22396533 | 0.28 |
ENST00000344041.6 |
RAPGEF5 |
Rap guanine nucleotide exchange factor (GEF) 5 |
| chr17_+_39394250 | 0.28 |
ENST00000254072.6 |
KRTAP9-8 |
keratin associated protein 9-8 |
| chr11_+_1860832 | 0.28 |
ENST00000252898.7 |
TNNI2 |
troponin I type 2 (skeletal, fast) |
| chr19_+_751122 | 0.28 |
ENST00000215582.6 |
MISP |
mitotic spindle positioning |
| chr11_-_118122996 | 0.27 |
ENST00000525386.1 ENST00000527472.1 ENST00000278949.4 |
MPZL3 |
myelin protein zero-like 3 |
| chr9_-_126030817 | 0.27 |
ENST00000348403.5 ENST00000447404.2 ENST00000360998.3 |
STRBP |
spermatid perinuclear RNA binding protein |
| chr1_-_55266926 | 0.27 |
ENST00000371276.4 |
TTC22 |
tetratricopeptide repeat domain 22 |
| chr2_+_68961934 | 0.27 |
ENST00000409202.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr8_-_144655141 | 0.27 |
ENST00000398882.3 |
MROH6 |
maestro heat-like repeat family member 6 |
| chr5_-_141257954 | 0.27 |
ENST00000456271.1 ENST00000394536.3 ENST00000503492.1 ENST00000287008.3 |
PCDH1 |
protocadherin 1 |
| chr2_+_68961905 | 0.26 |
ENST00000295381.3 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr4_+_75310851 | 0.26 |
ENST00000395748.3 ENST00000264487.2 |
AREG |
amphiregulin |
| chr2_-_238499725 | 0.26 |
ENST00000264601.3 |
RAB17 |
RAB17, member RAS oncogene family |
| chr20_-_23030296 | 0.26 |
ENST00000377103.2 |
THBD |
thrombomodulin |
| chr19_+_7660716 | 0.26 |
ENST00000160298.4 ENST00000446248.2 |
CAMSAP3 |
calmodulin regulated spectrin-associated protein family, member 3 |
| chr5_+_68711209 | 0.26 |
ENST00000512803.1 |
MARVELD2 |
MARVEL domain containing 2 |
| chr19_-_51472823 | 0.26 |
ENST00000310157.2 |
KLK6 |
kallikrein-related peptidase 6 |
| chr4_+_106816592 | 0.26 |
ENST00000379987.2 ENST00000453617.2 ENST00000427316.2 ENST00000514622.1 ENST00000305572.8 |
NPNT |
nephronectin |
| chr12_+_56473628 | 0.26 |
ENST00000549282.1 ENST00000549061.1 ENST00000267101.3 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr17_-_39296739 | 0.25 |
ENST00000345847.4 |
KRTAP4-6 |
keratin associated protein 4-6 |
| chr14_+_75746781 | 0.25 |
ENST00000555347.1 |
FOS |
FBJ murine osteosarcoma viral oncogene homolog |
| chr1_+_161676739 | 0.25 |
ENST00000236938.6 ENST00000367959.2 ENST00000546024.1 ENST00000540521.1 ENST00000367949.2 ENST00000350710.3 ENST00000540926.1 |
FCRLA |
Fc receptor-like A |
| chr5_+_68710906 | 0.25 |
ENST00000325631.5 ENST00000454295.2 |
MARVELD2 |
MARVEL domain containing 2 |
| chr4_+_75311019 | 0.25 |
ENST00000502307.1 |
AREG |
amphiregulin |
| chr9_+_132099158 | 0.25 |
ENST00000444125.1 |
RP11-65J3.1 |
RP11-65J3.1 |
| chr1_+_161676983 | 0.24 |
ENST00000367957.2 |
FCRLA |
Fc receptor-like A |
| chr2_+_220492373 | 0.24 |
ENST00000317151.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr1_+_13910194 | 0.24 |
ENST00000376057.4 ENST00000510906.1 |
PDPN |
podoplanin |
| chr1_+_161677034 | 0.24 |
ENST00000349527.4 ENST00000309691.6 ENST00000294796.4 ENST00000367953.3 ENST00000367950.1 |
FCRLA |
Fc receptor-like A |
| chr15_-_63674218 | 0.24 |
ENST00000178638.3 |
CA12 |
carbonic anhydrase XII |
| chrX_-_153141302 | 0.24 |
ENST00000361699.4 ENST00000543994.1 ENST00000370057.3 ENST00000538883.1 ENST00000361981.3 |
L1CAM |
L1 cell adhesion molecule |
| chr8_+_22438009 | 0.23 |
ENST00000409417.1 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr11_-_119991589 | 0.23 |
ENST00000526881.1 |
TRIM29 |
tripartite motif containing 29 |
| chr3_+_32433154 | 0.23 |
ENST00000334983.5 ENST00000349718.4 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
| chrX_-_34150428 | 0.23 |
ENST00000346193.3 |
FAM47A |
family with sequence similarity 47, member A |
| chr21_-_42219065 | 0.23 |
ENST00000400454.1 |
DSCAM |
Down syndrome cell adhesion molecule |
| chrX_+_105969893 | 0.23 |
ENST00000255499.2 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr3_+_32433363 | 0.23 |
ENST00000465248.1 |
CMTM7 |
CKLF-like MARVEL transmembrane domain containing 7 |
| chr1_-_184943610 | 0.22 |
ENST00000367511.3 |
FAM129A |
family with sequence similarity 129, member A |
| chr2_-_238499337 | 0.22 |
ENST00000411462.1 ENST00000409822.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr11_+_65779283 | 0.22 |
ENST00000312134.2 |
CST6 |
cystatin E/M |
| chr13_+_113656022 | 0.22 |
ENST00000423482.2 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chr17_-_39280419 | 0.22 |
ENST00000394014.1 |
KRTAP4-12 |
keratin associated protein 4-12 |
| chr11_-_115375107 | 0.22 |
ENST00000545380.1 ENST00000452722.3 ENST00000537058.1 ENST00000536727.1 ENST00000542447.2 ENST00000331581.6 |
CADM1 |
cell adhesion molecule 1 |
| chr18_+_55888767 | 0.22 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr9_-_23821842 | 0.22 |
ENST00000544538.1 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr9_+_101569944 | 0.21 |
ENST00000375011.3 |
GALNT12 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 12 (GalNAc-T12) |
| chr4_-_139163491 | 0.21 |
ENST00000280612.5 |
SLC7A11 |
solute carrier family 7 (anionic amino acid transporter light chain, xc- system), member 11 |
| chr4_+_75480629 | 0.21 |
ENST00000380846.3 |
AREGB |
amphiregulin B |
| chr2_-_238499131 | 0.21 |
ENST00000538644.1 |
RAB17 |
RAB17, member RAS oncogene family |
| chr7_+_128784712 | 0.21 |
ENST00000289407.4 |
TSPAN33 |
tetraspanin 33 |
| chr7_-_138666053 | 0.21 |
ENST00000440172.1 ENST00000422774.1 |
KIAA1549 |
KIAA1549 |
| chr14_+_65171315 | 0.21 |
ENST00000394691.1 |
PLEKHG3 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 3 |
| chr12_+_122459757 | 0.20 |
ENST00000261822.4 |
BCL7A |
B-cell CLL/lymphoma 7A |
| chr2_-_165477971 | 0.20 |
ENST00000446413.2 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr3_+_111717600 | 0.20 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr22_+_29469012 | 0.20 |
ENST00000400335.4 ENST00000400338.2 |
KREMEN1 |
kringle containing transmembrane protein 1 |
| chr5_-_1295104 | 0.20 |
ENST00000334602.6 ENST00000508104.2 ENST00000310581.5 ENST00000296820.5 |
TERT |
telomerase reverse transcriptase |
| chr2_-_160761179 | 0.20 |
ENST00000554112.1 ENST00000553424.1 ENST00000263636.4 ENST00000504764.1 ENST00000505052.1 |
LY75 LY75-CD302 |
lymphocyte antigen 75 LY75-CD302 readthrough |
| chr15_+_45722727 | 0.20 |
ENST00000396650.2 ENST00000558435.1 ENST00000344300.3 |
C15orf48 |
chromosome 15 open reading frame 48 |
| chr17_-_27503770 | 0.20 |
ENST00000533112.1 |
MYO18A |
myosin XVIIIA |
| chr13_-_52980263 | 0.20 |
ENST00000258613.4 ENST00000544466.1 |
THSD1 |
thrombospondin, type I, domain containing 1 |
| chr14_-_106781017 | 0.19 |
ENST00000390612.2 |
IGHV4-28 |
immunoglobulin heavy variable 4-28 |
| chr14_-_107170409 | 0.19 |
ENST00000390633.2 |
IGHV1-69 |
immunoglobulin heavy variable 1-69 |
| chr4_-_6202291 | 0.19 |
ENST00000282924.5 |
JAKMIP1 |
janus kinase and microtubule interacting protein 1 |
| chr1_-_156675368 | 0.18 |
ENST00000368222.3 |
CRABP2 |
cellular retinoic acid binding protein 2 |
| chr5_-_54281407 | 0.18 |
ENST00000381403.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr3_-_48470838 | 0.18 |
ENST00000358459.4 ENST00000358536.4 |
PLXNB1 |
plexin B1 |
| chr19_-_55660561 | 0.18 |
ENST00000587758.1 ENST00000356783.5 ENST00000291901.8 ENST00000588426.1 ENST00000588147.1 ENST00000536926.1 ENST00000588981.1 |
TNNT1 |
troponin T type 1 (skeletal, slow) |
| chr14_-_38064198 | 0.18 |
ENST00000250448.2 |
FOXA1 |
forkhead box A1 |
| chr3_+_140770183 | 0.18 |
ENST00000310546.2 |
SPSB4 |
splA/ryanodine receptor domain and SOCS box containing 4 |
| chr17_-_46507537 | 0.18 |
ENST00000336915.6 |
SKAP1 |
src kinase associated phosphoprotein 1 |
| chr6_-_30712313 | 0.18 |
ENST00000376377.2 ENST00000259874.5 |
IER3 |
immediate early response 3 |
| chr7_+_20370746 | 0.18 |
ENST00000222573.4 |
ITGB8 |
integrin, beta 8 |
| chr22_-_37882395 | 0.18 |
ENST00000416983.3 ENST00000424765.2 ENST00000356998.3 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr11_-_17035943 | 0.18 |
ENST00000355661.3 ENST00000532079.1 ENST00000448080.2 ENST00000531066.1 |
PLEKHA7 |
pleckstrin homology domain containing, family A member 7 |
| chr11_-_119993979 | 0.18 |
ENST00000524816.3 ENST00000525327.1 |
TRIM29 |
tripartite motif containing 29 |
| chr2_+_173292301 | 0.18 |
ENST00000264106.6 ENST00000375221.2 ENST00000343713.4 |
ITGA6 |
integrin, alpha 6 |
| chr2_-_20425158 | 0.17 |
ENST00000381150.1 |
SDC1 |
syndecan 1 |
| chr8_+_98881268 | 0.17 |
ENST00000254898.5 ENST00000524308.1 ENST00000522025.2 |
MATN2 |
matrilin 2 |
| chr15_-_31283618 | 0.17 |
ENST00000563714.1 |
MTMR10 |
myotubularin related protein 10 |
| chr12_-_6756559 | 0.17 |
ENST00000536350.1 ENST00000414226.2 ENST00000546114.1 |
ACRBP |
acrosin binding protein |
| chr5_-_16936340 | 0.17 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr9_-_136024721 | 0.17 |
ENST00000393160.3 |
RALGDS |
ral guanine nucleotide dissociation stimulator |
| chr11_-_44971702 | 0.17 |
ENST00000533940.1 ENST00000533937.1 |
TP53I11 |
tumor protein p53 inducible protein 11 |
| chr4_-_25865159 | 0.17 |
ENST00000502949.1 ENST00000264868.5 ENST00000513691.1 ENST00000514872.1 |
SEL1L3 |
sel-1 suppressor of lin-12-like 3 (C. elegans) |
| chr5_-_54281491 | 0.17 |
ENST00000381405.4 |
ESM1 |
endothelial cell-specific molecule 1 |
| chr14_+_67999999 | 0.17 |
ENST00000329153.5 |
PLEKHH1 |
pleckstrin homology domain containing, family H (with MyTH4 domain) member 1 |
| chr20_+_49348081 | 0.17 |
ENST00000371610.2 |
PARD6B |
par-6 family cell polarity regulator beta |
| chr2_+_173292390 | 0.17 |
ENST00000442250.1 ENST00000458358.1 ENST00000409080.1 |
ITGA6 |
integrin, alpha 6 |
| chr12_-_6756609 | 0.17 |
ENST00000229243.2 |
ACRBP |
acrosin binding protein |
| chr17_+_37894179 | 0.17 |
ENST00000577695.1 ENST00000309156.4 ENST00000309185.3 |
GRB7 |
growth factor receptor-bound protein 7 |
| chr13_+_88324870 | 0.17 |
ENST00000325089.6 |
SLITRK5 |
SLIT and NTRK-like family, member 5 |
| chr2_+_173292280 | 0.16 |
ENST00000264107.7 |
ITGA6 |
integrin, alpha 6 |
| chr1_-_156675535 | 0.16 |
ENST00000368221.1 |
CRABP2 |
cellular retinoic acid binding protein 2 |
| chr12_-_95044309 | 0.16 |
ENST00000261226.4 |
TMCC3 |
transmembrane and coiled-coil domain family 3 |
| chr22_+_29469100 | 0.16 |
ENST00000327813.5 ENST00000407188.1 |
KREMEN1 |
kringle containing transmembrane protein 1 |
| chr15_-_42186248 | 0.16 |
ENST00000320955.6 |
SPTBN5 |
spectrin, beta, non-erythrocytic 5 |
| chr9_-_117880477 | 0.16 |
ENST00000534839.1 ENST00000340094.3 ENST00000535648.1 ENST00000346706.3 ENST00000345230.3 ENST00000350763.4 |
TNC |
tenascin C |
| chr19_-_44174330 | 0.16 |
ENST00000340093.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr15_-_44487408 | 0.16 |
ENST00000402883.1 ENST00000417257.1 |
FRMD5 |
FERM domain containing 5 |
| chr16_-_3086927 | 0.16 |
ENST00000572449.1 |
CCDC64B |
coiled-coil domain containing 64B |
| chr17_+_73717407 | 0.16 |
ENST00000579662.1 |
ITGB4 |
integrin, beta 4 |
| chr3_+_111717511 | 0.16 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chr9_+_2622085 | 0.16 |
ENST00000382099.2 |
VLDLR |
very low density lipoprotein receptor |
| chr16_+_23847267 | 0.16 |
ENST00000321728.7 |
PRKCB |
protein kinase C, beta |
| chr2_-_75788038 | 0.15 |
ENST00000393913.3 ENST00000410113.1 |
EVA1A |
eva-1 homolog A (C. elegans) |
| chr17_-_39254391 | 0.15 |
ENST00000333822.4 |
KRTAP4-8 |
keratin associated protein 4-8 |
| chr19_+_39279838 | 0.15 |
ENST00000314980.4 |
LGALS7B |
lectin, galactoside-binding, soluble, 7B |
| chr19_-_39264072 | 0.15 |
ENST00000599035.1 ENST00000378626.4 |
LGALS7 |
lectin, galactoside-binding, soluble, 7 |
| chr9_-_5833027 | 0.15 |
ENST00000339450.5 |
ERMP1 |
endoplasmic reticulum metallopeptidase 1 |
| chr16_+_4845379 | 0.15 |
ENST00000588606.1 ENST00000586005.1 |
SMIM22 |
small integral membrane protein 22 |
| chr5_-_94620239 | 0.15 |
ENST00000515393.1 |
MCTP1 |
multiple C2 domains, transmembrane 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.6 | 1.9 | GO:0051796 | regulation of monophenol monooxygenase activity(GO:0032771) positive regulation of monophenol monooxygenase activity(GO:0032773) negative regulation of catagen(GO:0051796) regulation of hair cycle by canonical Wnt signaling pathway(GO:0060901) positive regulation of melanosome transport(GO:1902910) |
| 0.5 | 2.4 | GO:1904274 | tricellular tight junction assembly(GO:1904274) |
| 0.3 | 1.4 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.2 | 1.1 | GO:0002415 | immunoglobulin transcytosis in epithelial cells mediated by polymeric immunoglobulin receptor(GO:0002415) |
| 0.2 | 0.6 | GO:0048627 | myoblast development(GO:0048627) |
| 0.2 | 0.6 | GO:1904328 | regulation of myofibroblast contraction(GO:1904328) myofibroblast contraction(GO:1990764) |
| 0.2 | 0.9 | GO:2000048 | negative regulation of cell-cell adhesion mediated by cadherin(GO:2000048) |
| 0.2 | 0.5 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.6 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.1 | 0.6 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.1 | 0.4 | GO:1903348 | cellular response to lead ion(GO:0071284) positive regulation of bicellular tight junction assembly(GO:1903348) |
| 0.1 | 0.4 | GO:0043000 | Golgi to plasma membrane CFTR protein transport(GO:0043000) |
| 0.1 | 0.9 | GO:2001181 | positive regulation of interleukin-10 secretion(GO:2001181) |
| 0.1 | 1.1 | GO:0086073 | bundle of His cell-Purkinje myocyte adhesion involved in cell communication(GO:0086073) |
| 0.1 | 0.8 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.1 | 0.6 | GO:0097021 | Peyer's patch morphogenesis(GO:0061146) lymphocyte migration into lymphoid organs(GO:0097021) |
| 0.1 | 0.3 | GO:0048936 | neurofilament bundle assembly(GO:0033693) peripheral nervous system neuron axonogenesis(GO:0048936) |
| 0.1 | 0.4 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.1 | 0.3 | GO:1900220 | semaphorin-plexin signaling pathway involved in bone trabecula morphogenesis(GO:1900220) |
| 0.1 | 0.5 | GO:0055064 | chloride ion homeostasis(GO:0055064) |
| 0.1 | 0.3 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.1 | 0.5 | GO:0060744 | thelarche(GO:0042695) mammary gland branching involved in thelarche(GO:0060744) |
| 0.1 | 0.3 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.1 | 0.5 | GO:0002760 | positive regulation of antimicrobial peptide production(GO:0002225) positive regulation of antimicrobial humoral response(GO:0002760) positive regulation of antibacterial peptide production(GO:0002803) |
| 0.1 | 0.3 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.2 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.1 | 1.4 | GO:0090179 | planar cell polarity pathway involved in neural tube closure(GO:0090179) |
| 0.1 | 0.4 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.4 | GO:0045218 | zonula adherens maintenance(GO:0045218) |
| 0.1 | 0.3 | GO:2000721 | pilomotor reflex(GO:0097195) positive regulation of transcription from RNA polymerase II promoter involved in smooth muscle cell differentiation(GO:2000721) |
| 0.1 | 0.6 | GO:0001661 | conditioned taste aversion(GO:0001661) |
| 0.1 | 0.2 | GO:1900369 | transcription, RNA-templated(GO:0001172) negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.2 | GO:1903028 | positive regulation of opsonization(GO:1903028) |
| 0.1 | 0.3 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.2 | GO:0060738 | epithelial-mesenchymal signaling involved in prostate gland development(GO:0060738) |
| 0.1 | 0.9 | GO:0035878 | nail development(GO:0035878) |
| 0.1 | 0.4 | GO:0071681 | response to indole-3-methanol(GO:0071680) cellular response to indole-3-methanol(GO:0071681) |
| 0.1 | 0.2 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 1.3 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.1 | 0.2 | GO:0042271 | susceptibility to natural killer cell mediated cytotoxicity(GO:0042271) |
| 0.1 | 0.2 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.4 | GO:0033183 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.1 | 0.2 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.4 | GO:1902202 | regulation of hepatocyte growth factor receptor signaling pathway(GO:1902202) |
| 0.0 | 1.5 | GO:0060644 | mammary gland epithelial cell differentiation(GO:0060644) |
| 0.0 | 0.1 | GO:0043311 | positive regulation of eosinophil degranulation(GO:0043311) positive regulation of eosinophil activation(GO:1902568) |
| 0.0 | 0.1 | GO:1903572 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.2 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.1 | GO:0044771 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.7 | GO:0015871 | choline transport(GO:0015871) |
| 0.0 | 0.2 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.0 | 0.7 | GO:0090557 | establishment of endothelial intestinal barrier(GO:0090557) |
| 0.0 | 0.9 | GO:0016540 | protein autoprocessing(GO:0016540) |
| 0.0 | 0.3 | GO:0032439 | endosome localization(GO:0032439) |
| 0.0 | 0.1 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0072156 | distal tubule morphogenesis(GO:0072156) |
| 0.0 | 0.2 | GO:0003430 | growth plate cartilage chondrocyte growth(GO:0003430) Harderian gland development(GO:0070384) |
| 0.0 | 0.0 | GO:0072244 | renal filtration cell differentiation(GO:0061318) glomerular epithelium development(GO:0072010) glomerular visceral epithelial cell development(GO:0072015) glomerular visceral epithelial cell differentiation(GO:0072112) metanephric glomerular epithelium development(GO:0072244) metanephric glomerular visceral epithelial cell differentiation(GO:0072248) metanephric glomerular visceral epithelial cell development(GO:0072249) glomerular epithelial cell development(GO:0072310) glomerular epithelial cell differentiation(GO:0072311) metanephric glomerular epithelial cell differentiation(GO:0072312) metanephric glomerular epithelial cell development(GO:0072313) |
| 0.0 | 0.3 | GO:0048672 | positive regulation of collateral sprouting(GO:0048672) |
| 0.0 | 0.1 | GO:1904862 | inhibitory synapse assembly(GO:1904862) |
| 0.0 | 0.1 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.0 | 0.1 | GO:0036079 | GDP-fucose transport(GO:0015783) purine nucleotide-sugar transport(GO:0036079) |
| 0.0 | 0.1 | GO:1900108 | negative regulation of nodal signaling pathway(GO:1900108) |
| 0.0 | 0.5 | GO:0060670 | branching involved in labyrinthine layer morphogenesis(GO:0060670) |
| 0.0 | 0.0 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.7 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0003104 | positive regulation of glomerular filtration(GO:0003104) |
| 0.0 | 0.1 | GO:0035284 | rhombomere 5 development(GO:0021571) central nervous system segmentation(GO:0035283) brain segmentation(GO:0035284) |
| 0.0 | 0.1 | GO:2001302 | lipoxin biosynthetic process(GO:2001301) lipoxin A4 metabolic process(GO:2001302) lipoxin A4 biosynthetic process(GO:2001303) |
| 0.0 | 0.2 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.4 | GO:0070673 | response to interleukin-18(GO:0070673) |
| 0.0 | 0.1 | GO:2000567 | memory T cell activation(GO:0035709) regulation of memory T cell activation(GO:2000567) positive regulation of memory T cell activation(GO:2000568) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.2 | GO:0034436 | glycoprotein transport(GO:0034436) |
| 0.0 | 0.2 | GO:1900625 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.0 | 0.6 | GO:0045056 | transcytosis(GO:0045056) |
| 0.0 | 1.3 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.1 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.0 | GO:0003197 | endocardial cushion development(GO:0003197) |
| 0.0 | 0.1 | GO:0045014 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.0 | 0.1 | GO:0002913 | positive regulation of T cell anergy(GO:0002669) positive regulation of lymphocyte anergy(GO:0002913) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.1 | GO:0033861 | negative regulation of NAD(P)H oxidase activity(GO:0033861) |
| 0.0 | 0.3 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.3 | GO:0051918 | negative regulation of fibrinolysis(GO:0051918) |
| 0.0 | 0.2 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.0 | GO:0002125 | maternal aggressive behavior(GO:0002125) |
| 0.0 | 0.1 | GO:0090156 | cellular sphingolipid homeostasis(GO:0090156) |
| 0.0 | 0.3 | GO:0015693 | magnesium ion transport(GO:0015693) |
| 0.0 | 0.1 | GO:0035106 | operant conditioning(GO:0035106) |
| 0.0 | 0.2 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.1 | GO:0032911 | negative regulation of transforming growth factor beta1 production(GO:0032911) |
| 0.0 | 0.1 | GO:0010830 | regulation of myotube differentiation(GO:0010830) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) eosinophil differentiation(GO:0030222) |
| 0.0 | 0.1 | GO:0030167 | proteoglycan catabolic process(GO:0030167) heparan sulfate proteoglycan catabolic process(GO:0030200) |
| 0.0 | 0.1 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:0002325 | natural killer cell differentiation involved in immune response(GO:0002325) negative regulation of natural killer cell differentiation(GO:0032824) regulation of natural killer cell differentiation involved in immune response(GO:0032826) negative regulation of natural killer cell differentiation involved in immune response(GO:0032827) |
| 0.0 | 0.0 | GO:0038163 | thrombopoietin-mediated signaling pathway(GO:0038163) |
| 0.0 | 0.0 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.0 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 3.0 | GO:0031424 | keratinization(GO:0031424) |
| 0.0 | 1.4 | GO:0070830 | bicellular tight junction assembly(GO:0070830) |
| 0.0 | 0.2 | GO:0007157 | heterophilic cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0007157) |
| 0.0 | 0.2 | GO:0031444 | slow-twitch skeletal muscle fiber contraction(GO:0031444) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.1 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 1.1 | GO:0003009 | skeletal muscle contraction(GO:0003009) |
| 0.0 | 0.1 | GO:0006072 | glycerol-3-phosphate metabolic process(GO:0006072) |
| 0.0 | 0.0 | GO:0070103 | regulation of interleukin-6-mediated signaling pathway(GO:0070103) positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.1 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.0 | GO:0035544 | negative regulation of SNARE complex assembly(GO:0035544) |
| 0.0 | 0.0 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.1 | GO:0033277 | abortive mitotic cell cycle(GO:0033277) |
| 0.0 | 0.1 | GO:0035879 | plasma membrane lactate transport(GO:0035879) |
| 0.0 | 0.0 | GO:0072053 | renal inner medulla development(GO:0072053) renal outer medulla development(GO:0072054) |
| 0.0 | 0.1 | GO:0014859 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:0045163 | clustering of voltage-gated potassium channels(GO:0045163) |
| 0.0 | 0.1 | GO:0019348 | dolichol metabolic process(GO:0019348) |
| 0.0 | 0.0 | GO:0003221 | right ventricular cardiac muscle tissue morphogenesis(GO:0003221) |
| 0.0 | 0.1 | GO:0040009 | regulation of growth rate(GO:0040009) G-quadruplex DNA unwinding(GO:0044806) |
| 0.0 | 0.1 | GO:0010587 | miRNA catabolic process(GO:0010587) |
| 0.0 | 0.0 | GO:0035962 | response to interleukin-13(GO:0035962) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.7 | GO:0051602 | response to electrical stimulus(GO:0051602) |
| 0.0 | 0.2 | GO:0035269 | protein O-linked mannosylation(GO:0035269) |
| 0.0 | 0.2 | GO:0032074 | negative regulation of nuclease activity(GO:0032074) |
| 0.0 | 0.3 | GO:0015732 | prostaglandin transport(GO:0015732) |
| 0.0 | 0.0 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.1 | GO:1902961 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.3 | GO:0071539 | protein localization to centrosome(GO:0071539) |
| 0.0 | 0.0 | GO:0050773 | regulation of dendrite development(GO:0050773) |
| 0.0 | 0.1 | GO:0050976 | sensory perception of touch(GO:0050975) detection of mechanical stimulus involved in sensory perception of touch(GO:0050976) |
| 0.0 | 0.1 | GO:0045084 | positive regulation of interleukin-12 biosynthetic process(GO:0045084) |
| 0.0 | 0.0 | GO:0032286 | central nervous system myelin maintenance(GO:0032286) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0070424 | regulation of nucleotide-binding oligomerization domain containing signaling pathway(GO:0070424) |
| 0.0 | 0.1 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.0 | GO:0002541 | activation of plasma proteins involved in acute inflammatory response(GO:0002541) |
| 0.0 | 0.9 | GO:0048791 | calcium ion-regulated exocytosis of neurotransmitter(GO:0048791) |
| 0.0 | 0.2 | GO:0002315 | marginal zone B cell differentiation(GO:0002315) |
| 0.0 | 0.0 | GO:0006990 | positive regulation of transcription from RNA polymerase II promoter involved in unfolded protein response(GO:0006990) |
| 0.0 | 0.1 | GO:0006477 | protein sulfation(GO:0006477) |
| 0.0 | 0.2 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:2000503 | positive regulation of natural killer cell chemotaxis(GO:2000503) |
| 0.0 | 0.1 | GO:0097319 | fructose transport(GO:0015755) fructose import(GO:0032445) carbohydrate import into cell(GO:0097319) carbohydrate import across plasma membrane(GO:0098704) fructose import across plasma membrane(GO:1990539) |
| 0.0 | 0.1 | GO:2000348 | regulation of CD40 signaling pathway(GO:2000348) |
| 0.0 | 0.0 | GO:0052151 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:0098870 | neuronal action potential propagation(GO:0019227) action potential propagation(GO:0098870) |
| 0.0 | 0.1 | GO:0097151 | positive regulation of inhibitory postsynaptic potential(GO:0097151) modulation of inhibitory postsynaptic potential(GO:0098828) |
| 0.0 | 0.1 | GO:2000860 | positive regulation of mineralocorticoid secretion(GO:2000857) positive regulation of aldosterone secretion(GO:2000860) |
| 0.0 | 0.2 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.1 | GO:0007042 | lysosomal lumen acidification(GO:0007042) |
| 0.0 | 0.0 | GO:0010193 | response to ozone(GO:0010193) |
| 0.0 | 0.1 | GO:1990416 | cellular response to brain-derived neurotrophic factor stimulus(GO:1990416) |
| 0.0 | 0.1 | GO:0045793 | positive regulation of cell size(GO:0045793) |
| 0.0 | 0.0 | GO:0072428 | signal transduction involved in intra-S DNA damage checkpoint(GO:0072428) response to bisphenol A(GO:1903925) cellular response to bisphenol A(GO:1903926) |
| 0.0 | 0.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.1 | GO:1901552 | positive regulation of endothelial cell development(GO:1901552) positive regulation of establishment of endothelial barrier(GO:1903142) |
| 0.0 | 0.0 | GO:2000078 | glandular epithelial cell maturation(GO:0002071) regulation of transcription from RNA polymerase II promoter involved in ventral spinal cord interneuron specification(GO:0021913) type B pancreatic cell maturation(GO:0072560) positive regulation of type B pancreatic cell development(GO:2000078) |
| 0.0 | 0.0 | GO:0051892 | negative regulation of cardioblast differentiation(GO:0051892) regulation of cardiac muscle cell myoblast differentiation(GO:2000690) negative regulation of cardiac muscle cell myoblast differentiation(GO:2000691) |
| 0.0 | 0.1 | GO:1901374 | acetate ester transport(GO:1901374) |
| 0.0 | 0.2 | GO:0089711 | L-glutamate transmembrane transport(GO:0089711) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 2.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.2 | 0.6 | GO:0005607 | laminin-2 complex(GO:0005607) |
| 0.2 | 1.3 | GO:0070695 | FHF complex(GO:0070695) |
| 0.2 | 0.5 | GO:0034676 | integrin alpha6-beta4 complex(GO:0034676) |
| 0.1 | 1.2 | GO:0045179 | apical cortex(GO:0045179) |
| 0.1 | 0.8 | GO:0036057 | filtration diaphragm(GO:0036056) slit diaphragm(GO:0036057) |
| 0.1 | 0.8 | GO:0097197 | tetraspanin-enriched microdomain(GO:0097197) |
| 0.1 | 0.5 | GO:0097209 | epidermal lamellar body(GO:0097209) |
| 0.1 | 0.2 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.1 | 0.1 | GO:0034667 | integrin alpha3-beta1 complex(GO:0034667) |
| 0.1 | 0.2 | GO:0034686 | integrin alphav-beta8 complex(GO:0034686) |
| 0.1 | 0.4 | GO:0016342 | catenin complex(GO:0016342) |
| 0.1 | 0.9 | GO:0005916 | fascia adherens(GO:0005916) |
| 0.1 | 1.8 | GO:0016327 | apicolateral plasma membrane(GO:0016327) |
| 0.1 | 0.3 | GO:0034678 | integrin alpha8-beta1 complex(GO:0034678) |
| 0.1 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.1 | 1.2 | GO:0005922 | connexon complex(GO:0005922) |
| 0.1 | 0.4 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.1 | 0.4 | GO:0035976 | AP1 complex(GO:0035976) |
| 0.0 | 1.1 | GO:0005861 | troponin complex(GO:0005861) |
| 0.0 | 0.5 | GO:0005915 | zonula adherens(GO:0005915) |
| 0.0 | 3.0 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 2.1 | GO:0016328 | lateral plasma membrane(GO:0016328) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 2.5 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.2 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.0 | 0.4 | GO:0030056 | hemidesmosome(GO:0030056) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.8 | GO:0030673 | axolemma(GO:0030673) |
| 0.0 | 0.1 | GO:0038039 | G-protein coupled receptor heterodimeric complex(GO:0038039) |
| 0.0 | 0.2 | GO:0097418 | neurofibrillary tangle(GO:0097418) |
| 0.0 | 0.1 | GO:0002139 | stereocilia coupling link(GO:0002139) |
| 0.0 | 0.0 | GO:0017109 | glutamate-cysteine ligase complex(GO:0017109) |
| 0.0 | 0.1 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0035339 | SPOTS complex(GO:0035339) |
| 0.0 | 0.0 | GO:0005606 | laminin-1 complex(GO:0005606) laminin-10 complex(GO:0043259) |
| 0.0 | 0.6 | GO:0001772 | immunological synapse(GO:0001772) |
| 0.0 | 0.1 | GO:0032389 | MutLalpha complex(GO:0032389) |
| 0.0 | 0.1 | GO:0032039 | integrator complex(GO:0032039) |
| 0.0 | 0.8 | GO:0055038 | recycling endosome membrane(GO:0055038) |
| 0.0 | 0.0 | GO:0034657 | GID complex(GO:0034657) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.1 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.1 | 0.4 | GO:0005171 | hepatocyte growth factor receptor binding(GO:0005171) |
| 0.1 | 0.3 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 1.2 | GO:0031014 | troponin T binding(GO:0031014) |
| 0.1 | 0.8 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.1 | 1.0 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.1 | 0.7 | GO:0015220 | choline transmembrane transporter activity(GO:0015220) |
| 0.1 | 0.2 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.1 | 1.1 | GO:0008970 | phosphatidylcholine 1-acylhydrolase activity(GO:0008970) |
| 0.1 | 0.2 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.1 | 0.1 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.1 | 0.3 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.1 | 0.2 | GO:0034189 | very-low-density lipoprotein particle binding(GO:0034189) |
| 0.1 | 1.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.2 | GO:0097363 | protein O-GlcNAc transferase activity(GO:0097363) |
| 0.0 | 0.2 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.0 | 0.6 | GO:0008517 | folic acid transporter activity(GO:0008517) |
| 0.0 | 0.2 | GO:0005294 | neutral L-amino acid secondary active transmembrane transporter activity(GO:0005294) |
| 0.0 | 0.9 | GO:0043274 | phospholipase binding(GO:0043274) |
| 0.0 | 0.2 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.9 | GO:0032794 | GTPase activating protein binding(GO:0032794) |
| 0.0 | 0.2 | GO:1990430 | extracellular matrix protein binding(GO:1990430) |
| 0.0 | 0.5 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 1.4 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0003839 | gamma-glutamylcyclotransferase activity(GO:0003839) |
| 0.0 | 0.1 | GO:0036080 | GDP-fucose transmembrane transporter activity(GO:0005457) purine nucleotide-sugar transmembrane transporter activity(GO:0036080) |
| 0.0 | 0.2 | GO:0045505 | dynein intermediate chain binding(GO:0045505) |
| 0.0 | 0.3 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.5 | GO:0004089 | carbonate dehydratase activity(GO:0004089) |
| 0.0 | 0.1 | GO:0047977 | hepoxilin-epoxide hydrolase activity(GO:0047977) |
| 0.0 | 0.2 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.0 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.0 | 0.2 | GO:0070097 | delta-catenin binding(GO:0070097) |
| 0.0 | 0.3 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.1 | GO:0043183 | vascular endothelial growth factor receptor 1 binding(GO:0043183) |
| 0.0 | 0.1 | GO:0086006 | voltage-gated sodium channel activity involved in cardiac muscle cell action potential(GO:0086006) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.4 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.0 | GO:0017098 | sulfonylurea receptor binding(GO:0017098) |
| 0.0 | 0.1 | GO:0017060 | 3-galactosyl-N-acetylglucosaminide 4-alpha-L-fucosyltransferase activity(GO:0017060) |
| 0.0 | 0.1 | GO:0052595 | tryptamine:oxygen oxidoreductase (deaminating) activity(GO:0052593) aminoacetone:oxygen oxidoreductase(deaminating) activity(GO:0052594) aliphatic-amine oxidase activity(GO:0052595) phenethylamine:oxygen oxidoreductase (deaminating) activity(GO:0052596) |
| 0.0 | 0.1 | GO:0098782 | mechanically-gated potassium channel activity(GO:0098782) |
| 0.0 | 0.1 | GO:0097603 | temperature-gated ion channel activity(GO:0097603) |
| 0.0 | 0.1 | GO:0023030 | MHC class Ib protein complex binding(GO:0023025) MHC class Ib protein binding, via antigen binding groove(GO:0023030) |
| 0.0 | 1.2 | GO:0019894 | kinesin binding(GO:0019894) |
| 0.0 | 0.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) secondary active organic cation transmembrane transporter activity(GO:0008513) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.4 | GO:0034450 | ubiquitin-ubiquitin ligase activity(GO:0034450) |
| 0.0 | 0.1 | GO:0070051 | fibrinogen binding(GO:0070051) |
| 0.0 | 0.3 | GO:0051011 | microtubule minus-end binding(GO:0051011) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.1 | GO:0004706 | JUN kinase kinase kinase activity(GO:0004706) |
| 0.0 | 0.3 | GO:0001161 | intronic transcription regulatory region sequence-specific DNA binding(GO:0001161) intronic transcription regulatory region DNA binding(GO:0044213) |
| 0.0 | 0.1 | GO:0016941 | natriuretic peptide receptor activity(GO:0016941) |
| 0.0 | 0.1 | GO:0004771 | sterol esterase activity(GO:0004771) |
| 0.0 | 0.1 | GO:0005497 | androgen binding(GO:0005497) |
| 0.0 | 0.1 | GO:0050262 | ribosylnicotinamide kinase activity(GO:0050262) ribosylnicotinate kinase activity(GO:0061769) |
| 0.0 | 0.0 | GO:0004357 | glutamate-cysteine ligase activity(GO:0004357) |
| 0.0 | 0.1 | GO:0004995 | tachykinin receptor activity(GO:0004995) |
| 0.0 | 0.1 | GO:1990254 | keratin filament binding(GO:1990254) |
| 0.0 | 0.2 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.3 | GO:0019841 | retinol binding(GO:0019841) |
| 0.0 | 0.3 | GO:0015245 | fatty acid transporter activity(GO:0015245) |
| 0.0 | 0.1 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.0 | 0.3 | GO:0015095 | magnesium ion transmembrane transporter activity(GO:0015095) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.1 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.1 | GO:0008310 | single-stranded DNA 3'-5' exodeoxyribonuclease activity(GO:0008310) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0008745 | N-acetylmuramoyl-L-alanine amidase activity(GO:0008745) |
| 0.0 | 0.0 | GO:0047275 | glucosaminylgalactosylglucosylceramide beta-galactosyltransferase activity(GO:0047275) |
| 0.0 | 0.1 | GO:0004965 | G-protein coupled GABA receptor activity(GO:0004965) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 1.1 | GO:0019003 | GDP binding(GO:0019003) |
| 0.0 | 0.1 | GO:0015119 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.3 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.1 | GO:0003938 | IMP dehydrogenase activity(GO:0003938) |
| 0.0 | 0.1 | GO:0004873 | asialoglycoprotein receptor activity(GO:0004873) |
| 0.0 | 0.2 | GO:0004143 | diacylglycerol kinase activity(GO:0004143) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 1.0 | GO:0005544 | calcium-dependent phospholipid binding(GO:0005544) |
| 0.0 | 0.1 | GO:0005353 | fructose transmembrane transporter activity(GO:0005353) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
| 0.0 | 0.0 | GO:0034988 | Fc-gamma receptor I complex binding(GO:0034988) |
| 0.0 | 0.0 | GO:0016639 | oxidoreductase activity, acting on the CH-NH2 group of donors, NAD or NADP as acceptor(GO:0016639) |
| 0.0 | 0.1 | GO:0016901 | glycerol-3-phosphate dehydrogenase activity(GO:0004368) oxidoreductase activity, acting on the CH-OH group of donors, quinone or similar compound as acceptor(GO:0016901) |
| 0.0 | 0.2 | GO:0030506 | ankyrin binding(GO:0030506) |
| 0.0 | 0.3 | GO:0005545 | 1-phosphatidylinositol binding(GO:0005545) |
| 0.0 | 0.2 | GO:0019911 | structural constituent of myelin sheath(GO:0019911) |
| 0.0 | 1.4 | GO:0004867 | serine-type endopeptidase inhibitor activity(GO:0004867) |
| 0.0 | 0.6 | GO:0070412 | R-SMAD binding(GO:0070412) |
| 0.0 | 0.1 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.3 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.2 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 1.7 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.6 | PID NECTIN PATHWAY | Nectin adhesion pathway |
| 0.0 | 0.7 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.1 | ST GA12 PATHWAY | G alpha 12 Pathway |
| 0.0 | 0.0 | SA FAS SIGNALING | The TNF-type receptor Fas induces apoptosis on ligand binding. |
| 0.0 | 0.5 | PID WNT SIGNALING PATHWAY | Wnt signaling network |
| 0.0 | 0.9 | PID WNT NONCANONICAL PATHWAY | Noncanonical Wnt signaling pathway |
| 0.0 | 0.4 | PID ANTHRAX PATHWAY | Cellular roles of Anthrax toxin |
| 0.0 | 0.3 | PID ARF6 DOWNSTREAM PATHWAY | Arf6 downstream pathway |
| 0.0 | 0.8 | PID ALPHA SYNUCLEIN PATHWAY | Alpha-synuclein signaling |
| 0.0 | 0.1 | PID NEPHRIN NEPH1 PATHWAY | Nephrin/Neph1 signaling in the kidney podocyte |
| 0.0 | 0.2 | PID INTEGRIN3 PATHWAY | Beta3 integrin cell surface interactions |
| 0.0 | 0.8 | PID AJDISS 2PATHWAY | Posttranslational regulation of adherens junction stability and dissassembly |
| 0.0 | 0.2 | PID P38 GAMMA DELTA PATHWAY | Signaling mediated by p38-gamma and p38-delta |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME APOPTOTIC CLEAVAGE OF CELL ADHESION PROTEINS | Genes involved in Apoptotic cleavage of cell adhesion proteins |
| 0.1 | 2.6 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.1 | 1.2 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.1 | 1.6 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.7 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 1.1 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.5 | REACTOME DSCAM INTERACTIONS | Genes involved in DSCAM interactions |
| 0.0 | 0.5 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 0.8 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME ACYL CHAIN REMODELLING OF PS | Genes involved in Acyl chain remodelling of PS |
| 0.0 | 1.0 | REACTOME CELL JUNCTION ORGANIZATION | Genes involved in Cell junction organization |
| 0.0 | 1.9 | REACTOME INTERFERON GAMMA SIGNALING | Genes involved in Interferon gamma signaling |
| 0.0 | 0.3 | REACTOME COMMON PATHWAY | Genes involved in Common Pathway |
| 0.0 | 0.4 | REACTOME TRANSPORT OF ORGANIC ANIONS | Genes involved in Transport of organic anions |
| 0.0 | 0.4 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.4 | REACTOME TIE2 SIGNALING | Genes involved in Tie2 Signaling |
| 0.0 | 0.2 | REACTOME P38MAPK EVENTS | Genes involved in p38MAPK events |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |