Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
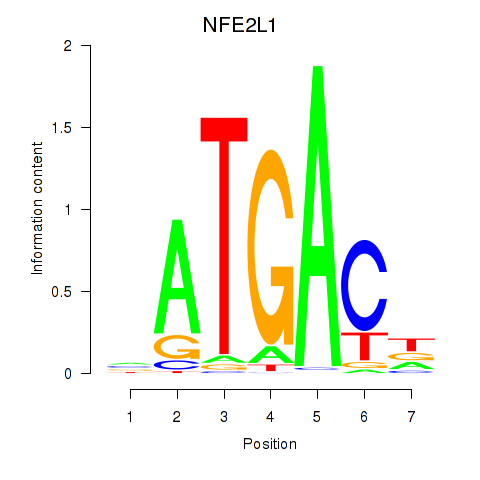
Results for NFE2L1
Z-value: 0.89
Transcription factors associated with NFE2L1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFE2L1
|
ENSG00000082641.11 | NFE2L1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFE2L1 | hg19_v2_chr17_+_46126135_46126152 | 0.17 | 6.8e-01 | Click! |
Activity profile of NFE2L1 motif
Sorted Z-values of NFE2L1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFE2L1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_-_52887034 | 2.63 |
ENST00000330722.6 |
KRT6A |
keratin 6A |
| chr2_-_161056802 | 1.88 |
ENST00000283249.2 ENST00000409872.1 |
ITGB6 |
integrin, beta 6 |
| chr2_-_161056762 | 1.82 |
ENST00000428609.2 ENST00000409967.2 |
ITGB6 |
integrin, beta 6 |
| chr10_-_123357598 | 1.20 |
ENST00000358487.5 ENST00000369058.3 ENST00000369060.4 ENST00000359354.2 |
FGFR2 |
fibroblast growth factor receptor 2 |
| chr11_+_117947724 | 1.15 |
ENST00000534111.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr1_+_35220613 | 1.06 |
ENST00000338513.1 |
GJB5 |
gap junction protein, beta 5, 31.1kDa |
| chr5_+_36608422 | 1.05 |
ENST00000381918.3 |
SLC1A3 |
solute carrier family 1 (glial high affinity glutamate transporter), member 3 |
| chr2_-_113594279 | 1.03 |
ENST00000416750.1 ENST00000418817.1 ENST00000263341.2 |
IL1B |
interleukin 1, beta |
| chr11_+_117947782 | 0.94 |
ENST00000522307.1 ENST00000523251.1 ENST00000437212.3 ENST00000522824.1 ENST00000522151.1 |
TMPRSS4 |
transmembrane protease, serine 4 |
| chr2_-_46385 | 0.90 |
ENST00000327669.4 |
FAM110C |
family with sequence similarity 110, member C |
| chr11_-_119999539 | 0.89 |
ENST00000541857.1 |
TRIM29 |
tripartite motif containing 29 |
| chr11_-_119999611 | 0.89 |
ENST00000529044.1 |
TRIM29 |
tripartite motif containing 29 |
| chr2_-_70781087 | 0.88 |
ENST00000394241.3 ENST00000295400.6 |
TGFA |
transforming growth factor, alpha |
| chr19_-_35992780 | 0.88 |
ENST00000593342.1 ENST00000601650.1 ENST00000408915.2 |
DMKN |
dermokine |
| chr5_+_140248518 | 0.82 |
ENST00000398640.2 |
PCDHA11 |
protocadherin alpha 11 |
| chr1_-_242612779 | 0.78 |
ENST00000427495.1 |
PLD5 |
phospholipase D family, member 5 |
| chr3_-_56809685 | 0.77 |
ENST00000413728.2 |
ARHGEF3 |
Rho guanine nucleotide exchange factor (GEF) 3 |
| chr16_+_76587314 | 0.75 |
ENST00000563764.1 |
RP11-58C22.1 |
Uncharacterized protein |
| chr10_-_123357910 | 0.74 |
ENST00000336553.6 ENST00000457416.2 ENST00000360144.3 ENST00000369059.1 ENST00000356226.4 ENST00000351936.6 |
FGFR2 |
fibroblast growth factor receptor 2 |
| chr12_+_41086297 | 0.72 |
ENST00000551295.2 |
CNTN1 |
contactin 1 |
| chr6_-_29343068 | 0.71 |
ENST00000396806.3 |
OR12D3 |
olfactory receptor, family 12, subfamily D, member 3 |
| chr11_-_119991589 | 0.69 |
ENST00000526881.1 |
TRIM29 |
tripartite motif containing 29 |
| chr1_+_43735646 | 0.69 |
ENST00000439858.1 |
TMEM125 |
transmembrane protein 125 |
| chr6_-_52109335 | 0.67 |
ENST00000336123.4 |
IL17F |
interleukin 17F |
| chr11_-_125366089 | 0.65 |
ENST00000366139.3 ENST00000278919.3 |
FEZ1 |
fasciculation and elongation protein zeta 1 (zygin I) |
| chr12_-_123201337 | 0.61 |
ENST00000528880.2 |
HCAR3 |
hydroxycarboxylic acid receptor 3 |
| chr12_-_123187890 | 0.59 |
ENST00000328880.5 |
HCAR2 |
hydroxycarboxylic acid receptor 2 |
| chr18_+_34124507 | 0.59 |
ENST00000591635.1 |
FHOD3 |
formin homology 2 domain containing 3 |
| chr6_-_32784687 | 0.59 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr7_-_121944491 | 0.57 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr7_-_41742697 | 0.57 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr7_+_150811705 | 0.56 |
ENST00000335367.3 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr16_+_57662138 | 0.55 |
ENST00000562414.1 ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr11_-_87908600 | 0.54 |
ENST00000531138.1 ENST00000526372.1 ENST00000243662.6 |
RAB38 |
RAB38, member RAS oncogene family |
| chr11_+_59480899 | 0.54 |
ENST00000300150.7 |
STX3 |
syntaxin 3 |
| chr19_+_15052301 | 0.54 |
ENST00000248072.3 |
OR7C2 |
olfactory receptor, family 7, subfamily C, member 2 |
| chr1_+_43735678 | 0.54 |
ENST00000432792.2 |
TMEM125 |
transmembrane protein 125 |
| chr17_-_8021710 | 0.53 |
ENST00000380149.1 ENST00000448843.2 |
ALOXE3 |
arachidonate lipoxygenase 3 |
| chr6_+_143381979 | 0.52 |
ENST00000367598.5 ENST00000447498.1 ENST00000357847.4 ENST00000344492.5 ENST00000367596.1 ENST00000494282.2 ENST00000275235.4 |
AIG1 |
androgen-induced 1 |
| chr6_-_41130914 | 0.49 |
ENST00000373113.3 ENST00000338469.3 |
TREM2 |
triggering receptor expressed on myeloid cells 2 |
| chr16_+_57662419 | 0.49 |
ENST00000388812.4 ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56 |
G protein-coupled receptor 56 |
| chr18_+_55888767 | 0.47 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr5_-_16509101 | 0.44 |
ENST00000399793.2 |
FAM134B |
family with sequence similarity 134, member B |
| chr4_-_123542224 | 0.44 |
ENST00000264497.3 |
IL21 |
interleukin 21 |
| chr1_+_62439037 | 0.44 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr11_+_7626950 | 0.44 |
ENST00000530181.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr3_-_98241358 | 0.43 |
ENST00000503004.1 ENST00000506575.1 ENST00000513452.1 ENST00000515620.1 |
CLDND1 |
claudin domain containing 1 |
| chr17_-_7165662 | 0.41 |
ENST00000571881.2 ENST00000360325.7 |
CLDN7 |
claudin 7 |
| chr10_+_5566916 | 0.38 |
ENST00000315238.1 |
CALML3 |
calmodulin-like 3 |
| chr6_-_155776966 | 0.37 |
ENST00000159060.2 |
NOX3 |
NADPH oxidase 3 |
| chr3_-_27764190 | 0.35 |
ENST00000537516.1 |
EOMES |
eomesodermin |
| chr9_+_72002837 | 0.35 |
ENST00000377216.3 |
FAM189A2 |
family with sequence similarity 189, member A2 |
| chr9_-_117150243 | 0.35 |
ENST00000374088.3 |
AKNA |
AT-hook transcription factor |
| chr9_-_127263265 | 0.34 |
ENST00000373587.3 |
NR5A1 |
nuclear receptor subfamily 5, group A, member 1 |
| chr11_+_63137251 | 0.33 |
ENST00000310969.4 ENST00000279178.3 |
SLC22A9 |
solute carrier family 22 (organic anion transporter), member 9 |
| chr19_-_44160768 | 0.33 |
ENST00000593447.1 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr12_-_39734783 | 0.33 |
ENST00000552961.1 |
KIF21A |
kinesin family member 21A |
| chr8_+_104831472 | 0.33 |
ENST00000262231.10 ENST00000507740.1 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr9_-_39239171 | 0.33 |
ENST00000358144.2 |
CNTNAP3 |
contactin associated protein-like 3 |
| chr15_+_89181974 | 0.32 |
ENST00000306072.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chrX_-_33229636 | 0.32 |
ENST00000357033.4 |
DMD |
dystrophin |
| chr15_+_89182178 | 0.31 |
ENST00000559876.1 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr1_-_153113927 | 0.31 |
ENST00000368752.4 |
SPRR2B |
small proline-rich protein 2B |
| chr2_+_113735575 | 0.31 |
ENST00000376489.2 ENST00000259205.4 |
IL36G |
interleukin 36, gamma |
| chr9_-_117853297 | 0.29 |
ENST00000542877.1 ENST00000537320.1 ENST00000341037.4 |
TNC |
tenascin C |
| chr11_-_7847519 | 0.29 |
ENST00000328375.1 |
OR5P3 |
olfactory receptor, family 5, subfamily P, member 3 |
| chr19_-_51530916 | 0.27 |
ENST00000594768.1 |
KLK11 |
kallikrein-related peptidase 11 |
| chr9_-_132515302 | 0.27 |
ENST00000340607.4 |
PTGES |
prostaglandin E synthase |
| chr17_+_39394250 | 0.26 |
ENST00000254072.6 |
KRTAP9-8 |
keratin associated protein 9-8 |
| chr3_-_143567262 | 0.26 |
ENST00000474151.1 ENST00000316549.6 |
SLC9A9 |
solute carrier family 9, subfamily A (NHE9, cation proton antiporter 9), member 9 |
| chr17_-_27418537 | 0.26 |
ENST00000408971.2 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr16_+_19467772 | 0.25 |
ENST00000219821.5 ENST00000561503.1 ENST00000564959.1 |
TMC5 |
transmembrane channel-like 5 |
| chr11_+_60197069 | 0.25 |
ENST00000528905.1 ENST00000528093.1 |
MS4A5 |
membrane-spanning 4-domains, subfamily A, member 5 |
| chr10_+_127661942 | 0.25 |
ENST00000417114.1 ENST00000445510.1 ENST00000368691.1 |
FANK1 |
fibronectin type III and ankyrin repeat domains 1 |
| chr9_+_133971863 | 0.25 |
ENST00000372309.3 |
AIF1L |
allograft inflammatory factor 1-like |
| chr5_+_147582387 | 0.24 |
ENST00000325630.2 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr6_+_144185573 | 0.24 |
ENST00000237275.6 ENST00000539295.1 |
ZC2HC1B |
zinc finger, C2HC-type containing 1B |
| chr9_+_124103625 | 0.24 |
ENST00000594963.1 |
AL161784.1 |
Uncharacterized protein |
| chr14_+_22977587 | 0.24 |
ENST00000390504.1 |
TRAJ33 |
T cell receptor alpha joining 33 |
| chr12_+_20963632 | 0.24 |
ENST00000540853.1 ENST00000261196.2 |
SLCO1B3 |
solute carrier organic anion transporter family, member 1B3 |
| chr6_+_155585147 | 0.24 |
ENST00000367165.3 |
CLDN20 |
claudin 20 |
| chr1_-_113258090 | 0.23 |
ENST00000309276.6 |
PPM1J |
protein phosphatase, Mg2+/Mn2+ dependent, 1J |
| chr15_+_89182156 | 0.23 |
ENST00000379224.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr11_+_128563652 | 0.23 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr4_+_71494461 | 0.22 |
ENST00000396073.3 |
ENAM |
enamelin |
| chr12_-_95510743 | 0.22 |
ENST00000551521.1 |
FGD6 |
FYVE, RhoGEF and PH domain containing 6 |
| chr3_-_98241760 | 0.22 |
ENST00000507874.1 ENST00000502299.1 ENST00000508659.1 ENST00000510545.1 ENST00000511667.1 ENST00000394185.2 ENST00000394181.2 ENST00000508902.1 ENST00000341181.6 ENST00000437922.1 ENST00000394180.2 |
CLDND1 |
claudin domain containing 1 |
| chr2_-_105030466 | 0.21 |
ENST00000449772.1 |
AC068535.3 |
AC068535.3 |
| chr11_+_55578854 | 0.21 |
ENST00000333973.2 |
OR5L1 |
olfactory receptor, family 5, subfamily L, member 1 |
| chr5_+_147582348 | 0.21 |
ENST00000514389.1 |
SPINK6 |
serine peptidase inhibitor, Kazal type 6 |
| chr6_+_160327974 | 0.21 |
ENST00000252660.4 |
MAS1 |
MAS1 oncogene |
| chr7_-_22539771 | 0.21 |
ENST00000406890.2 ENST00000424363.1 |
STEAP1B |
STEAP family member 1B |
| chr6_+_33043703 | 0.21 |
ENST00000418931.2 ENST00000535465.1 |
HLA-DPB1 |
major histocompatibility complex, class II, DP beta 1 |
| chr17_-_57784755 | 0.21 |
ENST00000537860.1 ENST00000393038.2 ENST00000409433.2 |
PTRH2 |
peptidyl-tRNA hydrolase 2 |
| chr2_-_99871570 | 0.20 |
ENST00000333017.2 ENST00000409679.1 ENST00000423306.1 |
LYG2 |
lysozyme G-like 2 |
| chr15_+_28624878 | 0.20 |
ENST00000450328.2 |
GOLGA8F |
golgin A8 family, member F |
| chr11_+_59522837 | 0.20 |
ENST00000437946.2 |
STX3 |
syntaxin 3 |
| chr14_+_22694606 | 0.20 |
ENST00000390463.3 |
TRAV36DV7 |
T cell receptor alpha variable 36/delta variable 7 |
| chr12_-_15104040 | 0.20 |
ENST00000541644.1 ENST00000545895.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr2_+_86669118 | 0.20 |
ENST00000427678.1 ENST00000542128.1 |
KDM3A |
lysine (K)-specific demethylase 3A |
| chr2_-_46769694 | 0.19 |
ENST00000522587.1 |
ATP6V1E2 |
ATPase, H+ transporting, lysosomal 31kDa, V1 subunit E2 |
| chr3_-_190167571 | 0.19 |
ENST00000354905.2 |
TMEM207 |
transmembrane protein 207 |
| chr4_-_155511887 | 0.19 |
ENST00000302053.3 ENST00000403106.3 |
FGA |
fibrinogen alpha chain |
| chrX_+_134654540 | 0.19 |
ENST00000370752.4 |
DDX26B |
DEAD/H (Asp-Glu-Ala-Asp/His) box polypeptide 26B |
| chr19_-_36019123 | 0.19 |
ENST00000588674.1 ENST00000452271.2 ENST00000518157.1 |
SBSN |
suprabasin |
| chr8_-_133123406 | 0.18 |
ENST00000434736.2 |
HHLA1 |
HERV-H LTR-associating 1 |
| chr11_+_60197040 | 0.18 |
ENST00000300190.2 |
MS4A5 |
membrane-spanning 4-domains, subfamily A, member 5 |
| chr11_+_100862811 | 0.18 |
ENST00000303130.2 |
TMEM133 |
transmembrane protein 133 |
| chr3_-_59035673 | 0.18 |
ENST00000491845.1 ENST00000472469.1 ENST00000471288.1 ENST00000295966.7 |
C3orf67 |
chromosome 3 open reading frame 67 |
| chr10_-_118765081 | 0.18 |
ENST00000392903.2 ENST00000355371.4 |
KIAA1598 |
KIAA1598 |
| chr12_-_8088871 | 0.18 |
ENST00000075120.7 |
SLC2A3 |
solute carrier family 2 (facilitated glucose transporter), member 3 |
| chr15_+_45406519 | 0.18 |
ENST00000323030.5 |
DUOXA2 |
dual oxidase maturation factor 2 |
| chr2_-_105882585 | 0.18 |
ENST00000595531.1 |
AC012360.2 |
LOC644617 protein; Uncharacterized protein |
| chr11_-_128894053 | 0.18 |
ENST00000392657.3 |
ARHGAP32 |
Rho GTPase activating protein 32 |
| chr6_+_111580508 | 0.18 |
ENST00000368847.4 |
KIAA1919 |
KIAA1919 |
| chr6_-_33037019 | 0.17 |
ENST00000437811.1 |
HLA-DPA1 |
major histocompatibility complex, class II, DP alpha 1 |
| chr6_-_49604545 | 0.17 |
ENST00000371175.4 ENST00000229810.7 |
RHAG |
Rh-associated glycoprotein |
| chr11_+_35198118 | 0.17 |
ENST00000525211.1 ENST00000526000.1 ENST00000279452.6 ENST00000527889.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr12_+_41086215 | 0.17 |
ENST00000547702.1 ENST00000551424.1 |
CNTN1 |
contactin 1 |
| chr1_+_29241027 | 0.17 |
ENST00000373797.1 |
EPB41 |
erythrocyte membrane protein band 4.1 (elliptocytosis 1, RH-linked) |
| chr9_+_139847347 | 0.17 |
ENST00000371632.3 |
LCN12 |
lipocalin 12 |
| chr14_+_22739823 | 0.17 |
ENST00000390464.2 |
TRAV38-1 |
T cell receptor alpha variable 38-1 |
| chr6_-_56507586 | 0.17 |
ENST00000439203.1 ENST00000518935.1 ENST00000446842.2 ENST00000370765.6 ENST00000244364.6 |
DST |
dystonin |
| chr19_-_12807422 | 0.17 |
ENST00000380339.3 ENST00000544494.1 ENST00000393261.3 |
FBXW9 |
F-box and WD repeat domain containing 9 |
| chr17_+_28268623 | 0.17 |
ENST00000394835.3 ENST00000320856.5 ENST00000394832.2 ENST00000378738.3 |
EFCAB5 |
EF-hand calcium binding domain 5 |
| chr14_+_102276192 | 0.17 |
ENST00000557714.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chr11_+_35211429 | 0.17 |
ENST00000525688.1 ENST00000278385.6 ENST00000533222.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr8_+_104831554 | 0.16 |
ENST00000408894.2 |
RIMS2 |
regulating synaptic membrane exocytosis 2 |
| chr9_-_117692697 | 0.16 |
ENST00000223795.2 |
TNFSF8 |
tumor necrosis factor (ligand) superfamily, member 8 |
| chr6_-_29324054 | 0.16 |
ENST00000543825.1 |
OR5V1 |
olfactory receptor, family 5, subfamily V, member 1 |
| chr2_+_182850743 | 0.16 |
ENST00000409702.1 |
PPP1R1C |
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr19_+_9361606 | 0.16 |
ENST00000456448.1 |
OR7E24 |
olfactory receptor, family 7, subfamily E, member 24 |
| chrX_-_30993201 | 0.16 |
ENST00000288422.2 ENST00000378932.2 |
TAB3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr10_+_81272287 | 0.16 |
ENST00000520547.2 |
EIF5AL1 |
eukaryotic translation initiation factor 5A-like 1 |
| chr11_-_107729887 | 0.16 |
ENST00000525815.1 |
SLC35F2 |
solute carrier family 35, member F2 |
| chr3_+_100120441 | 0.16 |
ENST00000489752.1 |
LNP1 |
leukemia NUP98 fusion partner 1 |
| chr22_-_38349552 | 0.16 |
ENST00000422191.1 ENST00000249079.2 ENST00000418863.1 ENST00000403305.1 ENST00000403026.1 |
C22orf23 |
chromosome 22 open reading frame 23 |
| chr2_+_182850551 | 0.15 |
ENST00000452904.1 ENST00000409137.3 ENST00000280295.3 |
PPP1R1C |
protein phosphatase 1, regulatory (inhibitor) subunit 1C |
| chr12_+_28343365 | 0.15 |
ENST00000545336.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr11_+_58390132 | 0.15 |
ENST00000361987.4 |
CNTF |
ciliary neurotrophic factor |
| chr11_+_35211511 | 0.15 |
ENST00000524922.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr17_-_73663168 | 0.15 |
ENST00000578201.1 ENST00000423245.2 |
RECQL5 |
RecQ protein-like 5 |
| chr10_+_32873190 | 0.15 |
ENST00000375025.4 |
C10orf68 |
Homo sapiens coiled-coil domain containing 7 (CCDC7), transcript variant 5, mRNA. |
| chr16_+_81348528 | 0.15 |
ENST00000568107.2 |
GAN |
gigaxonin |
| chr1_+_84630367 | 0.15 |
ENST00000370680.1 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr4_+_144303093 | 0.15 |
ENST00000505913.1 |
GAB1 |
GRB2-associated binding protein 1 |
| chr4_+_71200681 | 0.15 |
ENST00000273936.5 |
CABS1 |
calcium-binding protein, spermatid-specific 1 |
| chr16_-_48281305 | 0.15 |
ENST00000356608.2 |
ABCC11 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 11 |
| chr11_+_55594695 | 0.15 |
ENST00000378397.1 |
OR5L2 |
olfactory receptor, family 5, subfamily L, member 2 |
| chr1_-_39395165 | 0.14 |
ENST00000372985.3 |
RHBDL2 |
rhomboid, veinlet-like 2 (Drosophila) |
| chr18_+_55862622 | 0.14 |
ENST00000456173.2 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr3_+_113616317 | 0.14 |
ENST00000440446.2 ENST00000488680.1 |
GRAMD1C |
GRAM domain containing 1C |
| chr12_+_28343353 | 0.14 |
ENST00000539107.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr8_-_7287870 | 0.14 |
ENST00000318124.3 |
DEFB103B |
defensin, beta 103B |
| chr12_-_15103621 | 0.14 |
ENST00000536592.1 |
ARHGDIB |
Rho GDP dissociation inhibitor (GDI) beta |
| chr5_+_142149932 | 0.14 |
ENST00000274498.4 |
ARHGAP26 |
Rho GTPase activating protein 26 |
| chr22_+_32439019 | 0.14 |
ENST00000266088.4 |
SLC5A1 |
solute carrier family 5 (sodium/glucose cotransporter), member 1 |
| chr12_-_18890940 | 0.14 |
ENST00000543242.1 ENST00000539072.1 ENST00000541966.1 ENST00000266505.7 ENST00000447925.2 ENST00000435379.1 |
PLCZ1 |
phospholipase C, zeta 1 |
| chr15_+_68582544 | 0.14 |
ENST00000566008.1 |
FEM1B |
fem-1 homolog b (C. elegans) |
| chr22_+_30805086 | 0.14 |
ENST00000439838.1 ENST00000439023.3 |
RP4-539M6.19 |
Uncharacterized protein |
| chr2_-_220119280 | 0.14 |
ENST00000392088.2 |
TUBA4A |
tubulin, alpha 4a |
| chr9_-_128246769 | 0.14 |
ENST00000444226.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr2_+_159651821 | 0.14 |
ENST00000309950.3 ENST00000409042.1 |
DAPL1 |
death associated protein-like 1 |
| chrX_-_99891796 | 0.14 |
ENST00000373020.4 |
TSPAN6 |
tetraspanin 6 |
| chr11_+_7534999 | 0.13 |
ENST00000528947.1 ENST00000299492.4 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr3_-_52443799 | 0.13 |
ENST00000470173.1 ENST00000296288.5 |
BAP1 |
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr10_+_70939983 | 0.13 |
ENST00000359655.4 ENST00000422378.1 |
SUPV3L1 |
suppressor of var1, 3-like 1 (S. cerevisiae) |
| chr4_-_819901 | 0.13 |
ENST00000304062.6 |
CPLX1 |
complexin 1 |
| chr14_+_23842018 | 0.13 |
ENST00000397242.2 ENST00000329715.2 |
IL25 |
interleukin 25 |
| chr11_+_113779704 | 0.13 |
ENST00000537778.1 |
HTR3B |
5-hydroxytryptamine (serotonin) receptor 3B, ionotropic |
| chr7_+_42971799 | 0.13 |
ENST00000223324.2 |
MRPL32 |
mitochondrial ribosomal protein L32 |
| chr1_-_11865982 | 0.13 |
ENST00000418034.1 |
MTHFR |
methylenetetrahydrofolate reductase (NAD(P)H) |
| chr4_-_177190364 | 0.13 |
ENST00000296525.3 |
ASB5 |
ankyrin repeat and SOCS box containing 5 |
| chr8_-_20040638 | 0.13 |
ENST00000519026.1 ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr15_-_81616446 | 0.13 |
ENST00000302824.6 |
STARD5 |
StAR-related lipid transfer (START) domain containing 5 |
| chr4_-_819880 | 0.13 |
ENST00000505203.1 |
CPLX1 |
complexin 1 |
| chr11_+_123986069 | 0.13 |
ENST00000456829.2 ENST00000361352.5 ENST00000449321.1 ENST00000392748.1 ENST00000360334.4 ENST00000392744.4 |
VWA5A |
von Willebrand factor A domain containing 5A |
| chr6_+_52051171 | 0.13 |
ENST00000340057.1 |
IL17A |
interleukin 17A |
| chr14_-_21490958 | 0.13 |
ENST00000554104.1 |
NDRG2 |
NDRG family member 2 |
| chr14_-_24768913 | 0.13 |
ENST00000288111.7 |
DHRS1 |
dehydrogenase/reductase (SDR family) member 1 |
| chr2_+_234545148 | 0.13 |
ENST00000373445.1 |
UGT1A10 |
UDP glucuronosyltransferase 1 family, polypeptide A10 |
| chr8_+_7738726 | 0.12 |
ENST00000314357.3 |
DEFB103A |
defensin, beta 103A |
| chr1_+_152784447 | 0.12 |
ENST00000360090.3 |
LCE1B |
late cornified envelope 1B |
| chr19_-_12807395 | 0.12 |
ENST00000587955.1 |
FBXW9 |
F-box and WD repeat domain containing 9 |
| chr3_+_52813932 | 0.12 |
ENST00000537050.1 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr4_+_8442496 | 0.12 |
ENST00000389737.4 ENST00000504134.1 |
TRMT44 |
tRNA methyltransferase 44 homolog (S. cerevisiae) |
| chr2_+_234602305 | 0.12 |
ENST00000406651.1 |
UGT1A6 |
UDP glucuronosyltransferase 1 family, polypeptide A6 |
| chr6_+_29274403 | 0.12 |
ENST00000377160.2 |
OR14J1 |
olfactory receptor, family 14, subfamily J, member 1 |
| chr17_-_39324424 | 0.12 |
ENST00000391356.2 |
KRTAP4-3 |
keratin associated protein 4-3 |
| chr4_-_151936416 | 0.12 |
ENST00000510413.1 ENST00000507224.1 |
LRBA |
LPS-responsive vesicle trafficking, beach and anchor containing |
| chr15_+_80364901 | 0.12 |
ENST00000560228.1 ENST00000559835.1 ENST00000559775.1 ENST00000558688.1 ENST00000560392.1 ENST00000560976.1 ENST00000558272.1 ENST00000558390.1 |
ZFAND6 |
zinc finger, AN1-type domain 6 |
| chr11_+_35201826 | 0.12 |
ENST00000531873.1 |
CD44 |
CD44 molecule (Indian blood group) |
| chr6_-_10838710 | 0.12 |
ENST00000313243.2 |
MAK |
male germ cell-associated kinase |
| chr9_+_87283430 | 0.12 |
ENST00000376214.1 ENST00000376213.1 |
NTRK2 |
neurotrophic tyrosine kinase, receptor, type 2 |
| chr1_+_45265897 | 0.12 |
ENST00000372201.4 |
PLK3 |
polo-like kinase 3 |
| chr9_+_27524283 | 0.12 |
ENST00000276943.2 |
IFNK |
interferon, kappa |
| chr6_-_10838736 | 0.12 |
ENST00000536370.1 ENST00000474039.1 |
MAK |
male germ cell-associated kinase |
| chr13_+_48611665 | 0.12 |
ENST00000258662.2 |
NUDT15 |
nudix (nucleoside diphosphate linked moiety X)-type motif 15 |
| chr5_+_142149955 | 0.12 |
ENST00000378004.3 |
ARHGAP26 |
Rho GTPase activating protein 26 |
| chr4_+_123300121 | 0.12 |
ENST00000446706.1 ENST00000296513.2 |
ADAD1 |
adenosine deaminase domain containing 1 (testis-specific) |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.7 | 2.6 | GO:0051710 | cytolysis by symbiont of host cells(GO:0001897) regulation of cytolysis in other organism(GO:0051710) |
| 0.6 | 1.9 | GO:0035603 | fibroblast growth factor receptor signaling pathway involved in negative regulation of apoptotic process in bone marrow(GO:0035602) fibroblast growth factor receptor signaling pathway involved in hemopoiesis(GO:0035603) fibroblast growth factor receptor signaling pathway involved in positive regulation of cell proliferation in bone marrow(GO:0035604) |
| 0.3 | 1.0 | GO:0060559 | positive regulation of vitamin metabolic process(GO:0046136) positive regulation of vitamin D biosynthetic process(GO:0060557) positive regulation of calcidiol 1-monooxygenase activity(GO:0060559) |
| 0.3 | 3.7 | GO:0038044 | transforming growth factor-beta secretion(GO:0038044) |
| 0.3 | 1.1 | GO:0060708 | spongiotrophoblast differentiation(GO:0060708) |
| 0.2 | 1.1 | GO:0002586 | regulation of antigen processing and presentation of peptide antigen via MHC class II(GO:0002586) |
| 0.2 | 0.6 | GO:0060278 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.2 | 0.4 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.2 | 0.9 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.2 | 0.7 | GO:0032641 | lymphotoxin A production(GO:0032641) lymphotoxin A biosynthetic process(GO:0042109) |
| 0.2 | 1.1 | GO:0006537 | glutamate biosynthetic process(GO:0006537) |
| 0.1 | 0.9 | GO:1903575 | cornified envelope assembly(GO:1903575) |
| 0.1 | 0.6 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.7 | GO:0098967 | exocytic insertion of neurotransmitter receptor to plasma membrane(GO:0098881) exocytic insertion of neurotransmitter receptor to postsynaptic membrane(GO:0098967) |
| 0.1 | 0.6 | GO:0033031 | positive regulation of neutrophil apoptotic process(GO:0033031) |
| 0.1 | 0.6 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.1 | 0.7 | GO:1900623 | regulation of monocyte aggregation(GO:1900623) positive regulation of monocyte aggregation(GO:1900625) |
| 0.1 | 0.3 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.1 | 0.6 | GO:1903232 | melanosome assembly(GO:1903232) |
| 0.1 | 0.3 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.1 | 0.2 | GO:1990414 | replication-born double-strand break repair via sister chromatid exchange(GO:1990414) |
| 0.1 | 0.5 | GO:0051121 | hepoxilin metabolic process(GO:0051121) hepoxilin biosynthetic process(GO:0051122) |
| 0.1 | 0.5 | GO:0098814 | spontaneous neurotransmitter secretion(GO:0061669) spontaneous synaptic transmission(GO:0098814) |
| 0.1 | 0.2 | GO:0006425 | glutaminyl-tRNA aminoacylation(GO:0006425) |
| 0.1 | 0.2 | GO:0039689 | negative stranded viral RNA replication(GO:0039689) multi-organism biosynthetic process(GO:0044034) |
| 0.1 | 0.6 | GO:1902902 | negative regulation of autophagosome assembly(GO:1902902) |
| 0.1 | 0.3 | GO:0060447 | bud outgrowth involved in lung branching(GO:0060447) |
| 0.1 | 0.2 | GO:0035378 | carbon dioxide transmembrane transport(GO:0035378) |
| 0.1 | 0.9 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.3 | GO:0071461 | cellular response to redox state(GO:0071461) |
| 0.0 | 0.3 | GO:0070827 | chromatin maintenance(GO:0070827) |
| 0.0 | 0.1 | GO:0046586 | regulation of calcium-dependent cell-cell adhesion(GO:0046586) |
| 0.0 | 0.4 | GO:0048840 | otolith development(GO:0048840) |
| 0.0 | 0.2 | GO:0000270 | peptidoglycan metabolic process(GO:0000270) peptidoglycan catabolic process(GO:0009253) |
| 0.0 | 0.1 | GO:0007113 | endomitotic cell cycle(GO:0007113) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
| 0.0 | 0.2 | GO:0046292 | formaldehyde metabolic process(GO:0046292) |
| 0.0 | 0.2 | GO:0042488 | positive regulation of odontogenesis of dentin-containing tooth(GO:0042488) |
| 0.0 | 0.6 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0060133 | somatotropin secreting cell development(GO:0060133) |
| 0.0 | 0.1 | GO:0007343 | egg activation(GO:0007343) |
| 0.0 | 0.1 | GO:0060743 | epithelial cell maturation involved in prostate gland development(GO:0060743) |
| 0.0 | 0.4 | GO:0045078 | positive regulation of interferon-gamma biosynthetic process(GO:0045078) |
| 0.0 | 0.1 | GO:0035521 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.1 | GO:0032747 | positive regulation of interleukin-23 production(GO:0032747) |
| 0.0 | 0.1 | GO:0070837 | dehydroascorbic acid transport(GO:0070837) |
| 0.0 | 0.2 | GO:0046668 | regulation of retinal cell programmed cell death(GO:0046668) |
| 0.0 | 0.1 | GO:0006533 | aspartate catabolic process(GO:0006533) |
| 0.0 | 0.3 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.1 | GO:0036228 | protein targeting to nuclear inner membrane(GO:0036228) |
| 0.0 | 0.1 | GO:1990569 | UDP-N-acetylglucosamine transport(GO:0015788) UDP-N-acetylglucosamine transmembrane transport(GO:1990569) |
| 0.0 | 0.1 | GO:0001951 | intestinal D-glucose absorption(GO:0001951) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.2 | GO:0098706 | ferric iron import into cell(GO:0097461) ferric iron import across plasma membrane(GO:0098706) |
| 0.0 | 0.1 | GO:0019085 | early viral transcription(GO:0019085) |
| 0.0 | 0.4 | GO:0090360 | platelet-derived growth factor production(GO:0090360) regulation of platelet-derived growth factor production(GO:0090361) |
| 0.0 | 0.1 | GO:0099551 | synaptic signaling via neuropeptide(GO:0099538) trans-synaptic signaling by neuropeptide(GO:0099540) trans-synaptic signaling by neuropeptide, modulating synaptic transmission(GO:0099551) |
| 0.0 | 0.3 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.3 | GO:0052696 | flavonoid glucuronidation(GO:0052696) xenobiotic glucuronidation(GO:0052697) |
| 0.0 | 0.1 | GO:0061357 | positive regulation of Wnt protein secretion(GO:0061357) |
| 0.0 | 2.2 | GO:0050911 | detection of chemical stimulus involved in sensory perception of smell(GO:0050911) |
| 0.0 | 0.2 | GO:0098700 | neurotransmitter loading into synaptic vesicle(GO:0098700) |
| 0.0 | 0.1 | GO:0006203 | dGTP catabolic process(GO:0006203) |
| 0.0 | 0.1 | GO:0070105 | positive regulation of interleukin-6-mediated signaling pathway(GO:0070105) |
| 0.0 | 0.3 | GO:0090160 | Golgi to lysosome transport(GO:0090160) |
| 0.0 | 0.2 | GO:2000609 | regulation of thyroid hormone generation(GO:2000609) |
| 0.0 | 0.7 | GO:0016338 | calcium-independent cell-cell adhesion via plasma membrane cell-adhesion molecules(GO:0016338) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0032972 | diaphragm contraction(GO:0002086) regulation of muscle filament sliding speed(GO:0032972) |
| 0.0 | 0.1 | GO:2000053 | regulation of Wnt signaling pathway involved in dorsal/ventral axis specification(GO:2000053) |
| 0.0 | 0.2 | GO:0006452 | translational frameshifting(GO:0006452) positive regulation of translational termination(GO:0045905) |
| 0.0 | 0.1 | GO:0071233 | cellular response to leucine(GO:0071233) |
| 0.0 | 0.1 | GO:0021763 | subthalamic nucleus development(GO:0021763) prolactin secreting cell differentiation(GO:0060127) superior vena cava morphogenesis(GO:0060578) |
| 0.0 | 0.1 | GO:0048850 | hypophysis morphogenesis(GO:0048850) |
| 0.0 | 0.0 | GO:0031914 | negative regulation of synaptic plasticity(GO:0031914) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) eosinophil differentiation(GO:0030222) |
| 0.0 | 0.2 | GO:0043152 | induction of bacterial agglutination(GO:0043152) |
| 0.0 | 2.3 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
| 0.0 | 0.1 | GO:0032468 | Golgi calcium ion homeostasis(GO:0032468) |
| 0.0 | 0.2 | GO:2000210 | positive regulation of anoikis(GO:2000210) |
| 0.0 | 0.1 | GO:0098502 | DNA dephosphorylation(GO:0098502) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.0 | GO:0070407 | oxidation-dependent protein catabolic process(GO:0070407) |
| 0.0 | 0.1 | GO:1901223 | negative regulation of NIK/NF-kappaB signaling(GO:1901223) |
| 0.0 | 0.1 | GO:0021937 | cerebellar Purkinje cell-granule cell precursor cell signaling involved in regulation of granule cell precursor cell proliferation(GO:0021937) |
| 0.0 | 0.2 | GO:0043374 | CD8-positive, alpha-beta T cell differentiation(GO:0043374) |
| 0.0 | 0.1 | GO:0038155 | interleukin-23-mediated signaling pathway(GO:0038155) |
| 0.0 | 0.1 | GO:0070124 | mitochondrial translational initiation(GO:0070124) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.3 | GO:0098719 | sodium ion import across plasma membrane(GO:0098719) sodium ion import into cell(GO:1990118) |
| 0.0 | 0.1 | GO:0032790 | ribosome disassembly(GO:0032790) |
| 0.0 | 0.0 | GO:1904247 | positive regulation of polynucleotide adenylyltransferase activity(GO:1904247) |
| 0.0 | 0.1 | GO:0043605 | allantoin metabolic process(GO:0000255) cellular amide catabolic process(GO:0043605) |
| 0.0 | 0.0 | GO:0033326 | cerebrospinal fluid secretion(GO:0033326) |
| 0.0 | 0.1 | GO:0070973 | protein localization to endoplasmic reticulum exit site(GO:0070973) |
| 0.0 | 0.2 | GO:0008090 | retrograde axonal transport(GO:0008090) |
| 0.0 | 0.1 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0038166 | angiotensin-activated signaling pathway(GO:0038166) |
| 0.0 | 0.1 | GO:0010793 | regulation of mRNA export from nucleus(GO:0010793) |
| 0.0 | 0.1 | GO:0007258 | JUN phosphorylation(GO:0007258) |
| 0.0 | 0.1 | GO:0070508 | sterol import(GO:0035376) cholesterol import(GO:0070508) |
| 0.0 | 0.0 | GO:0060474 | positive regulation of sperm motility involved in capacitation(GO:0060474) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 1.2 | 3.7 | GO:0034685 | integrin alphav-beta6 complex(GO:0034685) |
| 0.2 | 0.6 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 0.7 | GO:1990796 | photoreceptor cell terminal bouton(GO:1990796) |
| 0.1 | 0.5 | GO:0031905 | early endosome lumen(GO:0031905) |
| 0.1 | 0.7 | GO:0035692 | macrophage migration inhibitory factor receptor complex(GO:0035692) |
| 0.1 | 0.2 | GO:0097058 | CRLF-CLCF1 complex(GO:0097058) |
| 0.0 | 1.0 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 1.1 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.2 | GO:0031673 | H zone(GO:0031673) |
| 0.0 | 0.3 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.3 | GO:0070554 | synaptobrevin 2-SNAP-25-syntaxin-3-complexin complex(GO:0070554) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 2.4 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.1 | GO:0044611 | nuclear pore inner ring(GO:0044611) |
| 0.0 | 0.4 | GO:0043020 | NADPH oxidase complex(GO:0043020) |
| 0.0 | 0.1 | GO:1990851 | Wnt-Frizzled-LRP5/6 complex(GO:1990851) |
| 0.0 | 0.1 | GO:1990075 | periciliary membrane compartment(GO:1990075) |
| 0.0 | 0.3 | GO:0005641 | nuclear envelope lumen(GO:0005641) |
| 0.0 | 0.1 | GO:0008024 | cyclin/CDK positive transcription elongation factor complex(GO:0008024) |
| 0.0 | 2.2 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0072536 | interleukin-23 receptor complex(GO:0072536) |
| 0.0 | 0.2 | GO:0005577 | fibrinogen complex(GO:0005577) |
| 0.0 | 0.1 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.0 | 0.1 | GO:0001405 | presequence translocase-associated import motor(GO:0001405) |
| 0.0 | 0.1 | GO:0097513 | myosin II filament(GO:0097513) |
| 0.0 | 0.6 | GO:0005865 | striated muscle thin filament(GO:0005865) |
| 0.0 | 0.2 | GO:0072546 | ER membrane protein complex(GO:0072546) |
| 0.0 | 0.1 | GO:0019773 | proteasome core complex, alpha-subunit complex(GO:0019773) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.9 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.2 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.9 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.2 | 1.1 | GO:0015501 | glutamate:sodium symporter activity(GO:0015501) |
| 0.1 | 0.5 | GO:0035651 | AP-1 adaptor complex binding(GO:0035650) AP-3 adaptor complex binding(GO:0035651) |
| 0.1 | 0.5 | GO:0051120 | hepoxilin A3 synthase activity(GO:0051120) |
| 0.1 | 0.3 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.1 | 2.5 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.1 | 0.5 | GO:0070891 | lipoteichoic acid binding(GO:0070891) |
| 0.1 | 0.7 | GO:0050544 | arachidonic acid binding(GO:0050544) |
| 0.1 | 0.2 | GO:0004819 | glutamine-tRNA ligase activity(GO:0004819) |
| 0.1 | 0.3 | GO:0050220 | prostaglandin-E synthase activity(GO:0050220) |
| 0.1 | 0.2 | GO:0035379 | carbon dioxide transmembrane transporter activity(GO:0035379) |
| 0.1 | 1.3 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.1 | 0.4 | GO:0005134 | interleukin-2 receptor binding(GO:0005134) |
| 0.1 | 0.8 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.7 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.6 | GO:0019871 | sodium channel inhibitor activity(GO:0019871) |
| 0.0 | 0.1 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.0 | 0.2 | GO:0009378 | four-way junction helicase activity(GO:0009378) |
| 0.0 | 0.2 | GO:0004489 | methylenetetrahydrofolate reductase (NAD(P)H) activity(GO:0004489) |
| 0.0 | 0.1 | GO:0035539 | 8-oxo-7,8-dihydroguanosine triphosphate pyrophosphatase activity(GO:0008413) 8-oxo-7,8-dihydrodeoxyguanosine triphosphate pyrophosphatase activity(GO:0035539) |
| 0.0 | 0.2 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.1 | GO:0008502 | melatonin receptor activity(GO:0008502) |
| 0.0 | 3.6 | GO:0001618 | virus receptor activity(GO:0001618) |
| 0.0 | 0.1 | GO:0033300 | dehydroascorbic acid transporter activity(GO:0033300) |
| 0.0 | 0.2 | GO:0001595 | angiotensin receptor activity(GO:0001595) angiotensin type II receptor activity(GO:0004945) |
| 0.0 | 0.2 | GO:0052851 | cupric reductase activity(GO:0008823) ferric-chelate reductase (NADPH) activity(GO:0052851) |
| 0.0 | 0.1 | GO:0004366 | glycerol-3-phosphate O-acyltransferase activity(GO:0004366) |
| 0.0 | 0.3 | GO:0045545 | syndecan binding(GO:0045545) |
| 0.0 | 0.1 | GO:0005462 | UDP-N-acetylglucosamine transmembrane transporter activity(GO:0005462) |
| 0.0 | 0.3 | GO:0005094 | Rho GDP-dissociation inhibitor activity(GO:0005094) |
| 0.0 | 0.2 | GO:0030345 | extracellular matrix structural constituent conferring compression resistance(GO:0030021) structural constituent of tooth enamel(GO:0030345) |
| 0.0 | 0.2 | GO:0005127 | ciliary neurotrophic factor receptor binding(GO:0005127) |
| 0.0 | 0.1 | GO:0060175 | brain-derived neurotrophic factor-activated receptor activity(GO:0060175) |
| 0.0 | 0.9 | GO:0043014 | alpha-tubulin binding(GO:0043014) |
| 0.0 | 0.7 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.3 | GO:0004865 | protein serine/threonine phosphatase inhibitor activity(GO:0004865) |
| 0.0 | 0.1 | GO:0022850 | serotonin-gated cation channel activity(GO:0022850) |
| 0.0 | 0.3 | GO:0015143 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.3 | GO:0015386 | potassium:proton antiporter activity(GO:0015386) |
| 0.0 | 0.1 | GO:0004069 | L-aspartate:2-oxoglutarate aminotransferase activity(GO:0004069) kynurenine-oxoglutarate transaminase activity(GO:0016212) kynurenine aminotransferase activity(GO:0036137) L-phenylalanine aminotransferase activity(GO:0070546) L-phenylalanine:2-oxoglutarate aminotransferase activity(GO:0080130) |
| 0.0 | 0.1 | GO:0004531 | deoxyribonuclease II activity(GO:0004531) |
| 0.0 | 0.1 | GO:0051734 | ATP-dependent polydeoxyribonucleotide 5'-hydroxyl-kinase activity(GO:0046404) polydeoxyribonucleotide kinase activity(GO:0051733) ATP-dependent polynucleotide kinase activity(GO:0051734) polynucleotide phosphatase activity(GO:0098518) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.1 | GO:0043812 | phosphatidylinositol-4-phosphate phosphatase activity(GO:0043812) |
| 0.0 | 0.4 | GO:0016175 | superoxide-generating NADPH oxidase activity(GO:0016175) |
| 0.0 | 1.5 | GO:0004984 | olfactory receptor activity(GO:0004984) |
| 0.0 | 0.1 | GO:0070728 | leucine binding(GO:0070728) |
| 0.0 | 0.1 | GO:0016300 | tRNA (uracil) methyltransferase activity(GO:0016300) |
| 0.0 | 0.0 | GO:0034353 | RNA pyrophosphohydrolase activity(GO:0034353) |
| 0.0 | 0.6 | GO:0043015 | gamma-tubulin binding(GO:0043015) |
| 0.0 | 0.1 | GO:0031726 | CCR1 chemokine receptor binding(GO:0031726) |
| 0.0 | 0.1 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.0 | 0.0 | GO:0070361 | mitochondrial light strand promoter anti-sense binding(GO:0070361) mitochondrial heavy strand promoter anti-sense binding(GO:0070362) mitochondrial heavy strand promoter sense binding(GO:0070364) |
| 0.0 | 0.1 | GO:0016909 | JUN kinase activity(GO:0004705) SAP kinase activity(GO:0016909) |
| 0.0 | 2.8 | GO:0005200 | structural constituent of cytoskeleton(GO:0005200) |
| 0.0 | 0.1 | GO:0005047 | signal recognition particle binding(GO:0005047) |
| 0.0 | 0.1 | GO:0042020 | interleukin-23 binding(GO:0042019) interleukin-23 receptor activity(GO:0042020) |
| 0.0 | 0.8 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.2 | GO:0003796 | lysozyme activity(GO:0003796) |
| 0.0 | 0.1 | GO:0043023 | ribosomal large subunit binding(GO:0043023) |
| 0.0 | 0.0 | GO:0004937 | alpha1-adrenergic receptor activity(GO:0004937) |
| 0.0 | 0.2 | GO:0015125 | bile acid transmembrane transporter activity(GO:0015125) |
| 0.0 | 0.5 | GO:0001972 | retinoic acid binding(GO:0001972) |
| 0.0 | 0.8 | GO:0005044 | scavenger receptor activity(GO:0005044) |
| 0.0 | 0.2 | GO:0032454 | histone demethylase activity (H3-K9 specific)(GO:0032454) |
| 0.0 | 0.0 | GO:0001181 | transcription factor activity, core RNA polymerase I binding(GO:0001181) |
| 0.0 | 0.1 | GO:0005250 | A-type (transient outward) potassium channel activity(GO:0005250) |
| 0.0 | 0.1 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0043225 | anion transmembrane-transporting ATPase activity(GO:0043225) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 4.0 | PID INTEGRIN5 PATHWAY | Beta5 beta6 beta7 and beta8 integrin cell surface interactions |
| 0.1 | 1.9 | SA MMP CYTOKINE CONNECTION | Cytokines can induce activation of matrix metalloproteinases, which degrade extracellular matrix. |
| 0.0 | 0.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 0.9 | PID IL23 PATHWAY | IL23-mediated signaling events |
| 0.0 | 1.8 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.6 | PID ALK1 PATHWAY | ALK1 signaling events |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.9 | REACTOME FGFR2C LIGAND BINDING AND ACTIVATION | Genes involved in FGFR2c ligand binding and activation |
| 0.1 | 1.1 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.6 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.7 | REACTOME HYALURONAN UPTAKE AND DEGRADATION | Genes involved in Hyaluronan uptake and degradation |
| 0.0 | 1.2 | REACTOME TIGHT JUNCTION INTERACTIONS | Genes involved in Tight junction interactions |
| 0.0 | 3.8 | REACTOME INTEGRIN CELL SURFACE INTERACTIONS | Genes involved in Integrin cell surface interactions |
| 0.0 | 0.5 | REACTOME OTHER SEMAPHORIN INTERACTIONS | Genes involved in Other semaphorin interactions |
| 0.0 | 0.3 | REACTOME RECYCLING OF BILE ACIDS AND SALTS | Genes involved in Recycling of bile acids and salts |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.4 | REACTOME TRANSLOCATION OF ZAP 70 TO IMMUNOLOGICAL SYNAPSE | Genes involved in Translocation of ZAP-70 to Immunological synapse |
| 0.0 | 0.1 | REACTOME JNK C JUN KINASES PHOSPHORYLATION AND ACTIVATION MEDIATED BY ACTIVATED HUMAN TAK1 | Genes involved in JNK (c-Jun kinases) phosphorylation and activation mediated by activated human TAK1 |
| 0.0 | 0.7 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 1.4 | REACTOME OLFACTORY SIGNALING PATHWAY | Genes involved in Olfactory Signaling Pathway |
| 0.0 | 0.4 | REACTOME PROTEOLYTIC CLEAVAGE OF SNARE COMPLEX PROTEINS | Genes involved in Proteolytic cleavage of SNARE complex proteins |
| 0.0 | 1.1 | REACTOME AMINO ACID AND OLIGOPEPTIDE SLC TRANSPORTERS | Genes involved in Amino acid and oligopeptide SLC transporters |
| 0.0 | 0.3 | REACTOME AMINE COMPOUND SLC TRANSPORTERS | Genes involved in Amine compound SLC transporters |
| 0.0 | 0.3 | REACTOME BETA DEFENSINS | Genes involved in Beta defensins |