Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for MNT_HEY1_HEY2
Z-value: 0.94
Transcription factors associated with MNT_HEY1_HEY2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
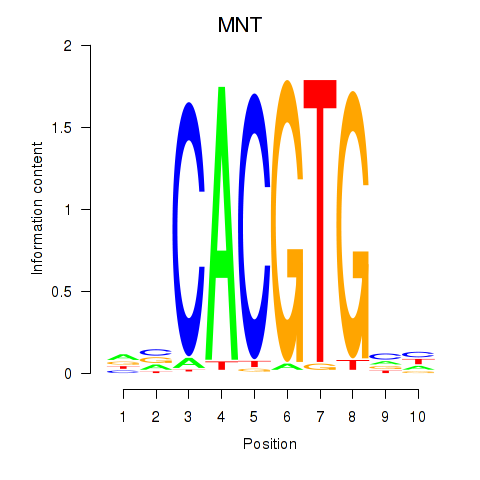
MNT
|
ENSG00000070444.10 | MNT |
|
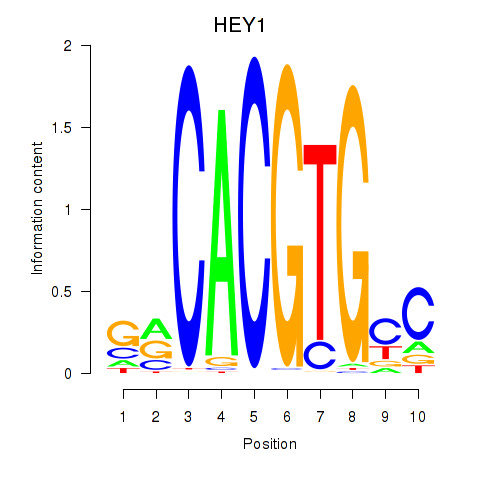
HEY1
|
ENSG00000164683.12 | HEY1 |
|
HEY2
|
ENSG00000135547.4 | HEY2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| MNT | hg19_v2_chr17_-_2304365_2304412 | 0.65 | 7.9e-02 | Click! |
| HEY1 | hg19_v2_chr8_-_80680078_80680101 | 0.18 | 6.7e-01 | Click! |
| HEY2 | hg19_v2_chr6_+_126070726_126070768 | -0.05 | 9.0e-01 | Click! |
Activity profile of MNT_HEY1_HEY2 motif
Sorted Z-values of MNT_HEY1_HEY2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of MNT_HEY1_HEY2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr16_+_66914264 | 1.42 |
ENST00000311765.2 ENST00000568869.1 ENST00000561704.1 ENST00000568398.1 ENST00000566776.1 |
PDP2 |
pyruvate dehyrogenase phosphatase catalytic subunit 2 |
| chr22_+_40390930 | 0.76 |
ENST00000333407.6 |
FAM83F |
family with sequence similarity 83, member F |
| chr11_+_72929402 | 0.73 |
ENST00000393596.2 |
P2RY2 |
purinergic receptor P2Y, G-protein coupled, 2 |
| chr11_+_72929319 | 0.72 |
ENST00000393597.2 ENST00000311131.2 |
P2RY2 |
purinergic receptor P2Y, G-protein coupled, 2 |
| chr18_+_33877654 | 0.62 |
ENST00000257209.4 ENST00000445677.1 ENST00000590592.1 ENST00000359247.4 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr5_-_158636512 | 0.61 |
ENST00000424310.2 |
RNF145 |
ring finger protein 145 |
| chr6_-_39197226 | 0.60 |
ENST00000359534.3 |
KCNK5 |
potassium channel, subfamily K, member 5 |
| chr1_-_159915386 | 0.58 |
ENST00000361509.3 ENST00000368094.1 |
IGSF9 |
immunoglobulin superfamily, member 9 |
| chr4_+_40198527 | 0.58 |
ENST00000381799.5 |
RHOH |
ras homolog family member H |
| chr3_+_10857885 | 0.53 |
ENST00000254488.2 ENST00000454147.1 |
SLC6A11 |
solute carrier family 6 (neurotransmitter transporter), member 11 |
| chr10_+_99344071 | 0.52 |
ENST00000370647.4 ENST00000370646.4 |
HOGA1 |
4-hydroxy-2-oxoglutarate aldolase 1 |
| chr19_-_19739007 | 0.51 |
ENST00000586703.1 ENST00000591042.1 ENST00000407877.3 |
LPAR2 |
lysophosphatidic acid receptor 2 |
| chr8_+_75896731 | 0.51 |
ENST00000262207.4 |
CRISPLD1 |
cysteine-rich secretory protein LCCL domain containing 1 |
| chr1_+_11866270 | 0.50 |
ENST00000376497.3 ENST00000376487.3 ENST00000376496.3 |
CLCN6 |
chloride channel, voltage-sensitive 6 |
| chrX_+_23685563 | 0.48 |
ENST00000379341.4 |
PRDX4 |
peroxiredoxin 4 |
| chr7_+_26191809 | 0.43 |
ENST00000056233.3 |
NFE2L3 |
nuclear factor, erythroid 2-like 3 |
| chr4_+_128651530 | 0.43 |
ENST00000281154.4 |
SLC25A31 |
solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 31 |
| chr7_+_116593433 | 0.42 |
ENST00000323984.3 ENST00000393449.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr17_-_17109579 | 0.41 |
ENST00000321560.3 |
PLD6 |
phospholipase D family, member 6 |
| chr7_+_116593568 | 0.41 |
ENST00000446490.1 |
ST7 |
suppression of tumorigenicity 7 |
| chr17_-_39093672 | 0.40 |
ENST00000209718.3 ENST00000436344.3 ENST00000485751.1 |
KRT23 |
keratin 23 (histone deacetylase inducible) |
| chrX_+_105937068 | 0.39 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr9_-_131709858 | 0.38 |
ENST00000372586.3 |
DOLK |
dolichol kinase |
| chr11_+_121322832 | 0.38 |
ENST00000260197.7 |
SORL1 |
sortilin-related receptor, L(DLR class) A repeats containing |
| chr6_+_7727030 | 0.36 |
ENST00000283147.6 |
BMP6 |
bone morphogenetic protein 6 |
| chr3_-_189840223 | 0.36 |
ENST00000427335.2 |
LEPREL1 |
leprecan-like 1 |
| chr17_-_38657849 | 0.35 |
ENST00000254051.6 |
TNS4 |
tensin 4 |
| chr3_-_184079382 | 0.34 |
ENST00000344937.7 ENST00000423355.2 ENST00000434054.2 ENST00000457512.1 ENST00000265593.4 |
CLCN2 |
chloride channel, voltage-sensitive 2 |
| chr6_-_13486369 | 0.34 |
ENST00000558378.1 |
AL583828.1 |
AL583828.1 |
| chr4_-_1400119 | 0.34 |
ENST00000422806.1 |
NKX1-1 |
NK1 homeobox 1 |
| chr17_+_54671047 | 0.34 |
ENST00000332822.4 |
NOG |
noggin |
| chr1_+_150254936 | 0.34 |
ENST00000447007.1 ENST00000369095.1 ENST00000369094.1 |
C1orf51 |
chromosome 1 open reading frame 51 |
| chr7_+_75024903 | 0.34 |
ENST00000323819.3 ENST00000430211.1 |
TRIM73 |
tripartite motif containing 73 |
| chr17_+_48503603 | 0.33 |
ENST00000502667.1 |
ACSF2 |
acyl-CoA synthetase family member 2 |
| chr20_-_6103666 | 0.33 |
ENST00000536936.1 |
FERMT1 |
fermitin family member 1 |
| chr7_-_138720763 | 0.33 |
ENST00000275766.1 |
ZC3HAV1L |
zinc finger CCCH-type, antiviral 1-like |
| chr2_+_64681641 | 0.32 |
ENST00000409537.2 |
LGALSL |
lectin, galactoside-binding-like |
| chr4_-_6202247 | 0.32 |
ENST00000409021.3 ENST00000409371.3 |
JAKMIP1 |
janus kinase and microtubule interacting protein 1 |
| chr16_+_68679193 | 0.31 |
ENST00000581171.1 |
CDH3 |
cadherin 3, type 1, P-cadherin (placental) |
| chr2_+_220042933 | 0.31 |
ENST00000430297.2 |
FAM134A |
family with sequence similarity 134, member A |
| chr4_+_164265035 | 0.31 |
ENST00000338566.3 |
NPY5R |
neuropeptide Y receptor Y5 |
| chr7_-_72439997 | 0.31 |
ENST00000285805.3 |
TRIM74 |
tripartite motif containing 74 |
| chr11_+_67159416 | 0.30 |
ENST00000307980.2 ENST00000544620.1 |
RAD9A |
RAD9 homolog A (S. pombe) |
| chr16_-_88923285 | 0.30 |
ENST00000542788.1 ENST00000569433.1 ENST00000268695.5 ENST00000568311.1 |
GALNS |
galactosamine (N-acetyl)-6-sulfate sulfatase |
| chr3_+_122785895 | 0.30 |
ENST00000316218.7 |
PDIA5 |
protein disulfide isomerase family A, member 5 |
| chr17_+_48503519 | 0.30 |
ENST00000300441.4 ENST00000541920.1 ENST00000506582.1 ENST00000504392.1 ENST00000427954.2 |
ACSF2 |
acyl-CoA synthetase family member 2 |
| chr7_-_72742085 | 0.30 |
ENST00000333149.2 |
TRIM50 |
tripartite motif containing 50 |
| chr6_+_138188551 | 0.30 |
ENST00000237289.4 ENST00000433680.1 |
TNFAIP3 |
tumor necrosis factor, alpha-induced protein 3 |
| chrX_+_54556633 | 0.29 |
ENST00000336470.4 ENST00000360845.2 |
GNL3L |
guanine nucleotide binding protein-like 3 (nucleolar)-like |
| chr1_-_154193091 | 0.29 |
ENST00000362076.4 ENST00000350592.3 ENST00000368516.1 |
C1orf43 |
chromosome 1 open reading frame 43 |
| chr15_-_83316254 | 0.29 |
ENST00000567678.1 ENST00000450751.2 |
CPEB1 |
cytoplasmic polyadenylation element binding protein 1 |
| chr17_+_30813576 | 0.28 |
ENST00000313401.3 |
CDK5R1 |
cyclin-dependent kinase 5, regulatory subunit 1 (p35) |
| chr5_-_175964366 | 0.28 |
ENST00000274811.4 |
RNF44 |
ring finger protein 44 |
| chr3_+_113666748 | 0.28 |
ENST00000330212.3 ENST00000498275.1 |
ZDHHC23 |
zinc finger, DHHC-type containing 23 |
| chr2_+_17721920 | 0.28 |
ENST00000295156.4 |
VSNL1 |
visinin-like 1 |
| chr1_-_154193009 | 0.28 |
ENST00000368518.1 ENST00000368519.1 ENST00000368521.5 |
C1orf43 |
chromosome 1 open reading frame 43 |
| chr7_+_2559399 | 0.28 |
ENST00000222725.5 ENST00000359574.3 |
LFNG |
LFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chrX_+_16804544 | 0.28 |
ENST00000380122.5 ENST00000398155.4 |
TXLNG |
taxilin gamma |
| chr5_+_150827143 | 0.28 |
ENST00000243389.3 ENST00000517945.1 ENST00000521925.1 |
SLC36A1 |
solute carrier family 36 (proton/amino acid symporter), member 1 |
| chr1_+_28261621 | 0.27 |
ENST00000549094.1 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr19_-_45909585 | 0.27 |
ENST00000593226.1 ENST00000418234.2 |
PPP1R13L |
protein phosphatase 1, regulatory subunit 13 like |
| chr17_-_2614927 | 0.27 |
ENST00000435359.1 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
| chr1_-_113498943 | 0.27 |
ENST00000369626.3 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr17_+_55163075 | 0.27 |
ENST00000571629.1 ENST00000570423.1 ENST00000575186.1 ENST00000573085.1 ENST00000572814.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr1_+_210111570 | 0.27 |
ENST00000367019.1 ENST00000472886.1 |
SYT14 |
synaptotagmin XIV |
| chr1_+_28261492 | 0.27 |
ENST00000373894.3 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr11_+_76494253 | 0.26 |
ENST00000333090.4 |
TSKU |
tsukushi, small leucine rich proteoglycan |
| chr5_-_1112141 | 0.26 |
ENST00000264930.5 |
SLC12A7 |
solute carrier family 12 (potassium/chloride transporter), member 7 |
| chr17_-_6915646 | 0.26 |
ENST00000574377.1 ENST00000399541.2 ENST00000399540.2 ENST00000575727.1 ENST00000573939.1 |
AC027763.2 |
Uncharacterized protein |
| chr1_-_113498616 | 0.26 |
ENST00000433570.4 ENST00000538576.1 ENST00000458229.1 |
SLC16A1 |
solute carrier family 16 (monocarboxylate transporter), member 1 |
| chr11_+_74951948 | 0.26 |
ENST00000562197.2 |
TPBGL |
trophoblast glycoprotein-like |
| chr1_+_28261533 | 0.25 |
ENST00000411604.1 ENST00000373888.4 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr11_+_7534999 | 0.25 |
ENST00000528947.1 ENST00000299492.4 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr12_-_121342170 | 0.25 |
ENST00000353487.2 |
SPPL3 |
signal peptide peptidase like 3 |
| chr15_-_72668805 | 0.24 |
ENST00000268097.5 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr16_+_5121814 | 0.24 |
ENST00000262374.5 ENST00000586840.1 |
ALG1 |
ALG1, chitobiosyldiphosphodolichol beta-mannosyltransferase |
| chr7_-_27135591 | 0.24 |
ENST00000343060.4 ENST00000355633.5 |
HOXA1 |
homeobox A1 |
| chr15_+_98503922 | 0.24 |
ENST00000268042.6 |
ARRDC4 |
arrestin domain containing 4 |
| chr14_+_39736582 | 0.23 |
ENST00000556148.1 ENST00000348007.3 |
CTAGE5 |
CTAGE family, member 5 |
| chr14_-_54908043 | 0.23 |
ENST00000556113.1 ENST00000553660.1 ENST00000395573.4 ENST00000557690.1 ENST00000216416.4 |
CNIH1 |
cornichon family AMPA receptor auxiliary protein 1 |
| chr6_+_123110465 | 0.23 |
ENST00000539041.1 |
SMPDL3A |
sphingomyelin phosphodiesterase, acid-like 3A |
| chr11_+_126225529 | 0.23 |
ENST00000227495.6 ENST00000444328.2 ENST00000356132.4 |
ST3GAL4 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 4 |
| chr16_+_84178874 | 0.23 |
ENST00000378553.5 |
DNAAF1 |
dynein, axonemal, assembly factor 1 |
| chr14_-_91526922 | 0.23 |
ENST00000418736.2 ENST00000261991.3 |
RPS6KA5 |
ribosomal protein S6 kinase, 90kDa, polypeptide 5 |
| chr11_-_125366089 | 0.23 |
ENST00000366139.3 ENST00000278919.3 |
FEZ1 |
fasciculation and elongation protein zeta 1 (zygin I) |
| chr7_+_23636992 | 0.23 |
ENST00000307471.3 ENST00000409765.1 |
CCDC126 |
coiled-coil domain containing 126 |
| chr11_-_66336060 | 0.23 |
ENST00000310325.5 |
CTSF |
cathepsin F |
| chr9_-_33447584 | 0.23 |
ENST00000297991.4 |
AQP3 |
aquaporin 3 (Gill blood group) |
| chr4_-_1107306 | 0.23 |
ENST00000433731.2 ENST00000333673.5 ENST00000382968.5 |
RNF212 |
ring finger protein 212 |
| chr10_-_46090334 | 0.22 |
ENST00000395771.3 ENST00000319836.3 |
MARCH8 |
membrane-associated ring finger (C3HC4) 8, E3 ubiquitin protein ligase |
| chr15_+_27112948 | 0.22 |
ENST00000555060.1 |
GABRA5 |
gamma-aminobutyric acid (GABA) A receptor, alpha 5 |
| chr17_-_8059638 | 0.22 |
ENST00000584202.1 ENST00000354903.5 ENST00000577253.1 |
PER1 |
period circadian clock 1 |
| chr6_-_44225231 | 0.22 |
ENST00000538577.1 ENST00000537814.1 ENST00000393810.1 ENST00000393812.3 |
SLC35B2 |
solute carrier family 35 (adenosine 3'-phospho 5'-phosphosulfate transporter), member B2 |
| chr1_+_152486950 | 0.22 |
ENST00000368790.3 |
CRCT1 |
cysteine-rich C-terminal 1 |
| chr10_-_50970322 | 0.22 |
ENST00000374103.4 |
OGDHL |
oxoglutarate dehydrogenase-like |
| chr12_+_50355647 | 0.22 |
ENST00000293599.6 |
AQP5 |
aquaporin 5 |
| chr21_-_38445470 | 0.22 |
ENST00000399098.1 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chrX_-_128977364 | 0.22 |
ENST00000371064.3 |
ZDHHC9 |
zinc finger, DHHC-type containing 9 |
| chr21_-_38445443 | 0.22 |
ENST00000360525.4 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr2_-_11810284 | 0.22 |
ENST00000306928.5 |
NTSR2 |
neurotensin receptor 2 |
| chr1_+_154193325 | 0.22 |
ENST00000428931.1 ENST00000441890.1 ENST00000271877.7 ENST00000412596.1 ENST00000368504.1 ENST00000437652.1 |
UBAP2L |
ubiquitin associated protein 2-like |
| chr15_-_72668185 | 0.22 |
ENST00000457859.2 ENST00000566304.1 ENST00000567159.1 ENST00000429918.2 |
HEXA |
hexosaminidase A (alpha polypeptide) |
| chr19_+_7587491 | 0.22 |
ENST00000264079.6 |
MCOLN1 |
mucolipin 1 |
| chr16_+_28986134 | 0.22 |
ENST00000352260.7 |
SPNS1 |
spinster homolog 1 (Drosophila) |
| chr6_-_91006627 | 0.22 |
ENST00000537989.1 |
BACH2 |
BTB and CNC homology 1, basic leucine zipper transcription factor 2 |
| chr6_-_167369612 | 0.22 |
ENST00000507747.1 |
RP11-514O12.4 |
RP11-514O12.4 |
| chr7_+_4815238 | 0.22 |
ENST00000348624.4 ENST00000401897.1 |
AP5Z1 |
adaptor-related protein complex 5, zeta 1 subunit |
| chrX_+_23685653 | 0.22 |
ENST00000379331.3 |
PRDX4 |
peroxiredoxin 4 |
| chr1_+_101361626 | 0.22 |
ENST00000370112.4 |
SLC30A7 |
solute carrier family 30 (zinc transporter), member 7 |
| chr4_-_170924888 | 0.21 |
ENST00000502832.1 ENST00000393704.3 |
MFAP3L |
microfibrillar-associated protein 3-like |
| chr16_+_67282853 | 0.21 |
ENST00000299798.11 |
SLC9A5 |
solute carrier family 9, subfamily A (NHE5, cation proton antiporter 5), member 5 |
| chr2_-_120980939 | 0.21 |
ENST00000426077.2 |
TMEM185B |
transmembrane protein 185B |
| chr1_+_45140360 | 0.21 |
ENST00000418644.1 ENST00000458657.2 ENST00000441519.1 ENST00000535358.1 ENST00000445071.1 |
C1orf228 |
chromosome 1 open reading frame 228 |
| chr2_+_10183651 | 0.21 |
ENST00000305883.1 |
KLF11 |
Kruppel-like factor 11 |
| chr2_+_17721230 | 0.21 |
ENST00000457525.1 |
VSNL1 |
visinin-like 1 |
| chr19_+_42259329 | 0.21 |
ENST00000199764.6 |
CEACAM6 |
carcinoembryonic antigen-related cell adhesion molecule 6 (non-specific cross reacting antigen) |
| chr9_-_139137648 | 0.21 |
ENST00000358701.5 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
| chr1_+_155099927 | 0.21 |
ENST00000368407.3 |
EFNA1 |
ephrin-A1 |
| chr14_+_93799556 | 0.20 |
ENST00000256339.4 |
UNC79 |
unc-79 homolog (C. elegans) |
| chr19_-_10047219 | 0.20 |
ENST00000264833.4 |
OLFM2 |
olfactomedin 2 |
| chr12_-_123849374 | 0.20 |
ENST00000602398.1 ENST00000602750.1 |
SBNO1 |
strawberry notch homolog 1 (Drosophila) |
| chr18_+_9136758 | 0.20 |
ENST00000383440.2 ENST00000262126.4 ENST00000577992.1 |
ANKRD12 |
ankyrin repeat domain 12 |
| chr17_+_28256874 | 0.20 |
ENST00000541045.1 ENST00000536908.2 |
EFCAB5 |
EF-hand calcium binding domain 5 |
| chr1_+_210111534 | 0.20 |
ENST00000422431.1 ENST00000534859.1 ENST00000399639.2 ENST00000537238.1 |
SYT14 |
synaptotagmin XIV |
| chr8_-_124553437 | 0.20 |
ENST00000517956.1 ENST00000443022.2 |
FBXO32 |
F-box protein 32 |
| chr7_+_150759634 | 0.19 |
ENST00000392826.2 ENST00000461735.1 |
SLC4A2 |
solute carrier family 4 (anion exchanger), member 2 |
| chr1_+_11333245 | 0.19 |
ENST00000376810.5 |
UBIAD1 |
UbiA prenyltransferase domain containing 1 |
| chr11_+_45944190 | 0.19 |
ENST00000401752.1 ENST00000389968.3 ENST00000325468.5 ENST00000536139.1 |
GYLTL1B |
glycosyltransferase-like 1B |
| chr19_+_45504688 | 0.19 |
ENST00000221452.8 ENST00000540120.1 ENST00000505236.1 |
RELB |
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr22_+_29279552 | 0.19 |
ENST00000544604.2 |
ZNRF3 |
zinc and ring finger 3 |
| chr4_+_79472888 | 0.19 |
ENST00000512884.1 ENST00000512542.1 ENST00000503570.2 |
ANXA3 |
annexin A3 |
| chr16_+_28986085 | 0.19 |
ENST00000565975.1 ENST00000311008.11 ENST00000323081.8 ENST00000334536.8 |
SPNS1 |
spinster homolog 1 (Drosophila) |
| chrX_+_55744228 | 0.19 |
ENST00000262850.7 |
RRAGB |
Ras-related GTP binding B |
| chr4_+_79472673 | 0.19 |
ENST00000264908.6 |
ANXA3 |
annexin A3 |
| chr12_-_45270151 | 0.19 |
ENST00000429094.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr1_+_228353495 | 0.19 |
ENST00000366711.3 |
IBA57 |
IBA57, iron-sulfur cluster assembly homolog (S. cerevisiae) |
| chr3_+_112280857 | 0.19 |
ENST00000492406.1 ENST00000468642.1 |
SLC35A5 |
solute carrier family 35, member A5 |
| chr2_-_165477971 | 0.19 |
ENST00000446413.2 |
GRB14 |
growth factor receptor-bound protein 14 |
| chr2_+_173420697 | 0.19 |
ENST00000282077.3 ENST00000392571.2 ENST00000410055.1 |
PDK1 |
pyruvate dehydrogenase kinase, isozyme 1 |
| chr20_+_42839600 | 0.19 |
ENST00000439943.1 ENST00000437730.1 |
OSER1-AS1 |
OSER1 antisense RNA 1 (head to head) |
| chr12_-_45270077 | 0.19 |
ENST00000551601.1 ENST00000549027.1 ENST00000452445.2 |
NELL2 |
NEL-like 2 (chicken) |
| chr13_+_113951532 | 0.18 |
ENST00000332556.4 |
LAMP1 |
lysosomal-associated membrane protein 1 |
| chr19_+_11546440 | 0.18 |
ENST00000589126.1 ENST00000588269.1 ENST00000587509.1 ENST00000592741.1 ENST00000593101.1 ENST00000587327.1 |
PRKCSH |
protein kinase C substrate 80K-H |
| chr20_+_42839722 | 0.18 |
ENST00000442383.1 ENST00000435163.1 |
OSER1-AS1 |
OSER1 antisense RNA 1 (head to head) |
| chr21_-_43373999 | 0.18 |
ENST00000380486.3 |
C2CD2 |
C2 calcium-dependent domain containing 2 |
| chr17_-_46688334 | 0.18 |
ENST00000239165.7 |
HOXB7 |
homeobox B7 |
| chr10_-_6019552 | 0.18 |
ENST00000379977.3 ENST00000397251.3 ENST00000397248.2 |
IL15RA |
interleukin 15 receptor, alpha |
| chr17_-_2304365 | 0.18 |
ENST00000575394.1 ENST00000174618.4 |
MNT |
MAX network transcriptional repressor |
| chr1_-_11866034 | 0.18 |
ENST00000376590.3 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr16_-_2246436 | 0.18 |
ENST00000343516.6 |
CASKIN1 |
CASK interacting protein 1 |
| chr1_+_92414952 | 0.18 |
ENST00000449584.1 ENST00000427104.1 ENST00000355011.3 ENST00000448194.1 ENST00000426141.1 ENST00000450792.1 ENST00000548992.1 ENST00000552654.1 ENST00000457265.1 |
BRDT |
bromodomain, testis-specific |
| chr7_+_23637118 | 0.18 |
ENST00000448353.1 |
CCDC126 |
coiled-coil domain containing 126 |
| chrX_+_55744166 | 0.18 |
ENST00000374941.4 ENST00000414239.1 |
RRAGB |
Ras-related GTP binding B |
| chr17_-_35969409 | 0.18 |
ENST00000394378.2 ENST00000585472.1 ENST00000591288.1 ENST00000502449.2 ENST00000345615.4 ENST00000346661.4 ENST00000585689.1 ENST00000339208.6 |
SYNRG |
synergin, gamma |
| chr11_+_33037401 | 0.18 |
ENST00000241051.3 |
DEPDC7 |
DEP domain containing 7 |
| chr3_-_69062764 | 0.18 |
ENST00000295571.5 |
EOGT |
EGF domain-specific O-linked N-acetylglucosamine (GlcNAc) transferase |
| chr16_-_1525016 | 0.18 |
ENST00000262318.8 ENST00000448525.1 |
CLCN7 |
chloride channel, voltage-sensitive 7 |
| chr2_+_203499901 | 0.18 |
ENST00000303116.6 ENST00000392238.2 |
FAM117B |
family with sequence similarity 117, member B |
| chr12_-_114404111 | 0.18 |
ENST00000545145.2 ENST00000392561.3 ENST00000261741.5 |
RBM19 |
RNA binding motif protein 19 |
| chr17_-_46507537 | 0.17 |
ENST00000336915.6 |
SKAP1 |
src kinase associated phosphoprotein 1 |
| chr17_+_78075498 | 0.17 |
ENST00000302262.3 |
GAA |
glucosidase, alpha; acid |
| chr12_-_15374343 | 0.17 |
ENST00000256953.2 ENST00000546331.1 |
RERG |
RAS-like, estrogen-regulated, growth inhibitor |
| chr9_+_130159504 | 0.17 |
ENST00000373352.1 ENST00000373360.3 |
SLC2A8 |
solute carrier family 2 (facilitated glucose transporter), member 8 |
| chr19_+_1067271 | 0.17 |
ENST00000536472.1 ENST00000590214.1 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr9_-_34637806 | 0.17 |
ENST00000477726.1 |
SIGMAR1 |
sigma non-opioid intracellular receptor 1 |
| chr16_+_76587314 | 0.17 |
ENST00000563764.1 |
RP11-58C22.1 |
Uncharacterized protein |
| chr8_-_54755459 | 0.17 |
ENST00000524234.1 ENST00000521275.1 ENST00000396774.2 |
ATP6V1H |
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr19_+_11546153 | 0.17 |
ENST00000591946.1 ENST00000252455.2 ENST00000412601.1 |
PRKCSH |
protein kinase C substrate 80K-H |
| chr11_-_66725837 | 0.17 |
ENST00000393958.2 ENST00000393960.1 ENST00000524491.1 ENST00000355677.3 |
PC |
pyruvate carboxylase |
| chr21_-_38445297 | 0.17 |
ENST00000430792.1 ENST00000399103.1 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr8_-_54755789 | 0.17 |
ENST00000359530.2 |
ATP6V1H |
ATPase, H+ transporting, lysosomal 50/57kDa, V1 subunit H |
| chr8_-_75233563 | 0.17 |
ENST00000342232.4 |
JPH1 |
junctophilin 1 |
| chr16_+_84002234 | 0.17 |
ENST00000305202.4 |
NECAB2 |
N-terminal EF-hand calcium binding protein 2 |
| chr12_-_49318715 | 0.17 |
ENST00000444214.2 |
FKBP11 |
FK506 binding protein 11, 19 kDa |
| chr3_-_178789220 | 0.17 |
ENST00000414084.1 |
ZMAT3 |
zinc finger, matrin-type 3 |
| chr5_+_78532003 | 0.17 |
ENST00000396137.4 |
JMY |
junction mediating and regulatory protein, p53 cofactor |
| chr7_-_95951432 | 0.17 |
ENST00000416240.2 |
SLC25A13 |
solute carrier family 25 (aspartate/glutamate carrier), member 13 |
| chr2_+_192543153 | 0.17 |
ENST00000425611.2 |
NABP1 |
nucleic acid binding protein 1 |
| chr21_-_38445011 | 0.17 |
ENST00000464265.1 ENST00000399102.1 |
PIGP |
phosphatidylinositol glycan anchor biosynthesis, class P |
| chr19_+_11546093 | 0.17 |
ENST00000591462.1 |
PRKCSH |
protein kinase C substrate 80K-H |
| chrX_-_100662881 | 0.17 |
ENST00000218516.3 |
GLA |
galactosidase, alpha |
| chr12_+_54674482 | 0.17 |
ENST00000547708.1 ENST00000340913.6 ENST00000551702.1 ENST00000330752.8 ENST00000547276.1 |
HNRNPA1 |
heterogeneous nuclear ribonucleoprotein A1 |
| chr7_-_95951334 | 0.17 |
ENST00000265631.5 |
SLC25A13 |
solute carrier family 25 (aspartate/glutamate carrier), member 13 |
| chr19_-_15090488 | 0.17 |
ENST00000594383.1 ENST00000598504.1 ENST00000597262.1 |
SLC1A6 |
solute carrier family 1 (high affinity aspartate/glutamate transporter), member 6 |
| chr20_-_44993012 | 0.17 |
ENST00000372229.1 ENST00000372230.5 ENST00000543605.1 ENST00000243896.2 ENST00000317734.8 |
SLC35C2 |
solute carrier family 35 (GDP-fucose transporter), member C2 |
| chr8_+_145734433 | 0.17 |
ENST00000301327.4 |
MFSD3 |
major facilitator superfamily domain containing 3 |
| chrX_+_153524024 | 0.17 |
ENST00000369915.3 ENST00000217905.7 |
TKTL1 |
transketolase-like 1 |
| chr14_-_105635090 | 0.17 |
ENST00000331782.3 ENST00000347004.2 |
JAG2 |
jagged 2 |
| chr4_-_103266626 | 0.17 |
ENST00000356736.4 |
SLC39A8 |
solute carrier family 39 (zinc transporter), member 8 |
| chr13_+_113548643 | 0.17 |
ENST00000375608.3 |
MCF2L |
MCF.2 cell line derived transforming sequence-like |
| chr17_-_37309480 | 0.17 |
ENST00000539608.1 |
PLXDC1 |
plexin domain containing 1 |
| chr10_+_46222648 | 0.16 |
ENST00000336378.4 ENST00000540872.1 ENST00000537517.1 ENST00000374362.2 ENST00000359860.4 ENST00000420848.1 |
FAM21C |
family with sequence similarity 21, member C |
| chr6_+_44191290 | 0.16 |
ENST00000371755.3 ENST00000371740.5 ENST00000371731.1 ENST00000393841.1 |
SLC29A1 |
solute carrier family 29 (equilibrative nucleoside transporter), member 1 |
| chr19_+_1067144 | 0.16 |
ENST00000313093.2 |
HMHA1 |
histocompatibility (minor) HA-1 |
| chr2_-_31361543 | 0.16 |
ENST00000349752.5 |
GALNT14 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 14 (GalNAc-T14) |
| chr17_+_4402133 | 0.16 |
ENST00000329078.3 |
SPNS2 |
spinster homolog 2 (Drosophila) |
| chr17_+_78075361 | 0.16 |
ENST00000577106.1 ENST00000390015.3 |
GAA |
glucosidase, alpha; acid |
| chr22_+_29469100 | 0.16 |
ENST00000327813.5 ENST00000407188.1 |
KREMEN1 |
kringle containing transmembrane protein 1 |
| chr13_-_103451307 | 0.16 |
ENST00000376004.4 |
KDELC1 |
KDEL (Lys-Asp-Glu-Leu) containing 1 |
| chr5_+_149737202 | 0.16 |
ENST00000451292.1 ENST00000377797.3 ENST00000445265.2 ENST00000323668.7 ENST00000439160.2 ENST00000394269.3 ENST00000427724.2 ENST00000504761.2 ENST00000513346.1 ENST00000515516.1 |
TCOF1 |
Treacher Collins-Franceschetti syndrome 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0015728 | mevalonate transport(GO:0015728) behavioral response to nutrient(GO:0051780) |
| 0.2 | 0.5 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 0.4 | GO:0051300 | spindle pole body duplication(GO:0030474) spindle pole body organization(GO:0051300) spindle pole body localization(GO:0070631) establishment of spindle pole body localization(GO:0070632) spindle pole body localization to nuclear envelope(GO:0071789) establishment of spindle pole body localization to nuclear envelope(GO:0071790) |
| 0.1 | 0.7 | GO:2000255 | negative regulation of male germ cell proliferation(GO:2000255) |
| 0.1 | 0.4 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.6 | GO:0048817 | negative regulation of hair follicle maturation(GO:0048817) |
| 0.1 | 0.4 | GO:1902955 | regulation of choline O-acetyltransferase activity(GO:1902769) positive regulation of choline O-acetyltransferase activity(GO:1902771) negative regulation of tau-protein kinase activity(GO:1902948) positive regulation of early endosome to recycling endosome transport(GO:1902955) negative regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902960) negative regulation of neurofibrillary tangle assembly(GO:1902997) negative regulation of aspartic-type peptidase activity(GO:1905246) |
| 0.1 | 0.5 | GO:0043490 | malate-aspartate shuttle(GO:0043490) |
| 0.1 | 1.4 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.1 | 1.8 | GO:0010510 | regulation of acetyl-CoA biosynthetic process from pyruvate(GO:0010510) |
| 0.1 | 0.3 | GO:0071947 | B-1 B cell homeostasis(GO:0001922) toll-like receptor 5 signaling pathway(GO:0034146) regulation of nucleotide-binding oligomerization domain containing 1 signaling pathway(GO:0070428) protein deubiquitination involved in ubiquitin-dependent protein catabolic process(GO:0071947) negative regulation of osteoclast proliferation(GO:0090291) |
| 0.1 | 0.3 | GO:0021718 | superior olivary nucleus development(GO:0021718) superior olivary nucleus maturation(GO:0021722) |
| 0.1 | 0.5 | GO:0015862 | uridine transport(GO:0015862) |
| 0.1 | 0.1 | GO:1900369 | negative regulation of RNA interference(GO:1900369) |
| 0.1 | 0.4 | GO:0014028 | notochord formation(GO:0014028) |
| 0.1 | 0.3 | GO:0089712 | L-aspartate transport(GO:0070778) L-aspartate transmembrane transport(GO:0089712) |
| 0.1 | 1.1 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.3 | GO:0002086 | diaphragm contraction(GO:0002086) |
| 0.1 | 0.2 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.1 | 0.3 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.1 | 0.1 | GO:0090149 | mitochondrial membrane fission(GO:0090149) |
| 0.1 | 0.2 | GO:0015670 | carbon dioxide transport(GO:0015670) |
| 0.1 | 0.4 | GO:0090031 | positive regulation of steroid hormone biosynthetic process(GO:0090031) |
| 0.1 | 0.2 | GO:1902490 | regulation of sperm capacitation(GO:1902490) |
| 0.1 | 0.3 | GO:0060825 | fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:0060825) regulation of fibroblast growth factor receptor signaling pathway involved in neural plate anterior/posterior pattern formation(GO:2000313) |
| 0.1 | 0.2 | GO:1904020 | regulation of G-protein coupled receptor internalization(GO:1904020) |
| 0.1 | 0.2 | GO:0001207 | histone displacement(GO:0001207) positive regulation of transcription involved in meiotic cell cycle(GO:0051039) |
| 0.1 | 0.1 | GO:0044828 | negative regulation by host of viral genome replication(GO:0044828) |
| 0.1 | 0.2 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.1 | 0.2 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.1 | 0.1 | GO:0016256 | N-glycan processing to lysosome(GO:0016256) |
| 0.1 | 0.1 | GO:0072034 | renal vesicle induction(GO:0072034) |
| 0.1 | 0.2 | GO:0018242 | protein O-linked glycosylation via serine(GO:0018242) |
| 0.1 | 0.2 | GO:0015772 | disaccharide transport(GO:0015766) sucrose transport(GO:0015770) oligosaccharide transport(GO:0015772) |
| 0.1 | 0.2 | GO:1902683 | regulation of receptor localization to synapse(GO:1902683) |
| 0.1 | 0.5 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0032240 | negative regulation of nucleobase-containing compound transport(GO:0032240) negative regulation of RNA export from nucleus(GO:0046832) |
| 0.0 | 0.1 | GO:0048627 | myoblast development(GO:0048627) |
| 0.0 | 0.2 | GO:2000051 | negative regulation of non-canonical Wnt signaling pathway(GO:2000051) |
| 0.0 | 0.4 | GO:0045716 | positive regulation of low-density lipoprotein particle receptor biosynthetic process(GO:0045716) |
| 0.0 | 0.9 | GO:0030322 | stabilization of membrane potential(GO:0030322) |
| 0.0 | 0.2 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.3 | GO:0035469 | determination of pancreatic left/right asymmetry(GO:0035469) |
| 0.0 | 0.2 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.0 | 0.5 | GO:0015860 | purine nucleoside transmembrane transport(GO:0015860) |
| 0.0 | 0.4 | GO:1990253 | cellular response to leucine starvation(GO:1990253) |
| 0.0 | 0.3 | GO:1902231 | positive regulation of intrinsic apoptotic signaling pathway in response to DNA damage(GO:1902231) |
| 0.0 | 0.3 | GO:0014050 | negative regulation of glutamate secretion(GO:0014050) negative regulation of synaptic transmission, GABAergic(GO:0032229) |
| 0.0 | 0.1 | GO:1901899 | positive regulation of relaxation of cardiac muscle(GO:1901899) regulation of calcium ion import into sarcoplasmic reticulum(GO:1902080) negative regulation of calcium ion import into sarcoplasmic reticulum(GO:1902081) |
| 0.0 | 0.5 | GO:0015691 | cadmium ion transport(GO:0015691) cadmium ion transmembrane transport(GO:0070574) |
| 0.0 | 0.2 | GO:0010157 | response to chlorate(GO:0010157) |
| 0.0 | 0.2 | GO:0015808 | L-alanine transport(GO:0015808) |
| 0.0 | 0.2 | GO:1903935 | response to sodium arsenite(GO:1903935) cellular response to sodium arsenite(GO:1903936) |
| 0.0 | 0.2 | GO:0090238 | positive regulation of arachidonic acid secretion(GO:0090238) |
| 0.0 | 0.4 | GO:0010635 | regulation of mitochondrial fusion(GO:0010635) |
| 0.0 | 0.1 | GO:1904933 | regulation of cell proliferation in midbrain(GO:1904933) |
| 0.0 | 0.2 | GO:0071400 | cellular response to oleic acid(GO:0071400) |
| 0.0 | 0.8 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0051182 | coenzyme transport(GO:0051182) |
| 0.0 | 0.3 | GO:0007386 | compartment pattern specification(GO:0007386) |
| 0.0 | 0.4 | GO:0006489 | dolichyl diphosphate biosynthetic process(GO:0006489) dolichyl diphosphate metabolic process(GO:0046465) |
| 0.0 | 0.1 | GO:0061054 | dermatome development(GO:0061054) regulation of dermatome development(GO:0061183) |
| 0.0 | 0.1 | GO:0046946 | hydroxylysine metabolic process(GO:0046946) hydroxylysine biosynthetic process(GO:0046947) |
| 0.0 | 0.3 | GO:0048312 | intracellular distribution of mitochondria(GO:0048312) |
| 0.0 | 0.2 | GO:0009233 | menaquinone metabolic process(GO:0009233) |
| 0.0 | 0.2 | GO:0070829 | response to vitamin B2(GO:0033274) heterochromatin maintenance(GO:0070829) |
| 0.0 | 0.2 | GO:0006311 | meiotic gene conversion(GO:0006311) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.0 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.0 | 0.1 | GO:0032679 | TRAIL production(GO:0032639) regulation of TRAIL production(GO:0032679) positive regulation of TRAIL production(GO:0032759) |
| 0.0 | 0.1 | GO:0010520 | regulation of reciprocal meiotic recombination(GO:0010520) positive regulation of reciprocal meiotic recombination(GO:0010845) |
| 0.0 | 0.3 | GO:0070544 | histone H3-K36 demethylation(GO:0070544) |
| 0.0 | 0.4 | GO:0018231 | peptidyl-L-cysteine S-palmitoylation(GO:0018230) peptidyl-S-diacylglycerol-L-cysteine biosynthetic process from peptidyl-cysteine(GO:0018231) |
| 0.0 | 0.1 | GO:0042489 | negative regulation of odontogenesis of dentin-containing tooth(GO:0042489) |
| 0.0 | 0.2 | GO:0021840 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0042360 | vitamin E metabolic process(GO:0042360) |
| 0.0 | 0.3 | GO:0060689 | cell differentiation involved in salivary gland development(GO:0060689) |
| 0.0 | 0.3 | GO:1904715 | negative regulation of chaperone-mediated autophagy(GO:1904715) |
| 0.0 | 0.2 | GO:0014718 | positive regulation of satellite cell activation involved in skeletal muscle regeneration(GO:0014718) |
| 0.0 | 0.2 | GO:0070358 | actin polymerization-dependent cell motility(GO:0070358) |
| 0.0 | 0.3 | GO:0019368 | fatty acid elongation, saturated fatty acid(GO:0019367) fatty acid elongation, unsaturated fatty acid(GO:0019368) fatty acid elongation, monounsaturated fatty acid(GO:0034625) fatty acid elongation, polyunsaturated fatty acid(GO:0034626) |
| 0.0 | 0.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.8 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.4 | GO:0015712 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.2 | GO:0033299 | secretion of lysosomal enzymes(GO:0033299) |
| 0.0 | 0.1 | GO:0042796 | snRNA transcription from RNA polymerase III promoter(GO:0042796) |
| 0.0 | 0.5 | GO:0015812 | gamma-aminobutyric acid transport(GO:0015812) |
| 0.0 | 0.2 | GO:0061484 | hematopoietic stem cell homeostasis(GO:0061484) |
| 0.0 | 0.1 | GO:0090107 | aminophospholipid transport(GO:0015917) regulation of high-density lipoprotein particle assembly(GO:0090107) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.1 | GO:0033341 | regulation of collagen binding(GO:0033341) |
| 0.0 | 0.1 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.4 | GO:0006488 | dolichol-linked oligosaccharide biosynthetic process(GO:0006488) |
| 0.0 | 0.1 | GO:0031133 | regulation of axon diameter(GO:0031133) |
| 0.0 | 0.1 | GO:1900454 | positive regulation of long term synaptic depression(GO:1900454) |
| 0.0 | 0.1 | GO:0006258 | UDP-glucose catabolic process(GO:0006258) galactose catabolic process via UDP-galactose(GO:0033499) |
| 0.0 | 0.1 | GO:1903565 | negative regulation of protein localization to cilium(GO:1903565) regulation of protein localization to ciliary membrane(GO:1903567) negative regulation of protein localization to ciliary membrane(GO:1903568) |
| 0.0 | 0.1 | GO:0002439 | chronic inflammatory response to antigenic stimulus(GO:0002439) |
| 0.0 | 0.1 | GO:0048205 | COPI-coated vesicle budding(GO:0035964) Golgi transport vesicle coating(GO:0048200) COPI coating of Golgi vesicle(GO:0048205) |
| 0.0 | 0.2 | GO:0060136 | embryonic process involved in female pregnancy(GO:0060136) |
| 0.0 | 0.0 | GO:0035927 | RNA import into mitochondrion(GO:0035927) |
| 0.0 | 0.2 | GO:0051533 | positive regulation of NFAT protein import into nucleus(GO:0051533) |
| 0.0 | 0.5 | GO:0090520 | sphingosine-1-phosphate signaling pathway(GO:0003376) sphingolipid mediated signaling pathway(GO:0090520) |
| 0.0 | 0.1 | GO:1904327 | protein localization to cytosolic proteasome complex(GO:1904327) protein localization to cytosolic proteasome complex involved in ERAD pathway(GO:1904379) |
| 0.0 | 0.4 | GO:0070050 | neuron cellular homeostasis(GO:0070050) |
| 0.0 | 0.0 | GO:0035494 | SNARE complex disassembly(GO:0035494) |
| 0.0 | 0.1 | GO:0017185 | peptidyl-lysine hydroxylation(GO:0017185) |
| 0.0 | 0.1 | GO:0060139 | positive regulation by symbiont of host apoptotic process(GO:0052151) positive regulation of apoptotic process by virus(GO:0060139) |
| 0.0 | 0.1 | GO:1903070 | negative regulation of ER-associated ubiquitin-dependent protein catabolic process(GO:1903070) |
| 0.0 | 0.0 | GO:0051885 | positive regulation of anagen(GO:0051885) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0002101 | tRNA wobble cytosine modification(GO:0002101) |
| 0.0 | 0.1 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.2 | GO:0061088 | regulation of sequestering of zinc ion(GO:0061088) |
| 0.0 | 0.4 | GO:0090073 | positive regulation of protein homodimerization activity(GO:0090073) |
| 0.0 | 0.0 | GO:1900368 | regulation of RNA interference(GO:1900368) |
| 0.0 | 0.2 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.1 | GO:1990834 | response to odorant(GO:1990834) |
| 0.0 | 0.2 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.0 | 0.1 | GO:0051029 | rRNA transport(GO:0051029) |
| 0.0 | 0.1 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.1 | GO:1901993 | meiotic cell cycle phase transition(GO:0044771) regulation of meiotic cell cycle phase transition(GO:1901993) negative regulation of meiotic cell cycle phase transition(GO:1901994) |
| 0.0 | 0.1 | GO:0044778 | meiotic DNA integrity checkpoint(GO:0044778) |
| 0.0 | 0.1 | GO:0032687 | negative regulation of interferon-alpha production(GO:0032687) |
| 0.0 | 0.4 | GO:0038003 | opioid receptor signaling pathway(GO:0038003) |
| 0.0 | 0.2 | GO:1903753 | negative regulation of p38MAPK cascade(GO:1903753) |
| 0.0 | 0.3 | GO:0021603 | cranial nerve formation(GO:0021603) |
| 0.0 | 0.0 | GO:1904378 | maintenance of unfolded protein(GO:0036506) maintenance of unfolded protein involved in ERAD pathway(GO:1904378) |
| 0.0 | 0.2 | GO:0006772 | thiamine metabolic process(GO:0006772) |
| 0.0 | 0.0 | GO:0016080 | synaptic vesicle targeting(GO:0016080) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:0090191 | negative regulation of branching involved in ureteric bud morphogenesis(GO:0090191) |
| 0.0 | 0.1 | GO:0070370 | heat acclimation(GO:0010286) cellular heat acclimation(GO:0070370) |
| 0.0 | 0.2 | GO:0048149 | behavioral response to ethanol(GO:0048149) |
| 0.0 | 0.1 | GO:0097477 | lateral motor column neuron migration(GO:0097477) |
| 0.0 | 0.1 | GO:0032223 | negative regulation of synaptic transmission, cholinergic(GO:0032223) neurotransmitter receptor biosynthetic process(GO:0045212) |
| 0.0 | 0.6 | GO:0006884 | cell volume homeostasis(GO:0006884) |
| 0.0 | 0.1 | GO:0043335 | protein unfolding(GO:0043335) |
| 0.0 | 0.6 | GO:0007035 | vacuolar acidification(GO:0007035) |
| 0.0 | 0.1 | GO:0071596 | ubiquitin-dependent protein catabolic process via the N-end rule pathway(GO:0071596) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.4 | GO:0034975 | protein folding in endoplasmic reticulum(GO:0034975) |
| 0.0 | 0.1 | GO:1901029 | negative regulation of mitochondrial outer membrane permeabilization involved in apoptotic signaling pathway(GO:1901029) |
| 0.0 | 0.4 | GO:1902236 | negative regulation of endoplasmic reticulum stress-induced intrinsic apoptotic signaling pathway(GO:1902236) |
| 0.0 | 0.3 | GO:0035878 | nail development(GO:0035878) |
| 0.0 | 0.2 | GO:0042492 | gamma-delta T cell differentiation(GO:0042492) |
| 0.0 | 0.1 | GO:1901895 | negative regulation of calcium-transporting ATPase activity(GO:1901895) |
| 0.0 | 0.1 | GO:1904885 | beta-catenin destruction complex assembly(GO:1904885) |
| 0.0 | 0.2 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0045661 | regulation of myoblast differentiation(GO:0045661) |
| 0.0 | 0.2 | GO:0014894 | response to muscle inactivity involved in regulation of muscle adaptation(GO:0014877) response to denervation involved in regulation of muscle adaptation(GO:0014894) |
| 0.0 | 0.4 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.1 | GO:2000664 | positive regulation of interleukin-5 secretion(GO:2000664) |
| 0.0 | 0.1 | GO:2000794 | positive regulation of epithelial cell proliferation involved in lung morphogenesis(GO:0060501) epithelial cell proliferation involved in lung morphogenesis(GO:0060502) regulation of epithelial cell proliferation involved in lung morphogenesis(GO:2000794) |
| 0.0 | 0.2 | GO:0044341 | sodium-dependent phosphate transport(GO:0044341) |
| 0.0 | 0.0 | GO:2000506 | negative regulation of energy homeostasis(GO:2000506) |
| 0.0 | 0.1 | GO:0060671 | epithelial cell differentiation involved in embryonic placenta development(GO:0060671) epithelial cell morphogenesis involved in placental branching(GO:0060672) |
| 0.0 | 0.1 | GO:0000472 | endonucleolytic cleavage to generate mature 5'-end of SSU-rRNA from (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000472) rRNA 5'-end processing(GO:0000967) ncRNA 5'-end processing(GO:0034471) |
| 0.0 | 0.5 | GO:0006491 | N-glycan processing(GO:0006491) |
| 0.0 | 0.0 | GO:2000662 | interleukin-5 secretion(GO:0072603) regulation of interleukin-5 secretion(GO:2000662) |
| 0.0 | 0.0 | GO:0035281 | pre-miRNA export from nucleus(GO:0035281) |
| 0.0 | 0.1 | GO:2000393 | negative regulation of lamellipodium morphogenesis(GO:2000393) |
| 0.0 | 0.2 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.1 | GO:0044387 | negative regulation of protein kinase activity by regulation of protein phosphorylation(GO:0044387) |
| 0.0 | 0.1 | GO:1903588 | negative regulation of blood vessel endothelial cell proliferation involved in sprouting angiogenesis(GO:1903588) |
| 0.0 | 0.0 | GO:0021898 | regulation of transcription from RNA polymerase II promoter involved in forebrain neuron fate commitment(GO:0021882) cerebral cortex GABAergic interneuron fate commitment(GO:0021893) commitment of multipotent stem cells to neuronal lineage in forebrain(GO:0021898) positive regulation of neural retina development(GO:0061075) positive regulation of retina development in camera-type eye(GO:1902868) regulation of amacrine cell differentiation(GO:1902869) positive regulation of amacrine cell differentiation(GO:1902871) |
| 0.0 | 0.2 | GO:1903025 | regulation of RNA polymerase II regulatory region sequence-specific DNA binding(GO:1903025) |
| 0.0 | 0.0 | GO:0007113 | endomitotic cell cycle(GO:0007113) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.0 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.1 | GO:1901222 | regulation of NIK/NF-kappaB signaling(GO:1901222) |
| 0.0 | 0.0 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0045964 | positive regulation of catecholamine metabolic process(GO:0045915) positive regulation of dopamine metabolic process(GO:0045964) |
| 0.0 | 0.1 | GO:0090074 | negative regulation of protein homodimerization activity(GO:0090074) |
| 0.0 | 0.1 | GO:0090070 | positive regulation of ribosome biogenesis(GO:0090070) positive regulation of rRNA processing(GO:2000234) |
| 0.0 | 0.1 | GO:1901314 | negative regulation of histone ubiquitination(GO:0033183) regulation of histone H2A K63-linked ubiquitination(GO:1901314) negative regulation of histone H2A K63-linked ubiquitination(GO:1901315) |
| 0.0 | 0.0 | GO:0048631 | regulation of skeletal muscle tissue growth(GO:0048631) |
| 0.0 | 0.1 | GO:0035751 | regulation of lysosomal lumen pH(GO:0035751) |
| 0.0 | 0.3 | GO:0051608 | histamine transport(GO:0051608) |
| 0.0 | 0.3 | GO:0018345 | protein palmitoylation(GO:0018345) |
| 0.0 | 0.1 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.3 | GO:0003215 | cardiac right ventricle morphogenesis(GO:0003215) |
| 0.0 | 0.1 | GO:0007290 | spermatid nucleus elongation(GO:0007290) |
| 0.0 | 0.1 | GO:0048254 | snoRNA localization(GO:0048254) |
| 0.0 | 0.5 | GO:0007202 | activation of phospholipase C activity(GO:0007202) |
| 0.0 | 0.1 | GO:0000447 | endonucleolytic cleavage in ITS1 to separate SSU-rRNA from 5.8S rRNA and LSU-rRNA from tricistronic rRNA transcript (SSU-rRNA, 5.8S rRNA, LSU-rRNA)(GO:0000447) |
| 0.0 | 0.2 | GO:1901838 | positive regulation of transcription of nuclear large rRNA transcript from RNA polymerase I promoter(GO:1901838) |
| 0.0 | 0.1 | GO:0072086 | specification of loop of Henle identity(GO:0072086) pattern specification involved in metanephros development(GO:0072268) |
| 0.0 | 0.1 | GO:0000395 | mRNA 5'-splice site recognition(GO:0000395) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.4 | GO:0007141 | male meiosis I(GO:0007141) |
| 0.0 | 0.2 | GO:0051152 | positive regulation of smooth muscle cell differentiation(GO:0051152) |
| 0.0 | 0.0 | GO:0002023 | reduction of food intake in response to dietary excess(GO:0002023) |
| 0.0 | 0.0 | GO:0097359 | UDP-glucosylation(GO:0097359) |
| 0.0 | 0.1 | GO:0080009 | mRNA methylation(GO:0080009) |
| 0.0 | 0.0 | GO:0021966 | corticospinal neuron axon guidance(GO:0021966) |
| 0.0 | 0.2 | GO:0015781 | pyrimidine nucleotide-sugar transport(GO:0015781) |
| 0.0 | 0.0 | GO:0019087 | transformation of host cell by virus(GO:0019087) |
| 0.0 | 0.0 | GO:2000619 | negative regulation of histone H4-K16 acetylation(GO:2000619) |
| 0.0 | 0.5 | GO:0060325 | face morphogenesis(GO:0060325) |
| 0.0 | 0.3 | GO:0061029 | eyelid development in camera-type eye(GO:0061029) |
| 0.0 | 0.1 | GO:0071638 | negative regulation of monocyte chemotactic protein-1 production(GO:0071638) |
| 0.0 | 0.0 | GO:1902630 | regulation of membrane hyperpolarization(GO:1902630) |
| 0.0 | 0.1 | GO:1904355 | positive regulation of telomere capping(GO:1904355) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0000056 | ribosomal small subunit export from nucleus(GO:0000056) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0030807 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.0 | GO:2000854 | positive regulation of corticosterone secretion(GO:2000854) |
| 0.0 | 0.0 | GO:0000960 | mitochondrial RNA catabolic process(GO:0000957) regulation of mitochondrial RNA catabolic process(GO:0000960) |
| 0.0 | 0.2 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0034773 | histone H4-K20 trimethylation(GO:0034773) |
| 0.0 | 0.0 | GO:0035526 | retrograde transport, plasma membrane to Golgi(GO:0035526) |
| 0.0 | 0.1 | GO:0035726 | common myeloid progenitor cell proliferation(GO:0035726) |
| 0.0 | 0.0 | GO:0070831 | basement membrane assembly(GO:0070831) |
| 0.0 | 0.0 | GO:2000824 | negative regulation of androgen receptor activity(GO:2000824) |
| 0.0 | 0.2 | GO:0034587 | piRNA metabolic process(GO:0034587) |
| 0.0 | 0.1 | GO:0019464 | glycine catabolic process(GO:0006546) glycine decarboxylation via glycine cleavage system(GO:0019464) |
| 0.0 | 0.1 | GO:0033484 | nitric oxide homeostasis(GO:0033484) |
| 0.0 | 0.1 | GO:0035457 | cellular response to interferon-alpha(GO:0035457) |
| 0.0 | 0.1 | GO:0032474 | otolith morphogenesis(GO:0032474) |
| 0.0 | 0.1 | GO:0034628 | nicotinamide nucleotide biosynthetic process from aspartate(GO:0019355) 'de novo' NAD biosynthetic process from aspartate(GO:0034628) |
| 0.0 | 0.1 | GO:0061343 | cell adhesion involved in heart morphogenesis(GO:0061343) |
| 0.0 | 0.1 | GO:0010499 | proteasomal ubiquitin-independent protein catabolic process(GO:0010499) |
| 0.0 | 1.1 | GO:0006487 | protein N-linked glycosylation(GO:0006487) |
| 0.0 | 0.2 | GO:0007196 | adenylate cyclase-inhibiting G-protein coupled glutamate receptor signaling pathway(GO:0007196) |
| 0.0 | 0.0 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.2 | GO:0009086 | methionine biosynthetic process(GO:0009086) |
| 0.0 | 0.1 | GO:0048619 | embryonic hindgut morphogenesis(GO:0048619) |
| 0.0 | 0.0 | GO:1990737 | response to manganese-induced endoplasmic reticulum stress(GO:1990737) |
| 0.0 | 0.0 | GO:0050747 | positive regulation of lipoprotein metabolic process(GO:0050747) |
| 0.0 | 0.2 | GO:0090190 | positive regulation of branching involved in ureteric bud morphogenesis(GO:0090190) |
| 0.0 | 0.0 | GO:0021979 | hypothalamus cell differentiation(GO:0021979) |
| 0.0 | 0.1 | GO:1903800 | positive regulation of production of miRNAs involved in gene silencing by miRNA(GO:1903800) |
| 0.0 | 0.1 | GO:0019236 | response to pheromone(GO:0019236) |
| 0.0 | 0.1 | GO:0046940 | nucleoside monophosphate phosphorylation(GO:0046940) |
| 0.0 | 0.1 | GO:0038145 | macrophage colony-stimulating factor signaling pathway(GO:0038145) |
| 0.0 | 0.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.0 | GO:0033625 | positive regulation of integrin activation(GO:0033625) positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0032815 | negative regulation of natural killer cell activation(GO:0032815) |
| 0.0 | 0.1 | GO:0000244 | spliceosomal tri-snRNP complex assembly(GO:0000244) |
| 0.0 | 0.1 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.2 | GO:0018027 | peptidyl-lysine dimethylation(GO:0018027) |
| 0.0 | 0.0 | GO:0045196 | establishment or maintenance of neuroblast polarity(GO:0045196) establishment of neuroblast polarity(GO:0045200) |
| 0.0 | 0.0 | GO:0032241 | positive regulation of nucleobase-containing compound transport(GO:0032241) positive regulation of RNA export from nucleus(GO:0046833) |
| 0.0 | 0.0 | GO:0071344 | diphosphate metabolic process(GO:0071344) |
| 0.0 | 0.2 | GO:0035721 | intraciliary retrograde transport(GO:0035721) |
| 0.0 | 0.1 | GO:0060081 | membrane hyperpolarization(GO:0060081) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0070762 | nuclear pore transmembrane ring(GO:0070762) |
| 0.1 | 0.3 | GO:0016533 | cyclin-dependent protein kinase 5 holoenzyme complex(GO:0016533) |
| 0.1 | 0.4 | GO:0034448 | EGO complex(GO:0034448) |
| 0.1 | 0.2 | GO:0005967 | mitochondrial pyruvate dehydrogenase complex(GO:0005967) |
| 0.1 | 0.7 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.1 | 0.3 | GO:0070876 | SOSS complex(GO:0070876) |
| 0.1 | 0.3 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.1 | 0.2 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.3 | GO:0098575 | lumenal side of lysosomal membrane(GO:0098575) |
| 0.0 | 0.2 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.4 | GO:0030896 | checkpoint clamp complex(GO:0030896) |
| 0.0 | 0.8 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.2 | GO:0043209 | myelin sheath(GO:0043209) |
| 0.0 | 0.1 | GO:0030849 | autosome(GO:0030849) |
| 0.0 | 0.1 | GO:0031379 | RNA-directed RNA polymerase complex(GO:0031379) |
| 0.0 | 0.1 | GO:0030689 | Noc complex(GO:0030689) |
| 0.0 | 0.1 | GO:0033596 | TSC1-TSC2 complex(GO:0033596) |
| 0.0 | 0.2 | GO:0005797 | Golgi medial cisterna(GO:0005797) |
| 0.0 | 0.3 | GO:0046581 | intercellular canaliculus(GO:0046581) |
| 0.0 | 0.4 | GO:0002178 | palmitoyltransferase complex(GO:0002178) |
| 0.0 | 0.2 | GO:0030121 | AP-1 adaptor complex(GO:0030121) |
| 0.0 | 0.1 | GO:0034666 | integrin alpha2-beta1 complex(GO:0034666) |
| 0.0 | 0.1 | GO:0098536 | deuterosome(GO:0098536) |
| 0.0 | 0.2 | GO:0044294 | dendritic growth cone(GO:0044294) |
| 0.0 | 0.3 | GO:0016342 | catenin complex(GO:0016342) |
| 0.0 | 0.2 | GO:0070847 | core mediator complex(GO:0070847) |
| 0.0 | 0.2 | GO:0000220 | vacuolar proton-transporting V-type ATPase, V0 domain(GO:0000220) |
| 0.0 | 0.1 | GO:0042565 | RNA nuclear export complex(GO:0042565) |
| 0.0 | 0.1 | GO:0097447 | dendritic tree(GO:0097447) |
| 0.0 | 0.1 | GO:0005960 | glycine cleavage complex(GO:0005960) |
| 0.0 | 0.4 | GO:0005671 | Ada2/Gcn5/Ada3 transcription activator complex(GO:0005671) |
| 0.0 | 0.2 | GO:0030314 | junctional membrane complex(GO:0030314) |
| 0.0 | 0.1 | GO:1990037 | Lewy body core(GO:1990037) |
| 0.0 | 0.1 | GO:0019034 | viral replication complex(GO:0019034) |
| 0.0 | 1.0 | GO:0030173 | integral component of Golgi membrane(GO:0030173) |
| 0.0 | 0.1 | GO:0030893 | meiotic cohesin complex(GO:0030893) |
| 0.0 | 0.1 | GO:1990761 | growth cone lamellipodium(GO:1990761) |
| 0.0 | 0.0 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0031428 | box C/D snoRNP complex(GO:0031428) |
| 0.0 | 0.0 | GO:0034455 | t-UTP complex(GO:0034455) |
| 0.0 | 2.2 | GO:0043202 | lysosomal lumen(GO:0043202) |
| 0.0 | 0.2 | GO:0005642 | annulate lamellae(GO:0005642) |
| 0.0 | 0.1 | GO:0005853 | eukaryotic translation elongation factor 1 complex(GO:0005853) |
| 0.0 | 0.1 | GO:0005787 | signal peptidase complex(GO:0005787) |
| 0.0 | 0.3 | GO:0042405 | nuclear inclusion body(GO:0042405) |
| 0.0 | 0.2 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0005672 | transcription factor TFIIA complex(GO:0005672) |
| 0.0 | 0.1 | GO:0033018 | sarcoplasmic reticulum lumen(GO:0033018) |
| 0.0 | 0.1 | GO:0070695 | FHF complex(GO:0070695) |
| 0.0 | 0.3 | GO:0070971 | endoplasmic reticulum exit site(GO:0070971) |
| 0.0 | 0.5 | GO:0005790 | smooth endoplasmic reticulum(GO:0005790) |
| 0.0 | 0.3 | GO:0031045 | dense core granule(GO:0031045) |
| 0.0 | 0.4 | GO:0005640 | nuclear outer membrane(GO:0005640) |
| 0.0 | 0.0 | GO:0048179 | activin receptor complex(GO:0048179) |
| 0.0 | 0.1 | GO:0045179 | apical cortex(GO:0045179) |
| 0.0 | 0.2 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.2 | GO:0097225 | sperm midpiece(GO:0097225) |
| 0.0 | 0.1 | GO:0097025 | MPP7-DLG1-LIN7 complex(GO:0097025) |
| 0.0 | 0.1 | GO:0033503 | HULC complex(GO:0033503) |
| 0.0 | 0.9 | GO:0033116 | endoplasmic reticulum-Golgi intermediate compartment membrane(GO:0033116) |
| 0.0 | 0.0 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.0 | GO:0097362 | MCM8-MCM9 complex(GO:0097362) |
| 0.0 | 0.2 | GO:0005675 | holo TFIIH complex(GO:0005675) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.1 | GO:0044308 | axonal spine(GO:0044308) |
| 0.0 | 0.1 | GO:0030056 | hemidesmosome(GO:0030056) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.6 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.2 | 0.5 | GO:0015130 | mevalonate transmembrane transporter activity(GO:0015130) |
| 0.2 | 0.5 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.1 | 0.6 | GO:0046624 | sphingolipid transporter activity(GO:0046624) |
| 0.1 | 0.3 | GO:0004574 | oligo-1,6-glucosidase activity(GO:0004574) |
| 0.1 | 0.7 | GO:0015183 | L-aspartate transmembrane transporter activity(GO:0015183) |
| 0.1 | 0.3 | GO:0016534 | cyclin-dependent protein kinase 5 activator activity(GO:0016534) |
| 0.1 | 0.5 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.1 | 1.1 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.3 | GO:0072345 | NAADP-sensitive calcium-release channel activity(GO:0072345) |
| 0.1 | 0.5 | GO:0016833 | oxo-acid-lyase activity(GO:0016833) |
| 0.1 | 0.7 | GO:0008379 | thioredoxin peroxidase activity(GO:0008379) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 0.4 | GO:0030144 | alpha-1,6-mannosylglycoprotein 6-beta-N-acetylglucosaminyltransferase activity(GO:0030144) |
| 0.1 | 0.3 | GO:0047288 | monosialoganglioside sialyltransferase activity(GO:0047288) |
| 0.1 | 0.8 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.1 | 0.4 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.1 | 0.4 | GO:0051185 | coenzyme transporter activity(GO:0051185) |
| 0.1 | 0.2 | GO:0004740 | pyruvate dehydrogenase (acetyl-transferring) kinase activity(GO:0004740) |
| 0.1 | 1.4 | GO:0001608 | G-protein coupled nucleotide receptor activity(GO:0001608) G-protein coupled purinergic nucleotide receptor activity(GO:0045028) |
| 0.1 | 0.6 | GO:0003996 | acyl-CoA ligase activity(GO:0003996) |
| 0.1 | 0.5 | GO:0070915 | lysophosphatidic acid receptor activity(GO:0070915) |
| 0.1 | 0.2 | GO:0031685 | adenosine receptor binding(GO:0031685) |
| 0.1 | 0.3 | GO:0005280 | hydrogen:amino acid symporter activity(GO:0005280) |
| 0.1 | 0.2 | GO:0050252 | retinol O-fatty-acyltransferase activity(GO:0050252) |
| 0.1 | 0.4 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.1 | 0.2 | GO:0008515 | sucrose:proton symporter activity(GO:0008506) sucrose transmembrane transporter activity(GO:0008515) disaccharide transmembrane transporter activity(GO:0015154) oligosaccharide transmembrane transporter activity(GO:0015157) |
| 0.1 | 0.3 | GO:0001602 | pancreatic polypeptide receptor activity(GO:0001602) |
| 0.1 | 0.5 | GO:0004563 | beta-N-acetylhexosaminidase activity(GO:0004563) |
| 0.1 | 0.3 | GO:0008853 | exodeoxyribonuclease III activity(GO:0008853) |
| 0.0 | 0.2 | GO:0070815 | peptidyl-lysine 5-dioxygenase activity(GO:0070815) |
| 0.0 | 0.3 | GO:0004308 | exo-alpha-sialidase activity(GO:0004308) alpha-sialidase activity(GO:0016997) |
| 0.0 | 0.1 | GO:0004103 | choline kinase activity(GO:0004103) |
| 0.0 | 0.2 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.2 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.1 | GO:0030627 | pre-mRNA 5'-splice site binding(GO:0030627) |
| 0.0 | 0.2 | GO:0016936 | galactoside binding(GO:0016936) |
| 0.0 | 0.1 | GO:0090556 | apolipoprotein A-I receptor activity(GO:0034188) phosphatidylserine-translocating ATPase activity(GO:0090556) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.2 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.8 | GO:0022841 | potassium ion leak channel activity(GO:0022841) |
| 0.0 | 0.2 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.2 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.3 | GO:0004565 | beta-galactosidase activity(GO:0004565) |
| 0.0 | 0.1 | GO:0052810 | 1-phosphatidylinositol-5-kinase activity(GO:0052810) |
| 0.0 | 0.1 | GO:0019961 | interferon binding(GO:0019961) |
| 0.0 | 0.3 | GO:0051864 | histone demethylase activity (H3-K36 specific)(GO:0051864) |
| 0.0 | 0.1 | GO:0003976 | UDP-N-acetylglucosamine-lysosomal-enzyme N-acetylglucosaminephosphotransferase activity(GO:0003976) |
| 0.0 | 0.3 | GO:0005329 | dopamine transmembrane transporter activity(GO:0005329) |
| 0.0 | 0.3 | GO:0102338 | fatty acid elongase activity(GO:0009922) 3-oxo-arachidoyl-CoA synthase activity(GO:0102336) 3-oxo-cerotoyl-CoA synthase activity(GO:0102337) 3-oxo-lignoceronyl-CoA synthase activity(GO:0102338) |
| 0.0 | 0.1 | GO:0000179 | rRNA (adenine-N6,N6-)-dimethyltransferase activity(GO:0000179) |
| 0.0 | 0.9 | GO:0005247 | voltage-gated chloride channel activity(GO:0005247) |
| 0.0 | 0.1 | GO:0003845 | 11-beta-hydroxysteroid dehydrogenase [NAD(P)] activity(GO:0003845) |
| 0.0 | 0.4 | GO:0030976 | thiamine pyrophosphate binding(GO:0030976) |
| 0.0 | 0.1 | GO:0003968 | RNA-directed RNA polymerase activity(GO:0003968) |
| 0.0 | 0.5 | GO:0005402 | sugar:proton symporter activity(GO:0005351) cation:sugar symporter activity(GO:0005402) |
| 0.0 | 0.2 | GO:0005119 | smoothened binding(GO:0005119) |
| 0.0 | 0.4 | GO:0070700 | BMP receptor binding(GO:0070700) |
| 0.0 | 0.4 | GO:0001206 | transcriptional repressor activity, RNA polymerase II distal enhancer sequence-specific binding(GO:0001206) |
| 0.0 | 0.1 | GO:0030984 | kininogen binding(GO:0030984) |
| 0.0 | 0.8 | GO:0005337 | nucleoside transmembrane transporter activity(GO:0005337) |
| 0.0 | 0.4 | GO:0004985 | opioid receptor activity(GO:0004985) |
| 0.0 | 0.6 | GO:0050811 | GABA receptor binding(GO:0050811) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.6 | GO:0016864 | protein disulfide isomerase activity(GO:0003756) intramolecular oxidoreductase activity, transposing S-S bonds(GO:0016864) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.3 | GO:0048406 | nerve growth factor binding(GO:0048406) |
| 0.0 | 0.1 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.4 | GO:0061578 | Lys63-specific deubiquitinase activity(GO:0061578) |
| 0.0 | 0.1 | GO:1904288 | BAT3 complex binding(GO:1904288) |
| 0.0 | 0.1 | GO:0022865 | transmembrane electron transfer carrier(GO:0022865) |
| 0.0 | 0.2 | GO:0005176 | ErbB-2 class receptor binding(GO:0005176) |
| 0.0 | 0.7 | GO:0019707 | protein-cysteine S-palmitoyltransferase activity(GO:0019706) protein-cysteine S-acyltransferase activity(GO:0019707) |
| 0.0 | 0.6 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.1 | GO:0044547 | DNA topoisomerase binding(GO:0044547) |
| 0.0 | 0.1 | GO:0032422 | purine-rich negative regulatory element binding(GO:0032422) |
| 0.0 | 0.1 | GO:0090599 | alpha-glucosidase activity(GO:0090599) |
| 0.0 | 0.1 | GO:0004802 | transketolase activity(GO:0004802) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.2 | GO:0046974 | histone methyltransferase activity (H3-K9 specific)(GO:0046974) |
| 0.0 | 0.1 | GO:0046899 | nucleoside triphosphate adenylate kinase activity(GO:0046899) |
| 0.0 | 0.1 | GO:0090631 | pre-miRNA transporter activity(GO:0090631) |
| 0.0 | 0.6 | GO:0017056 | structural constituent of nuclear pore(GO:0017056) |
| 0.0 | 0.1 | GO:0004487 | methylenetetrahydrofolate dehydrogenase (NAD+) activity(GO:0004487) |
| 0.0 | 0.1 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.2 | GO:0047499 | calcium-independent phospholipase A2 activity(GO:0047499) |
| 0.0 | 0.2 | GO:0015929 | hexosaminidase activity(GO:0015929) |
| 0.0 | 0.5 | GO:0004653 | polypeptide N-acetylgalactosaminyltransferase activity(GO:0004653) |
| 0.0 | 0.6 | GO:0005385 | zinc ion transmembrane transporter activity(GO:0005385) |
| 0.0 | 0.2 | GO:0004075 | biotin carboxylase activity(GO:0004075) |
| 0.0 | 0.2 | GO:0015250 | water channel activity(GO:0015250) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.2 | GO:0015295 | solute:proton symporter activity(GO:0015295) |
| 0.0 | 0.0 | GO:0000035 | acyl binding(GO:0000035) phosphopantetheine binding(GO:0031177) |
| 0.0 | 0.1 | GO:0047894 | flavonol 3-sulfotransferase activity(GO:0047894) steroid sulfotransferase activity(GO:0050294) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0004047 | aminomethyltransferase activity(GO:0004047) |
| 0.0 | 0.1 | GO:0001641 | group II metabotropic glutamate receptor activity(GO:0001641) |
| 0.0 | 0.1 | GO:0098639 | collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.1 | GO:0043208 | glycosphingolipid binding(GO:0043208) |
| 0.0 | 0.7 | GO:0098632 | protein binding involved in cell-cell adhesion(GO:0098632) |
| 0.0 | 0.0 | GO:0047192 | 1-alkylglycerophosphocholine O-acetyltransferase activity(GO:0047192) |
| 0.0 | 0.1 | GO:0004459 | L-lactate dehydrogenase activity(GO:0004459) |
| 0.0 | 0.1 | GO:0016520 | growth hormone-releasing hormone receptor activity(GO:0016520) |
| 0.0 | 0.0 | GO:0032093 | SAM domain binding(GO:0032093) |
| 0.0 | 0.0 | GO:0000773 | phosphatidyl-N-methylethanolamine N-methyltransferase activity(GO:0000773) phosphatidylethanolamine N-methyltransferase activity(GO:0004608) phosphatidyl-N-dimethylethanolamine N-methyltransferase activity(GO:0080101) |
| 0.0 | 0.4 | GO:0000030 | mannosyltransferase activity(GO:0000030) |
| 0.0 | 0.2 | GO:0001042 | RNA polymerase I core binding(GO:0001042) |
| 0.0 | 0.1 | GO:0004104 | acetylcholinesterase activity(GO:0003990) cholinesterase activity(GO:0004104) |
| 0.0 | 0.3 | GO:0030306 | ADP-ribosylation factor binding(GO:0030306) |
| 0.0 | 0.0 | GO:0003980 | UDP-glucose:glycoprotein glucosyltransferase activity(GO:0003980) |
| 0.0 | 0.0 | GO:0086062 | voltage-gated sodium channel activity involved in Purkinje myocyte action potential(GO:0086062) |
| 0.0 | 0.5 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.1 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.5 | GO:0070840 | dynein complex binding(GO:0070840) |
| 0.0 | 0.1 | GO:0097016 | L27 domain binding(GO:0097016) |
| 0.0 | 0.1 | GO:0008174 | mRNA methyltransferase activity(GO:0008174) |
| 0.0 | 0.3 | GO:0005112 | Notch binding(GO:0005112) |
| 0.0 | 0.1 | GO:0035939 | microsatellite binding(GO:0035939) |
| 0.0 | 0.0 | GO:0050211 | procollagen galactosyltransferase activity(GO:0050211) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.1 | GO:0035374 | chondroitin sulfate binding(GO:0035374) |
| 0.0 | 0.4 | GO:0046961 | proton-transporting ATPase activity, rotational mechanism(GO:0046961) |
| 0.0 | 0.3 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.2 | GO:0031404 | chloride ion binding(GO:0031404) |
| 0.0 | 0.1 | GO:0008823 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0000309 | nicotinamide-nucleotide adenylyltransferase activity(GO:0000309) |
| 0.0 | 0.1 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.2 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.0 | GO:0030621 | U4 snRNA binding(GO:0030621) |
| 0.0 | 0.2 | GO:0005326 | neurotransmitter transporter activity(GO:0005326) |
| 0.0 | 0.2 | GO:0003688 | DNA replication origin binding(GO:0003688) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.0 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0004430 | 1-phosphatidylinositol 4-kinase activity(GO:0004430) |
| 0.0 | 0.1 | GO:0016774 | phosphotransferase activity, carboxyl group as acceptor(GO:0016774) |
| 0.0 | 0.0 | GO:0015235 | cobalamin transporter activity(GO:0015235) |
| 0.0 | 0.0 | GO:0008808 | cardiolipin synthase activity(GO:0008808) phosphatidyltransferase activity(GO:0030572) CDP-diacylglycerol-phosphatidylglycerol phosphatidyltransferase activity(GO:0043337) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID MET PATHWAY | Signaling events mediated by Hepatocyte Growth Factor Receptor (c-Met) |
| 0.0 | 0.4 | PID LPA4 PATHWAY | LPA4-mediated signaling events |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.1 | PID HEDGEHOG 2PATHWAY | Signaling events mediated by the Hedgehog family |
| 0.0 | 0.1 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.8 | REACTOME REGULATION OF PYRUVATE DEHYDROGENASE PDH COMPLEX | Genes involved in Regulation of pyruvate dehydrogenase (PDH) complex |
| 0.1 | 1.4 | REACTOME P2Y RECEPTORS | Genes involved in P2Y receptors |
| 0.1 | 0.6 | REACTOME PYRUVATE METABOLISM | Genes involved in Pyruvate metabolism |
| 0.0 | 0.6 | REACTOME CALNEXIN CALRETICULIN CYCLE | Genes involved in Calnexin/calreticulin cycle |
| 0.0 | 0.4 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.9 | REACTOME BIOSYNTHESIS OF THE N GLYCAN PRECURSOR DOLICHOL LIPID LINKED OLIGOSACCHARIDE LLO AND TRANSFER TO A NASCENT PROTEIN | Genes involved in Biosynthesis of the N-glycan precursor (dolichol lipid-linked oligosaccharide, LLO) and transfer to a nascent protein |
| 0.0 | 0.8 | REACTOME ZINC TRANSPORTERS | Genes involved in Zinc transporters |
| 0.0 | 0.7 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.7 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME RECEPTOR LIGAND BINDING INITIATES THE SECOND PROTEOLYTIC CLEAVAGE OF NOTCH RECEPTOR | Genes involved in Receptor-ligand binding initiates the second proteolytic cleavage of Notch receptor |
| 0.0 | 0.0 | REACTOME INSULIN RECEPTOR RECYCLING | Genes involved in Insulin receptor recycling |
| 0.0 | 0.3 | REACTOME ABACAVIR TRANSPORT AND METABOLISM | Genes involved in Abacavir transport and metabolism |
| 0.0 | 0.7 | REACTOME TRANSFERRIN ENDOCYTOSIS AND RECYCLING | Genes involved in Transferrin endocytosis and recycling |
| 0.0 | 1.0 | REACTOME TRANSPORT OF VITAMINS NUCLEOSIDES AND RELATED MOLECULES | Genes involved in Transport of vitamins, nucleosides, and related molecules |
| 0.0 | 0.9 | REACTOME GLYCOSPHINGOLIPID METABOLISM | Genes involved in Glycosphingolipid metabolism |
| 0.0 | 0.3 | REACTOME FACILITATIVE NA INDEPENDENT GLUCOSE TRANSPORTERS | Genes involved in Facilitative Na+-independent glucose transporters |
| 0.0 | 0.3 | REACTOME E2F ENABLED INHIBITION OF PRE REPLICATION COMPLEX FORMATION | Genes involved in E2F-enabled inhibition of pre-replication complex formation |
| 0.0 | 0.2 | REACTOME ADENYLATE CYCLASE ACTIVATING PATHWAY | Genes involved in Adenylate cyclase activating pathway |
| 0.0 | 0.2 | REACTOME TANDEM PORE DOMAIN POTASSIUM CHANNELS | Genes involved in Tandem pore domain potassium channels |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.4 | REACTOME ADHERENS JUNCTIONS INTERACTIONS | Genes involved in Adherens junctions interactions |
| 0.0 | 0.3 | REACTOME TGF BETA RECEPTOR SIGNALING IN EMT EPITHELIAL TO MESENCHYMAL TRANSITION | Genes involved in TGF-beta receptor signaling in EMT (epithelial to mesenchymal transition) |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF VERY LONG CHAIN FATTY ACYL COAS | Genes involved in Synthesis of very long-chain fatty acyl-CoAs |
| 0.0 | 0.0 | REACTOME N GLYCAN TRIMMING IN THE ER AND CALNEXIN CALRETICULIN CYCLE | Genes involved in N-glycan trimming in the ER and Calnexin/Calreticulin cycle |
| 0.0 | 0.0 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |