Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
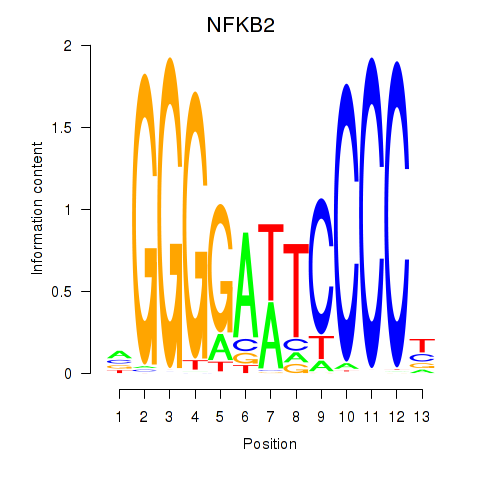
Results for NFKB2
Z-value: 0.67
Transcription factors associated with NFKB2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NFKB2
|
ENSG00000077150.13 | NFKB2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NFKB2 | hg19_v2_chr10_+_104154229_104154354, hg19_v2_chr10_+_104155450_104155479 | 0.24 | 5.7e-01 | Click! |
Activity profile of NFKB2 motif
Sorted Z-values of NFKB2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NFKB2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr18_+_21529811 | 0.38 |
ENST00000588004.1 |
LAMA3 |
laminin, alpha 3 |
| chr9_+_17579084 | 0.21 |
ENST00000380607.4 |
SH3GL2 |
SH3-domain GRB2-like 2 |
| chr19_+_46367518 | 0.20 |
ENST00000302177.2 |
FOXA3 |
forkhead box A3 |
| chr19_+_54371114 | 0.20 |
ENST00000448420.1 ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM |
myeloid-associated differentiation marker |
| chr8_-_145641864 | 0.20 |
ENST00000276833.5 |
SLC39A4 |
solute carrier family 39 (zinc transporter), member 4 |
| chr16_-_68269971 | 0.19 |
ENST00000565858.1 |
ESRP2 |
epithelial splicing regulatory protein 2 |
| chr1_-_186649543 | 0.16 |
ENST00000367468.5 |
PTGS2 |
prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) |
| chr1_-_209979375 | 0.16 |
ENST00000367021.3 |
IRF6 |
interferon regulatory factor 6 |
| chr9_-_139948487 | 0.15 |
ENST00000355097.2 |
ENTPD2 |
ectonucleoside triphosphate diphosphohydrolase 2 |
| chr22_-_25801333 | 0.15 |
ENST00000444995.3 |
LRP5L |
low density lipoprotein receptor-related protein 5-like |
| chr20_+_43803517 | 0.14 |
ENST00000243924.3 |
PI3 |
peptidase inhibitor 3, skin-derived |
| chr5_+_137203557 | 0.14 |
ENST00000515645.1 |
MYOT |
myotilin |
| chr5_+_137203541 | 0.14 |
ENST00000421631.2 |
MYOT |
myotilin |
| chr1_-_159915386 | 0.13 |
ENST00000361509.3 ENST00000368094.1 |
IGSF9 |
immunoglobulin superfamily, member 9 |
| chr2_-_70781087 | 0.13 |
ENST00000394241.3 ENST00000295400.6 |
TGFA |
transforming growth factor, alpha |
| chr16_+_57653854 | 0.13 |
ENST00000568908.1 ENST00000568909.1 ENST00000566778.1 ENST00000561988.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr16_+_57653989 | 0.13 |
ENST00000567835.1 ENST00000569372.1 ENST00000563548.1 ENST00000562003.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr11_-_18270182 | 0.12 |
ENST00000528349.1 ENST00000526900.1 ENST00000529528.1 ENST00000414546.2 ENST00000256733.4 |
SAA2 |
serum amyloid A2 |
| chr15_-_89438742 | 0.12 |
ENST00000562281.1 ENST00000562889.1 ENST00000359595.3 |
HAPLN3 |
hyaluronan and proteoglycan link protein 3 |
| chr4_+_74735102 | 0.12 |
ENST00000395761.3 |
CXCL1 |
chemokine (C-X-C motif) ligand 1 (melanoma growth stimulating activity, alpha) |
| chr6_+_80341000 | 0.11 |
ENST00000369838.4 |
SH3BGRL2 |
SH3 domain binding glutamic acid-rich protein like 2 |
| chr8_+_22436635 | 0.11 |
ENST00000452226.1 ENST00000397760.4 ENST00000339162.7 ENST00000397761.2 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr1_-_6545502 | 0.11 |
ENST00000535355.1 |
PLEKHG5 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 5 |
| chr22_-_20255212 | 0.10 |
ENST00000416372.1 |
RTN4R |
reticulon 4 receptor |
| chr2_-_70780770 | 0.10 |
ENST00000444975.1 ENST00000445399.1 ENST00000418333.2 |
TGFA |
transforming growth factor, alpha |
| chr21_+_27011899 | 0.10 |
ENST00000425221.2 |
JAM2 |
junctional adhesion molecule 2 |
| chr1_+_25943959 | 0.10 |
ENST00000374332.4 |
MAN1C1 |
mannosidase, alpha, class 1C, member 1 |
| chr16_+_75256507 | 0.10 |
ENST00000495583.1 |
CTRB1 |
chymotrypsinogen B1 |
| chr9_+_127539481 | 0.10 |
ENST00000373580.3 |
OLFML2A |
olfactomedin-like 2A |
| chr9_+_139377947 | 0.09 |
ENST00000354376.1 |
C9orf163 |
chromosome 9 open reading frame 163 |
| chr8_+_22436248 | 0.09 |
ENST00000308354.7 |
PDLIM2 |
PDZ and LIM domain 2 (mystique) |
| chr12_-_322504 | 0.09 |
ENST00000424061.2 |
SLC6A12 |
solute carrier family 6 (neurotransmitter transporter), member 12 |
| chr3_+_111718036 | 0.09 |
ENST00000455401.2 |
TAGLN3 |
transgelin 3 |
| chr7_-_100061869 | 0.09 |
ENST00000332375.3 |
C7orf61 |
chromosome 7 open reading frame 61 |
| chr12_-_57352103 | 0.09 |
ENST00000398138.3 |
RDH16 |
retinol dehydrogenase 16 (all-trans) |
| chr19_-_52227221 | 0.09 |
ENST00000222115.1 ENST00000540069.2 |
HAS1 |
hyaluronan synthase 1 |
| chr8_+_86376081 | 0.09 |
ENST00000285379.5 |
CA2 |
carbonic anhydrase II |
| chr15_+_90234028 | 0.09 |
ENST00000268130.7 ENST00000560294.1 ENST00000558000.1 |
WDR93 |
WD repeat domain 93 |
| chr3_+_111717600 | 0.09 |
ENST00000273368.4 |
TAGLN3 |
transgelin 3 |
| chr8_+_143916217 | 0.09 |
ENST00000220940.1 |
GML |
glycosylphosphatidylinositol anchored molecule like |
| chr6_+_14117872 | 0.08 |
ENST00000379153.3 |
CD83 |
CD83 molecule |
| chr17_-_26127525 | 0.08 |
ENST00000313735.6 |
NOS2 |
nitric oxide synthase 2, inducible |
| chr21_+_27011584 | 0.08 |
ENST00000400532.1 ENST00000480456.1 ENST00000312957.5 |
JAM2 |
junctional adhesion molecule 2 |
| chr17_+_40440481 | 0.08 |
ENST00000590726.2 ENST00000452307.2 ENST00000444283.1 ENST00000588868.1 |
STAT5A |
signal transducer and activator of transcription 5A |
| chr10_+_124030819 | 0.08 |
ENST00000260723.4 ENST00000368994.2 |
BTBD16 |
BTB (POZ) domain containing 16 |
| chr17_-_7193711 | 0.08 |
ENST00000571464.1 |
YBX2 |
Y box binding protein 2 |
| chr19_+_45504688 | 0.08 |
ENST00000221452.8 ENST00000540120.1 ENST00000505236.1 |
RELB |
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr3_-_187455680 | 0.08 |
ENST00000438077.1 |
BCL6 |
B-cell CLL/lymphoma 6 |
| chr9_+_103790991 | 0.08 |
ENST00000374874.3 |
LPPR1 |
Lipid phosphate phosphatase-related protein type 1 |
| chr1_-_183559693 | 0.07 |
ENST00000367535.3 ENST00000413720.1 ENST00000418089.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr1_-_183560011 | 0.07 |
ENST00000367536.1 |
NCF2 |
neutrophil cytosolic factor 2 |
| chr16_+_31539197 | 0.07 |
ENST00000564707.1 |
AHSP |
alpha hemoglobin stabilizing protein |
| chr12_+_56473628 | 0.07 |
ENST00000549282.1 ENST00000549061.1 ENST00000267101.3 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr8_+_32406179 | 0.07 |
ENST00000405005.3 |
NRG1 |
neuregulin 1 |
| chr17_+_9548845 | 0.07 |
ENST00000570475.1 ENST00000285199.7 |
USP43 |
ubiquitin specific peptidase 43 |
| chr17_+_12569306 | 0.07 |
ENST00000425538.1 |
MYOCD |
myocardin |
| chr17_-_39928106 | 0.07 |
ENST00000540235.1 |
JUP |
junction plakoglobin |
| chr19_-_49401990 | 0.07 |
ENST00000221399.3 |
TULP2 |
tubby like protein 2 |
| chr11_+_706113 | 0.07 |
ENST00000318562.8 ENST00000533256.1 ENST00000534755.1 |
EPS8L2 |
EPS8-like 2 |
| chr17_-_6459802 | 0.07 |
ENST00000262483.8 |
PITPNM3 |
PITPNM family member 3 |
| chr7_-_38403077 | 0.07 |
ENST00000426402.2 |
TRGV2 |
T cell receptor gamma variable 2 |
| chr2_+_61108771 | 0.07 |
ENST00000394479.3 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr3_+_96533413 | 0.07 |
ENST00000470610.2 ENST00000389672.5 |
EPHA6 |
EPH receptor A6 |
| chr14_+_23842018 | 0.07 |
ENST00000397242.2 ENST00000329715.2 |
IL25 |
interleukin 25 |
| chr9_+_82187630 | 0.07 |
ENST00000265284.6 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr9_+_82187487 | 0.07 |
ENST00000435650.1 ENST00000414465.1 ENST00000376537.4 ENST00000376534.4 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr17_+_8924837 | 0.07 |
ENST00000173229.2 |
NTN1 |
netrin 1 |
| chr1_+_151735431 | 0.06 |
ENST00000321531.5 ENST00000315067.8 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr17_+_16318850 | 0.06 |
ENST00000338560.7 |
TRPV2 |
transient receptor potential cation channel, subfamily V, member 2 |
| chrX_-_24665208 | 0.06 |
ENST00000356768.4 |
PCYT1B |
phosphate cytidylyltransferase 1, choline, beta |
| chr2_+_86947296 | 0.06 |
ENST00000283632.4 |
RMND5A |
required for meiotic nuclear division 5 homolog A (S. cerevisiae) |
| chr1_-_42921915 | 0.06 |
ENST00000372565.3 ENST00000433602.2 |
ZMYND12 |
zinc finger, MYND-type containing 12 |
| chr7_+_142031986 | 0.06 |
ENST00000547918.2 |
TRBV7-1 |
T cell receptor beta variable 7-1 (non-functional) |
| chr16_-_57318566 | 0.06 |
ENST00000569059.1 ENST00000219207.5 |
PLLP |
plasmolipin |
| chr1_+_244624678 | 0.06 |
ENST00000366534.4 ENST00000366533.4 ENST00000428042.1 ENST00000366531.3 |
C1orf101 |
chromosome 1 open reading frame 101 |
| chr6_-_109762344 | 0.06 |
ENST00000521072.2 ENST00000424445.2 ENST00000440797.2 |
PPIL6 |
peptidylprolyl isomerase (cyclophilin)-like 6 |
| chr17_+_16318909 | 0.06 |
ENST00000577397.1 |
TRPV2 |
transient receptor potential cation channel, subfamily V, member 2 |
| chr19_-_33555780 | 0.06 |
ENST00000254260.3 ENST00000400226.4 |
RHPN2 |
rhophilin, Rho GTPase binding protein 2 |
| chr19_+_45842445 | 0.06 |
ENST00000598357.1 |
L47234.1 |
Uncharacterized protein |
| chr4_+_86396321 | 0.06 |
ENST00000503995.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr17_+_25799008 | 0.06 |
ENST00000583370.1 ENST00000398988.3 ENST00000268763.6 |
KSR1 |
kinase suppressor of ras 1 |
| chr12_+_120740119 | 0.06 |
ENST00000536460.1 ENST00000202967.4 |
SIRT4 |
sirtuin 4 |
| chr16_-_46864955 | 0.06 |
ENST00000565112.1 |
C16orf87 |
chromosome 16 open reading frame 87 |
| chr12_+_56473939 | 0.06 |
ENST00000450146.2 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chrX_+_130192318 | 0.06 |
ENST00000370922.1 |
ARHGAP36 |
Rho GTPase activating protein 36 |
| chr1_-_24513737 | 0.06 |
ENST00000374421.3 ENST00000374418.3 ENST00000327535.1 ENST00000327575.2 |
IFNLR1 |
interferon, lambda receptor 1 |
| chr19_-_38747172 | 0.06 |
ENST00000347262.4 ENST00000591585.1 ENST00000301242.4 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr9_+_33524240 | 0.06 |
ENST00000290943.6 |
ANKRD18B |
ankyrin repeat domain 18B |
| chr17_-_6459768 | 0.06 |
ENST00000421306.3 |
PITPNM3 |
PITPNM family member 3 |
| chr11_+_64358686 | 0.06 |
ENST00000473690.1 |
SLC22A12 |
solute carrier family 22 (organic anion/urate transporter), member 12 |
| chr1_+_32083301 | 0.05 |
ENST00000403528.2 |
HCRTR1 |
hypocretin (orexin) receptor 1 |
| chr19_-_4338783 | 0.05 |
ENST00000601482.1 ENST00000600324.1 |
STAP2 |
signal transducing adaptor family member 2 |
| chr21_+_17443521 | 0.05 |
ENST00000456342.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr16_+_2867228 | 0.05 |
ENST00000005995.3 ENST00000574813.1 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr18_+_43914159 | 0.05 |
ENST00000588679.1 ENST00000269439.7 ENST00000543885.1 |
RNF165 |
ring finger protein 165 |
| chr19_-_8567478 | 0.05 |
ENST00000255612.3 |
PRAM1 |
PML-RARA regulated adaptor molecule 1 |
| chr11_+_60691924 | 0.05 |
ENST00000544065.1 ENST00000453848.2 ENST00000005286.4 |
TMEM132A |
transmembrane protein 132A |
| chr8_-_66754172 | 0.05 |
ENST00000401827.3 |
PDE7A |
phosphodiesterase 7A |
| chr21_+_17443434 | 0.05 |
ENST00000400178.2 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr17_-_34207295 | 0.05 |
ENST00000463941.1 ENST00000293272.3 |
CCL5 |
chemokine (C-C motif) ligand 5 |
| chr9_+_130911770 | 0.05 |
ENST00000372998.1 |
LCN2 |
lipocalin 2 |
| chr9_+_35792151 | 0.05 |
ENST00000342694.2 |
NPR2 |
natriuretic peptide receptor B/guanylate cyclase B (atrionatriuretic peptide receptor B) |
| chr17_+_63133587 | 0.05 |
ENST00000449996.3 ENST00000262406.9 |
RGS9 |
regulator of G-protein signaling 9 |
| chrX_+_103357202 | 0.05 |
ENST00000537356.3 |
ZCCHC18 |
zinc finger, CCHC domain containing 18 |
| chr9_-_123691047 | 0.05 |
ENST00000373887.3 |
TRAF1 |
TNF receptor-associated factor 1 |
| chr16_+_2867164 | 0.05 |
ENST00000455114.1 ENST00000450020.3 |
PRSS21 |
protease, serine, 21 (testisin) |
| chr1_+_26503894 | 0.05 |
ENST00000361530.6 ENST00000374253.5 |
CNKSR1 |
connector enhancer of kinase suppressor of Ras 1 |
| chr1_-_85462762 | 0.05 |
ENST00000284027.5 |
MCOLN2 |
mucolipin 2 |
| chr6_-_117086873 | 0.05 |
ENST00000368557.4 |
FAM162B |
family with sequence similarity 162, member B |
| chr15_+_77287426 | 0.05 |
ENST00000558012.1 ENST00000267939.5 ENST00000379595.3 |
PSTPIP1 |
proline-serine-threonine phosphatase interacting protein 1 |
| chr11_+_64358110 | 0.05 |
ENST00000377567.2 |
SLC22A12 |
solute carrier family 22 (organic anion/urate transporter), member 12 |
| chr19_-_47734448 | 0.05 |
ENST00000439096.2 |
BBC3 |
BCL2 binding component 3 |
| chr7_+_150811705 | 0.05 |
ENST00000335367.3 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr9_+_140145713 | 0.05 |
ENST00000388931.3 ENST00000412566.1 |
C9orf173 |
chromosome 9 open reading frame 173 |
| chr19_-_4338838 | 0.05 |
ENST00000594605.1 |
STAP2 |
signal transducing adaptor family member 2 |
| chr6_-_24358264 | 0.05 |
ENST00000378454.3 |
DCDC2 |
doublecortin domain containing 2 |
| chr1_-_47082495 | 0.05 |
ENST00000545730.1 ENST00000531769.1 ENST00000319928.3 |
MKNK1 MOB3C |
MAP kinase interacting serine/threonine kinase 1 MOB kinase activator 3C |
| chr12_-_57504069 | 0.05 |
ENST00000543873.2 ENST00000554663.1 ENST00000557635.1 |
STAT6 |
signal transducer and activator of transcription 6, interleukin-4 induced |
| chr12_+_57998595 | 0.05 |
ENST00000337737.3 ENST00000548198.1 ENST00000551632.1 |
DTX3 |
deltex homolog 3 (Drosophila) |
| chr11_+_706219 | 0.05 |
ENST00000533500.1 |
EPS8L2 |
EPS8-like 2 |
| chr6_+_132891461 | 0.05 |
ENST00000275198.1 |
TAAR6 |
trace amine associated receptor 6 |
| chr19_-_38746979 | 0.05 |
ENST00000591291.1 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr15_-_52587945 | 0.05 |
ENST00000443683.2 ENST00000558479.1 ENST00000261839.7 |
MYO5C |
myosin VC |
| chr9_+_115913222 | 0.05 |
ENST00000259392.3 |
SLC31A2 |
solute carrier family 31 (copper transporter), member 2 |
| chr19_+_35940486 | 0.05 |
ENST00000246549.2 |
FFAR2 |
free fatty acid receptor 2 |
| chr10_+_99609996 | 0.05 |
ENST00000370602.1 |
GOLGA7B |
golgin A7 family, member B |
| chr9_+_130911723 | 0.04 |
ENST00000277480.2 ENST00000373013.2 ENST00000540948.1 |
LCN2 |
lipocalin 2 |
| chr1_+_24117627 | 0.04 |
ENST00000400061.1 |
LYPLA2 |
lysophospholipase II |
| chr4_+_74702214 | 0.04 |
ENST00000226317.5 ENST00000515050.1 |
CXCL6 |
chemokine (C-X-C motif) ligand 6 |
| chr17_-_34195889 | 0.04 |
ENST00000311880.2 |
C17orf66 |
chromosome 17 open reading frame 66 |
| chr17_-_40333150 | 0.04 |
ENST00000264661.3 |
KCNH4 |
potassium voltage-gated channel, subfamily H (eag-related), member 4 |
| chr10_-_86001210 | 0.04 |
ENST00000372105.3 |
LRIT1 |
leucine-rich repeat, immunoglobulin-like and transmembrane domains 1 |
| chr19_-_14640005 | 0.04 |
ENST00000596853.1 ENST00000596075.1 ENST00000595992.1 ENST00000396969.4 ENST00000601533.1 ENST00000598692.1 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr5_+_60241020 | 0.04 |
ENST00000511107.1 ENST00000502658.1 |
NDUFAF2 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
| chr11_-_104840093 | 0.04 |
ENST00000417440.2 ENST00000444739.2 |
CASP4 |
caspase 4, apoptosis-related cysteine peptidase |
| chr1_-_85462623 | 0.04 |
ENST00000370608.3 |
MCOLN2 |
mucolipin 2 |
| chr1_-_209824643 | 0.04 |
ENST00000391911.1 ENST00000415782.1 |
LAMB3 |
laminin, beta 3 |
| chr2_-_211036051 | 0.04 |
ENST00000418791.1 ENST00000452086.1 ENST00000281772.9 |
KANSL1L |
KAT8 regulatory NSL complex subunit 1-like |
| chr17_-_34195862 | 0.04 |
ENST00000592980.1 ENST00000587626.1 |
C17orf66 |
chromosome 17 open reading frame 66 |
| chr1_+_42921761 | 0.04 |
ENST00000372562.1 |
PPCS |
phosphopantothenoylcysteine synthetase |
| chr19_-_33716750 | 0.04 |
ENST00000253188.4 |
SLC7A10 |
solute carrier family 7 (neutral amino acid transporter light chain, asc system), member 10 |
| chr20_-_22559211 | 0.04 |
ENST00000564492.1 |
LINC00261 |
long intergenic non-protein coding RNA 261 |
| chr2_-_208490027 | 0.04 |
ENST00000458426.1 ENST00000406927.2 ENST00000425132.1 |
METTL21A |
methyltransferase like 21A |
| chr7_-_27213893 | 0.04 |
ENST00000283921.4 |
HOXA10 |
homeobox A10 |
| chr11_-_2924720 | 0.04 |
ENST00000455942.2 |
SLC22A18AS |
solute carrier family 22 (organic cation transporter), member 18 antisense |
| chr1_+_45965725 | 0.04 |
ENST00000401061.4 |
MMACHC |
methylmalonic aciduria (cobalamin deficiency) cblC type, with homocystinuria |
| chr19_-_4831701 | 0.04 |
ENST00000248244.5 |
TICAM1 |
toll-like receptor adaptor molecule 1 |
| chr6_+_18155560 | 0.04 |
ENST00000546309.2 ENST00000388870.2 ENST00000397244.1 |
KDM1B |
lysine (K)-specific demethylase 1B |
| chr19_-_14628645 | 0.04 |
ENST00000598235.1 |
DNAJB1 |
DnaJ (Hsp40) homolog, subfamily B, member 1 |
| chr4_-_122854612 | 0.04 |
ENST00000264811.5 |
TRPC3 |
transient receptor potential cation channel, subfamily C, member 3 |
| chr14_+_32798462 | 0.04 |
ENST00000280979.4 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr6_+_18155632 | 0.04 |
ENST00000297792.5 |
KDM1B |
lysine (K)-specific demethylase 1B |
| chr1_+_111770278 | 0.04 |
ENST00000369748.4 |
CHI3L2 |
chitinase 3-like 2 |
| chr1_+_111770232 | 0.04 |
ENST00000369744.2 |
CHI3L2 |
chitinase 3-like 2 |
| chr5_+_60240943 | 0.04 |
ENST00000296597.5 |
NDUFAF2 |
NADH dehydrogenase (ubiquinone) complex I, assembly factor 2 |
| chr6_+_135502408 | 0.04 |
ENST00000341911.5 ENST00000442647.2 ENST00000316528.8 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr9_+_139877445 | 0.04 |
ENST00000408973.2 |
LCNL1 |
lipocalin-like 1 |
| chr2_+_61108650 | 0.04 |
ENST00000295025.8 |
REL |
v-rel avian reticuloendotheliosis viral oncogene homolog |
| chr14_+_92789498 | 0.04 |
ENST00000531433.1 |
SLC24A4 |
solute carrier family 24 (sodium/potassium/calcium exchanger), member 4 |
| chr10_+_104154229 | 0.04 |
ENST00000428099.1 ENST00000369966.3 |
NFKB2 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 2 (p49/p100) |
| chr10_+_112257596 | 0.04 |
ENST00000369583.3 |
DUSP5 |
dual specificity phosphatase 5 |
| chr3_+_23847394 | 0.04 |
ENST00000306627.3 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
| chr4_+_86396265 | 0.04 |
ENST00000395184.1 |
ARHGAP24 |
Rho GTPase activating protein 24 |
| chr6_+_90142884 | 0.04 |
ENST00000369408.5 ENST00000339746.4 ENST00000447838.2 |
ANKRD6 |
ankyrin repeat domain 6 |
| chr14_+_32798547 | 0.04 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr2_-_109605663 | 0.04 |
ENST00000409271.1 ENST00000258443.2 ENST00000376651.1 |
EDAR |
ectodysplasin A receptor |
| chr7_+_100547156 | 0.04 |
ENST00000379458.4 |
MUC3A |
Protein LOC100131514 |
| chr1_-_33283754 | 0.04 |
ENST00000373477.4 |
YARS |
tyrosyl-tRNA synthetase |
| chr16_-_10674528 | 0.04 |
ENST00000359543.3 |
EMP2 |
epithelial membrane protein 2 |
| chr7_+_156433396 | 0.04 |
ENST00000432459.2 |
RNF32 |
ring finger protein 32 |
| chr17_+_28884130 | 0.03 |
ENST00000580161.1 |
TBC1D29 |
TBC1 domain family, member 29 |
| chr2_+_42795745 | 0.03 |
ENST00000406911.1 |
MTA3 |
metastasis associated 1 family, member 3 |
| chr2_+_242127924 | 0.03 |
ENST00000402530.3 ENST00000274979.8 ENST00000402430.3 |
ANO7 |
anoctamin 7 |
| chr17_+_76164639 | 0.03 |
ENST00000225777.3 ENST00000585591.1 ENST00000589711.1 ENST00000588282.1 ENST00000589168.1 |
SYNGR2 |
synaptogyrin 2 |
| chr2_-_136743169 | 0.03 |
ENST00000264161.4 |
DARS |
aspartyl-tRNA synthetase |
| chr12_+_7055631 | 0.03 |
ENST00000543115.1 ENST00000399448.1 |
PTPN6 |
protein tyrosine phosphatase, non-receptor type 6 |
| chr12_+_124457746 | 0.03 |
ENST00000392404.3 ENST00000538932.2 ENST00000337815.4 ENST00000540762.2 |
ZNF664 FAM101A |
zinc finger protein 664 family with sequence similarity 101, member A |
| chr12_-_54673871 | 0.03 |
ENST00000209875.4 |
CBX5 |
chromobox homolog 5 |
| chr3_-_50340996 | 0.03 |
ENST00000266031.4 ENST00000395143.2 ENST00000457214.2 ENST00000447605.2 ENST00000418723.1 ENST00000395144.2 |
HYAL1 |
hyaluronoglucosaminidase 1 |
| chrX_-_1572629 | 0.03 |
ENST00000534940.1 |
ASMTL |
acetylserotonin O-methyltransferase-like |
| chr1_-_21978312 | 0.03 |
ENST00000359708.4 ENST00000290101.4 |
RAP1GAP |
RAP1 GTPase activating protein |
| chrX_+_68048803 | 0.03 |
ENST00000204961.4 |
EFNB1 |
ephrin-B1 |
| chr5_-_178772424 | 0.03 |
ENST00000251582.7 ENST00000274609.5 |
ADAMTS2 |
ADAM metallopeptidase with thrombospondin type 1 motif, 2 |
| chr1_-_205744574 | 0.03 |
ENST00000367139.3 ENST00000235932.4 ENST00000437324.2 ENST00000414729.1 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr1_-_205744205 | 0.03 |
ENST00000446390.2 |
RAB7L1 |
RAB7, member RAS oncogene family-like 1 |
| chr21_+_17442799 | 0.03 |
ENST00000602580.1 ENST00000458468.1 ENST00000602935.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr11_-_14913190 | 0.03 |
ENST00000532378.1 |
CYP2R1 |
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr4_+_8271471 | 0.03 |
ENST00000307358.2 ENST00000382512.3 |
HTRA3 |
HtrA serine peptidase 3 |
| chr3_-_50329835 | 0.03 |
ENST00000429673.2 |
IFRD2 |
interferon-related developmental regulator 2 |
| chr17_+_60536002 | 0.03 |
ENST00000582809.1 |
TLK2 |
tousled-like kinase 2 |
| chr6_+_149068464 | 0.03 |
ENST00000367463.4 |
UST |
uronyl-2-sulfotransferase |
| chr3_+_111718173 | 0.03 |
ENST00000494932.1 |
TAGLN3 |
transgelin 3 |
| chr1_-_247171347 | 0.03 |
ENST00000339986.7 ENST00000487338.2 |
ZNF695 |
zinc finger protein 695 |
| chr3_-_50329990 | 0.03 |
ENST00000417626.2 |
IFRD2 |
interferon-related developmental regulator 2 |
| chrX_-_20134990 | 0.03 |
ENST00000379651.3 ENST00000443379.3 ENST00000379643.5 |
MAP7D2 |
MAP7 domain containing 2 |
| chr10_+_89622870 | 0.03 |
ENST00000371953.3 |
PTEN |
phosphatase and tensin homolog |
| chr16_-_88878305 | 0.03 |
ENST00000569616.1 ENST00000563655.1 ENST00000567713.1 ENST00000426324.2 ENST00000378364.3 |
APRT |
adenine phosphoribosyltransferase |
| chr1_+_154474689 | 0.03 |
ENST00000368482.4 |
TDRD10 |
tudor domain containing 10 |
| chr2_+_26915584 | 0.03 |
ENST00000302909.3 |
KCNK3 |
potassium channel, subfamily K, member 3 |
| chr6_-_32157947 | 0.03 |
ENST00000375050.4 |
PBX2 |
pre-B-cell leukemia homeobox 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0090271 | positive regulation of fibroblast growth factor production(GO:0090271) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.2 | GO:0034224 | cellular response to zinc ion starvation(GO:0034224) |
| 0.0 | 0.1 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.0 | 0.1 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.0 | 0.1 | GO:0032764 | negative regulation of mast cell cytokine production(GO:0032764) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) negative regulation of beta-amyloid clearance(GO:1900222) |
| 0.0 | 0.1 | GO:0009137 | purine nucleoside diphosphate catabolic process(GO:0009137) purine ribonucleoside diphosphate catabolic process(GO:0009181) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:0000255 | allantoin metabolic process(GO:0000255) |
| 0.0 | 0.1 | GO:0006422 | aspartyl-tRNA aminoacylation(GO:0006422) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.1 | GO:0015891 | iron chelate transport(GO:0015688) siderophore transport(GO:0015891) |
| 0.0 | 0.1 | GO:1904381 | Golgi apparatus mannose trimming(GO:1904381) |
| 0.0 | 0.0 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.0 | 0.1 | GO:1903215 | negative regulation of protein targeting to mitochondrion(GO:1903215) |
| 0.0 | 0.4 | GO:0031581 | hemidesmosome assembly(GO:0031581) |
| 0.0 | 0.1 | GO:0002159 | desmosome assembly(GO:0002159) endothelial cell-cell adhesion(GO:0071603) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0002268 | follicular dendritic cell differentiation(GO:0002268) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0033634 | positive regulation of cell-cell adhesion mediated by integrin(GO:0033634) |
| 0.0 | 0.1 | GO:1900194 | negative regulation of oocyte maturation(GO:1900194) |
| 0.0 | 0.1 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.1 | GO:0046122 | purine deoxyribonucleoside metabolic process(GO:0046122) |
| 0.0 | 0.0 | GO:1990922 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.0 | 0.1 | GO:1902261 | positive regulation of delayed rectifier potassium channel activity(GO:1902261) |
| 0.0 | 0.2 | GO:0032688 | negative regulation of interferon-beta production(GO:0032688) |
| 0.0 | 0.0 | GO:0002296 | T-helper 1 cell lineage commitment(GO:0002296) |
| 0.0 | 0.1 | GO:0009624 | response to nematode(GO:0009624) |
| 0.0 | 0.0 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.0 | GO:1903697 | negative regulation of microvillus assembly(GO:1903697) |
| 0.0 | 0.1 | GO:1900029 | positive regulation of ruffle assembly(GO:1900029) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0005610 | laminin-5 complex(GO:0005610) |
| 0.0 | 0.1 | GO:0034657 | GID complex(GO:0034657) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:0032002 | interleukin-28 receptor complex(GO:0032002) |
| 0.0 | 0.1 | GO:0032010 | phagolysosome(GO:0032010) |
| 0.0 | 0.1 | GO:0032584 | growth cone membrane(GO:0032584) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0038131 | neuregulin receptor activity(GO:0038131) |
| 0.0 | 0.1 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.1 | GO:0005332 | gamma-aminobutyric acid:sodium symporter activity(GO:0005332) |
| 0.0 | 0.1 | GO:0047708 | biotinidase activity(GO:0047708) |
| 0.0 | 0.1 | GO:0008526 | phosphatidylinositol transporter activity(GO:0008526) |
| 0.0 | 0.1 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.1 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.1 | GO:0030492 | hemoglobin binding(GO:0030492) |
| 0.0 | 0.1 | GO:0004105 | choline-phosphate cytidylyltransferase activity(GO:0004105) |
| 0.0 | 0.1 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.0 | 0.2 | GO:0045236 | CXCR chemokine receptor binding(GO:0045236) |
| 0.0 | 0.1 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.0 | 0.1 | GO:0016176 | superoxide-generating NADPH oxidase activator activity(GO:0016176) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.4 | PID INTEGRIN4 PATHWAY | Alpha6 beta4 integrin-ligand interactions |
| 0.0 | 0.4 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |