Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for NKX1-1
Z-value: 0.23
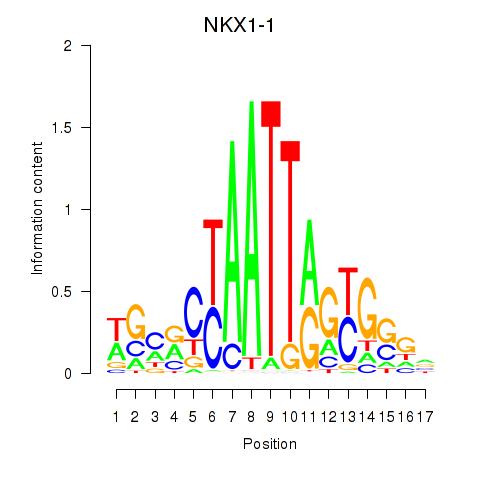
Transcription factors associated with NKX1-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX1-1
|
ENSG00000235608.1 | NKX1-1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX1-1 | hg19_v2_chr4_-_1400119_1400119 | 0.51 | 1.9e-01 | Click! |
Activity profile of NKX1-1 motif
Sorted Z-values of NKX1-1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX1-1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr6_+_26501449 | 0.20 |
ENST00000244513.6 |
BTN1A1 |
butyrophilin, subfamily 1, member A1 |
| chr1_-_159915386 | 0.19 |
ENST00000361509.3 ENST00000368094.1 |
IGSF9 |
immunoglobulin superfamily, member 9 |
| chr3_+_186560462 | 0.16 |
ENST00000412955.2 |
ADIPOQ |
adiponectin, C1Q and collagen domain containing |
| chr3_+_186560476 | 0.16 |
ENST00000320741.2 ENST00000444204.2 |
ADIPOQ |
adiponectin, C1Q and collagen domain containing |
| chr7_+_26331541 | 0.12 |
ENST00000416246.1 ENST00000338523.4 ENST00000412416.1 |
SNX10 |
sorting nexin 10 |
| chr5_+_66300446 | 0.11 |
ENST00000261569.7 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr1_-_197115818 | 0.11 |
ENST00000367409.4 ENST00000294732.7 |
ASPM |
asp (abnormal spindle) homolog, microcephaly associated (Drosophila) |
| chr18_+_55888767 | 0.10 |
ENST00000431212.2 ENST00000586268.1 ENST00000587190.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr2_-_239197201 | 0.10 |
ENST00000254658.3 |
PER2 |
period circadian clock 2 |
| chr1_+_161691353 | 0.10 |
ENST00000367948.2 |
FCRLB |
Fc receptor-like B |
| chr2_-_239197238 | 0.10 |
ENST00000254657.3 |
PER2 |
period circadian clock 2 |
| chr1_+_62439037 | 0.09 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr12_-_91451758 | 0.09 |
ENST00000266719.3 |
KERA |
keratocan |
| chr2_-_97405775 | 0.09 |
ENST00000264963.4 ENST00000537039.1 ENST00000377079.4 ENST00000426463.2 ENST00000534882.1 |
LMAN2L |
lectin, mannose-binding 2-like |
| chr14_+_22670455 | 0.09 |
ENST00000390460.1 |
TRAV26-2 |
T cell receptor alpha variable 26-2 |
| chr4_+_87515454 | 0.09 |
ENST00000427191.2 ENST00000436978.1 ENST00000502971.1 |
PTPN13 |
protein tyrosine phosphatase, non-receptor type 13 (APO-1/CD95 (Fas)-associated phosphatase) |
| chr17_-_27418537 | 0.08 |
ENST00000408971.2 |
TIAF1 |
TGFB1-induced anti-apoptotic factor 1 |
| chr11_-_111741994 | 0.08 |
ENST00000398006.2 |
ALG9 |
ALG9, alpha-1,2-mannosyltransferase |
| chr3_-_59035673 | 0.08 |
ENST00000491845.1 ENST00000472469.1 ENST00000471288.1 ENST00000295966.7 |
C3orf67 |
chromosome 3 open reading frame 67 |
| chr3_+_158519654 | 0.08 |
ENST00000415822.2 ENST00000392813.4 ENST00000264266.8 |
MFSD1 |
major facilitator superfamily domain containing 1 |
| chr12_+_16500599 | 0.07 |
ENST00000535309.1 ENST00000540056.1 ENST00000396209.1 ENST00000540126.1 |
MGST1 |
microsomal glutathione S-transferase 1 |
| chr8_+_98656693 | 0.07 |
ENST00000519934.1 |
MTDH |
metadherin |
| chr8_+_94929168 | 0.06 |
ENST00000518107.1 ENST00000396200.3 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr4_-_54232144 | 0.06 |
ENST00000388940.4 ENST00000503450.1 ENST00000401642.3 |
SCFD2 |
sec1 family domain containing 2 |
| chr12_+_16500571 | 0.06 |
ENST00000543076.1 ENST00000396210.3 |
MGST1 |
microsomal glutathione S-transferase 1 |
| chr18_-_33709268 | 0.06 |
ENST00000269187.5 ENST00000590986.1 ENST00000440549.2 |
SLC39A6 |
solute carrier family 39 (zinc transporter), member 6 |
| chr5_+_63802109 | 0.06 |
ENST00000334025.2 |
RGS7BP |
regulator of G-protein signaling 7 binding protein |
| chr8_+_94929077 | 0.06 |
ENST00000297598.4 ENST00000520614.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr10_+_5238793 | 0.06 |
ENST00000263126.1 |
AKR1C4 |
aldo-keto reductase family 1, member C4 |
| chr19_+_45504688 | 0.05 |
ENST00000221452.8 ENST00000540120.1 ENST00000505236.1 |
RELB |
v-rel avian reticuloendotheliosis viral oncogene homolog B |
| chr12_+_16500037 | 0.05 |
ENST00000536371.1 ENST00000010404.2 |
MGST1 |
microsomal glutathione S-transferase 1 |
| chr20_-_17662705 | 0.05 |
ENST00000455029.2 |
RRBP1 |
ribosome binding protein 1 |
| chrY_-_26959519 | 0.05 |
ENST00000400212.1 ENST00000306737.8 ENST00000315357.7 ENST00000446723.1 ENST00000382365.2 |
DAZ3 |
deleted in azoospermia 3 |
| chr13_-_36050819 | 0.05 |
ENST00000379919.4 |
MAB21L1 |
mab-21-like 1 (C. elegans) |
| chr19_+_11071546 | 0.05 |
ENST00000358026.2 |
SMARCA4 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 4 |
| chr10_+_99205959 | 0.05 |
ENST00000352634.4 ENST00000353979.3 ENST00000370842.2 ENST00000345745.5 |
ZDHHC16 |
zinc finger, DHHC-type containing 16 |
| chr11_+_119205222 | 0.05 |
ENST00000311413.4 |
RNF26 |
ring finger protein 26 |
| chr10_+_99205894 | 0.05 |
ENST00000370854.3 ENST00000393760.1 ENST00000414567.1 ENST00000370846.4 |
ZDHHC16 |
zinc finger, DHHC-type containing 16 |
| chr8_+_98656336 | 0.05 |
ENST00000336273.3 |
MTDH |
metadherin |
| chr17_+_57232690 | 0.05 |
ENST00000262293.4 |
PRR11 |
proline rich 11 |
| chr4_+_128554081 | 0.05 |
ENST00000335251.6 ENST00000296461.5 |
INTU |
inturned planar cell polarity protein |
| chr7_+_6048856 | 0.04 |
ENST00000223029.3 ENST00000400479.2 ENST00000395236.2 |
AIMP2 |
aminoacyl tRNA synthetase complex-interacting multifunctional protein 2 |
| chr7_-_33102338 | 0.04 |
ENST00000610140.1 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
| chr7_-_33102399 | 0.04 |
ENST00000242210.7 |
NT5C3A |
5'-nucleotidase, cytosolic IIIA |
| chr1_+_240255166 | 0.04 |
ENST00000319653.9 |
FMN2 |
formin 2 |
| chr11_-_59383617 | 0.04 |
ENST00000263847.1 |
OSBP |
oxysterol binding protein |
| chr3_+_186288454 | 0.04 |
ENST00000265028.3 |
DNAJB11 |
DnaJ (Hsp40) homolog, subfamily B, member 11 |
| chr9_+_124329336 | 0.04 |
ENST00000394340.3 ENST00000436835.1 ENST00000259371.2 |
DAB2IP |
DAB2 interacting protein |
| chr22_-_39151463 | 0.04 |
ENST00000405510.1 ENST00000433561.1 |
SUN2 |
Sad1 and UNC84 domain containing 2 |
| chr7_+_144052381 | 0.04 |
ENST00000498580.1 ENST00000056217.5 |
ARHGEF5 |
Rho guanine nucleotide exchange factor (GEF) 5 |
| chr14_-_81425828 | 0.03 |
ENST00000555529.1 ENST00000556042.1 ENST00000556981.1 |
CEP128 |
centrosomal protein 128kDa |
| chr17_-_8113886 | 0.03 |
ENST00000577833.1 ENST00000534871.1 ENST00000583915.1 ENST00000316199.6 ENST00000581511.1 ENST00000585124.1 |
AURKB |
aurora kinase B |
| chr17_-_61777459 | 0.03 |
ENST00000578993.1 ENST00000583211.1 ENST00000259006.3 |
LIMD2 |
LIM domain containing 2 |
| chr19_+_36249057 | 0.03 |
ENST00000301165.5 ENST00000536950.1 ENST00000537459.1 ENST00000421853.2 |
C19orf55 |
chromosome 19 open reading frame 55 |
| chr1_-_221915418 | 0.03 |
ENST00000323825.3 ENST00000366899.3 |
DUSP10 |
dual specificity phosphatase 10 |
| chr7_-_27239703 | 0.03 |
ENST00000222753.4 |
HOXA13 |
homeobox A13 |
| chr2_+_105050794 | 0.02 |
ENST00000429464.1 ENST00000414442.1 ENST00000447380.1 |
AC013402.2 |
long intergenic non-protein coding RNA 1102 |
| chr19_+_41256764 | 0.02 |
ENST00000243563.3 ENST00000601253.1 ENST00000597353.1 ENST00000599362.1 |
SNRPA |
small nuclear ribonucleoprotein polypeptide A |
| chr12_+_2921788 | 0.02 |
ENST00000228799.2 ENST00000419778.2 ENST00000542548.1 |
ITFG2 |
integrin alpha FG-GAP repeat containing 2 |
| chr17_-_57232596 | 0.02 |
ENST00000581068.1 ENST00000330137.7 |
SKA2 |
spindle and kinetochore associated complex subunit 2 |
| chr8_+_94929110 | 0.02 |
ENST00000520728.1 |
PDP1 |
pyruvate dehyrogenase phosphatase catalytic subunit 1 |
| chr12_-_6961050 | 0.02 |
ENST00000538862.2 |
CDCA3 |
cell division cycle associated 3 |
| chr1_-_201140673 | 0.02 |
ENST00000367333.2 |
TMEM9 |
transmembrane protein 9 |
| chr16_-_66864806 | 0.02 |
ENST00000566336.1 ENST00000394074.2 ENST00000563185.2 ENST00000359087.4 ENST00000379463.2 ENST00000565535.1 ENST00000290810.3 |
NAE1 |
NEDD8 activating enzyme E1 subunit 1 |
| chr2_-_47142884 | 0.02 |
ENST00000409105.1 ENST00000409973.1 ENST00000409913.1 ENST00000319466.4 |
MCFD2 |
multiple coagulation factor deficiency 2 |
| chr2_-_47143160 | 0.02 |
ENST00000409800.1 ENST00000409218.1 |
MCFD2 |
multiple coagulation factor deficiency 2 |
| chr11_+_134201768 | 0.02 |
ENST00000535456.2 ENST00000339772.7 |
GLB1L2 |
galactosidase, beta 1-like 2 |
| chr9_+_116225999 | 0.02 |
ENST00000317613.6 |
RGS3 |
regulator of G-protein signaling 3 |
| chr12_-_23737534 | 0.02 |
ENST00000396007.2 |
SOX5 |
SRY (sex determining region Y)-box 5 |
| chr12_-_102513843 | 0.01 |
ENST00000551744.2 ENST00000552283.1 |
NUP37 |
nucleoporin 37kDa |
| chr8_+_22132847 | 0.01 |
ENST00000521356.1 |
PIWIL2 |
piwi-like RNA-mediated gene silencing 2 |
| chr19_+_54641444 | 0.01 |
ENST00000221232.5 ENST00000358389.3 |
CNOT3 |
CCR4-NOT transcription complex, subunit 3 |
| chr6_-_82957433 | 0.01 |
ENST00000306270.7 |
IBTK |
inhibitor of Bruton agammaglobulinemia tyrosine kinase |
| chr12_+_102513950 | 0.01 |
ENST00000378128.3 ENST00000327680.2 ENST00000541394.1 ENST00000543784.1 |
PARPBP |
PARP1 binding protein |
| chr1_-_190446759 | 0.01 |
ENST00000367462.3 |
BRINP3 |
bone morphogenetic protein/retinoic acid inducible neural-specific 3 |
| chr14_-_106066376 | 0.01 |
ENST00000412518.1 ENST00000428654.1 ENST00000427543.1 ENST00000420153.1 ENST00000577108.1 ENST00000576077.1 |
AL928742.12 IGHE |
AL928742.12 immunoglobulin heavy constant epsilon |
| chr1_+_100818156 | 0.01 |
ENST00000336454.3 |
CDC14A |
cell division cycle 14A |
| chr12_+_102514019 | 0.01 |
ENST00000537257.1 ENST00000358383.5 ENST00000392911.2 |
PARPBP |
PARP1 binding protein |
| chr15_-_78526942 | 0.01 |
ENST00000258873.4 |
ACSBG1 |
acyl-CoA synthetase bubblegum family member 1 |
| chr11_+_72983246 | 0.01 |
ENST00000393590.2 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr7_-_122339162 | 0.01 |
ENST00000340112.2 |
RNF133 |
ring finger protein 133 |
| chr19_-_6424783 | 0.01 |
ENST00000398148.3 |
KHSRP |
KH-type splicing regulatory protein |
| chr9_+_139780942 | 0.01 |
ENST00000247668.2 ENST00000359662.3 |
TRAF2 |
TNF receptor-associated factor 2 |
| chr17_-_45266542 | 0.01 |
ENST00000531206.1 ENST00000527547.1 ENST00000446365.2 ENST00000575483.1 ENST00000066544.3 |
CDC27 |
cell division cycle 27 |
| chr4_-_66536057 | 0.01 |
ENST00000273854.3 |
EPHA5 |
EPH receptor A5 |
| chr11_-_47198380 | 0.01 |
ENST00000419701.2 ENST00000526342.1 ENST00000528444.1 ENST00000530596.1 ENST00000525398.1 ENST00000319543.6 ENST00000426335.2 ENST00000527927.1 ENST00000525314.1 |
ARFGAP2 |
ADP-ribosylation factor GTPase activating protein 2 |
| chrX_+_19362011 | 0.01 |
ENST00000379806.5 ENST00000545074.1 ENST00000540249.1 ENST00000423505.1 ENST00000417819.1 ENST00000422285.2 ENST00000355808.5 ENST00000379805.3 |
PDHA1 |
pyruvate dehydrogenase (lipoamide) alpha 1 |
| chr12_+_28410128 | 0.00 |
ENST00000381259.1 ENST00000381256.1 |
CCDC91 |
coiled-coil domain containing 91 |
| chr11_-_65625678 | 0.00 |
ENST00000308162.5 |
CFL1 |
cofilin 1 (non-muscle) |
| chr7_+_50348268 | 0.00 |
ENST00000438033.1 ENST00000439701.1 |
IKZF1 |
IKAROS family zinc finger 1 (Ikaros) |
| chr22_+_41601209 | 0.00 |
ENST00000216237.5 |
L3MBTL2 |
l(3)mbt-like 2 (Drosophila) |
| chr12_+_6961279 | 0.00 |
ENST00000229268.8 ENST00000389231.5 ENST00000542087.1 |
USP5 |
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr1_+_100818009 | 0.00 |
ENST00000370125.2 ENST00000361544.6 ENST00000370124.3 |
CDC14A |
cell division cycle 14A |
| chr20_-_50722183 | 0.00 |
ENST00000371523.4 |
ZFP64 |
ZFP64 zinc finger protein |
| chr14_+_32798547 | 0.00 |
ENST00000557354.1 ENST00000557102.1 ENST00000557272.1 |
AKAP6 |
A kinase (PRKA) anchor protein 6 |
| chr7_+_70597109 | 0.00 |
ENST00000333538.5 |
WBSCR17 |
Williams-Beuren syndrome chromosome region 17 |
| chr8_+_22132810 | 0.00 |
ENST00000356766.6 |
PIWIL2 |
piwi-like RNA-mediated gene silencing 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2000532 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.1 | 0.2 | GO:0071449 | cellular response to lipid hydroperoxide(GO:0071449) |
| 0.0 | 0.2 | GO:0051946 | regulation of amino acid uptake involved in synaptic transmission(GO:0051941) regulation of glutamate uptake involved in transmission of nerve impulse(GO:0051946) regulation of L-glutamate import(GO:1900920) |
| 0.0 | 0.1 | GO:0009786 | regulation of asymmetric cell division(GO:0009786) |
| 0.0 | 0.1 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.0 | 0.0 | GO:1903630 | regulation of aminoacyl-tRNA ligase activity(GO:1903630) |
| 0.0 | 0.0 | GO:0051758 | homologous chromosome movement towards spindle pole involved in homologous chromosome segregation(GO:0051758) |
| 0.0 | 0.0 | GO:0002314 | germinal center B cell differentiation(GO:0002314) |
| 0.0 | 0.2 | GO:0035970 | peptidyl-threonine dephosphorylation(GO:0035970) |
| 0.0 | 0.1 | GO:0007070 | negative regulation of transcription during mitosis(GO:0007068) negative regulation of transcription from RNA polymerase II promoter during mitosis(GO:0007070) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031313 | extrinsic component of endosome membrane(GO:0031313) |
| 0.0 | 0.0 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.1 | GO:0036449 | microtubule minus-end(GO:0036449) |
| 0.0 | 0.2 | GO:0060077 | inhibitory synapse(GO:0060077) |
| 0.0 | 0.1 | GO:0046581 | intercellular canaliculus(GO:0046581) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0000026 | alpha-1,2-mannosyltransferase activity(GO:0000026) |
| 0.0 | 0.3 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.1 | GO:0004741 | [pyruvate dehydrogenase (lipoamide)] phosphatase activity(GO:0004741) |
| 0.0 | 0.1 | GO:0008665 | 2'-phosphotransferase activity(GO:0008665) |
| 0.0 | 0.1 | GO:0047023 | androsterone dehydrogenase activity(GO:0047023) |