Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
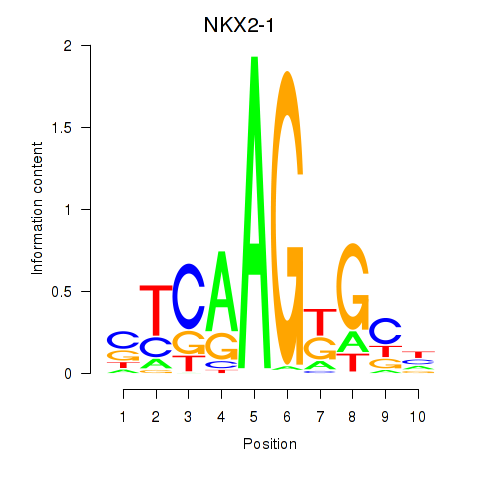
Results for NKX2-1
Z-value: 0.32
Transcription factors associated with NKX2-1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-1
|
ENSG00000136352.13 | NKX2-1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-1 | hg19_v2_chr14_-_36989336_36989354 | -0.30 | 4.7e-01 | Click! |
Activity profile of NKX2-1 motif
Sorted Z-values of NKX2-1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_38326119 | 0.21 |
ENST00000356207.5 ENST00000326324.6 |
FGFR1 |
fibroblast growth factor receptor 1 |
| chr16_+_12995614 | 0.20 |
ENST00000423335.2 |
SHISA9 |
shisa family member 9 |
| chr12_+_18414446 | 0.19 |
ENST00000433979.1 |
PIK3C2G |
phosphatidylinositol-4-phosphate 3-kinase, catalytic subunit type 2 gamma |
| chr1_+_180165672 | 0.18 |
ENST00000443059.1 |
QSOX1 |
quiescin Q6 sulfhydryl oxidase 1 |
| chr8_+_37654693 | 0.18 |
ENST00000412232.2 |
GPR124 |
G protein-coupled receptor 124 |
| chrX_+_86772787 | 0.16 |
ENST00000373114.4 |
KLHL4 |
kelch-like family member 4 |
| chr17_+_37784749 | 0.14 |
ENST00000394265.1 ENST00000394267.2 |
PPP1R1B |
protein phosphatase 1, regulatory (inhibitor) subunit 1B |
| chrX_+_86772707 | 0.14 |
ENST00000373119.4 |
KLHL4 |
kelch-like family member 4 |
| chr12_+_60058458 | 0.13 |
ENST00000548610.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr8_+_70404996 | 0.11 |
ENST00000402687.4 ENST00000419716.3 |
SULF1 |
sulfatase 1 |
| chr5_-_146889619 | 0.11 |
ENST00000343218.5 |
DPYSL3 |
dihydropyrimidinase-like 3 |
| chr11_-_86383650 | 0.11 |
ENST00000526944.1 ENST00000530335.1 ENST00000543262.1 ENST00000524826.1 |
ME3 |
malic enzyme 3, NADP(+)-dependent, mitochondrial |
| chr1_-_153517473 | 0.11 |
ENST00000368715.1 |
S100A4 |
S100 calcium binding protein A4 |
| chr1_-_153518270 | 0.11 |
ENST00000354332.4 ENST00000368716.4 |
S100A4 |
S100 calcium binding protein A4 |
| chr3_-_42744312 | 0.10 |
ENST00000416756.1 ENST00000441594.1 |
HHATL |
hedgehog acyltransferase-like |
| chr6_-_87804815 | 0.10 |
ENST00000369582.2 |
CGA |
glycoprotein hormones, alpha polypeptide |
| chr6_-_117747015 | 0.09 |
ENST00000368508.3 ENST00000368507.3 |
ROS1 |
c-ros oncogene 1 , receptor tyrosine kinase |
| chr2_-_175870085 | 0.09 |
ENST00000409156.3 |
CHN1 |
chimerin 1 |
| chr17_+_59489112 | 0.09 |
ENST00000335108.2 |
C17orf82 |
chromosome 17 open reading frame 82 |
| chr16_+_58533951 | 0.08 |
ENST00000566192.1 ENST00000565088.1 ENST00000568640.1 ENST00000563978.1 ENST00000569923.1 ENST00000356752.4 ENST00000563799.1 ENST00000562999.1 ENST00000570248.1 ENST00000562731.1 ENST00000568424.1 |
NDRG4 |
NDRG family member 4 |
| chr11_-_62313090 | 0.08 |
ENST00000528508.1 ENST00000533365.1 |
AHNAK |
AHNAK nucleoprotein |
| chr19_-_38720354 | 0.07 |
ENST00000416611.1 |
DPF1 |
D4, zinc and double PHD fingers family 1 |
| chr1_+_202431859 | 0.07 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr3_-_16524357 | 0.07 |
ENST00000432519.1 |
RFTN1 |
raftlin, lipid raft linker 1 |
| chr17_+_76210367 | 0.06 |
ENST00000592734.1 ENST00000587746.1 |
BIRC5 |
baculoviral IAP repeat containing 5 |
| chr8_+_62200509 | 0.06 |
ENST00000519846.1 ENST00000518592.1 ENST00000325897.4 |
CLVS1 |
clavesin 1 |
| chr21_-_35016231 | 0.06 |
ENST00000438788.1 |
CRYZL1 |
crystallin, zeta (quinone reductase)-like 1 |
| chr7_+_99156145 | 0.06 |
ENST00000452314.1 ENST00000252713.4 |
ZNF655 |
zinc finger protein 655 |
| chr19_+_39390320 | 0.06 |
ENST00000576510.1 |
NFKBIB |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chr5_-_58335281 | 0.06 |
ENST00000358923.6 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr12_+_93772402 | 0.06 |
ENST00000546925.1 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr17_+_4613918 | 0.06 |
ENST00000574954.1 ENST00000346341.2 ENST00000572457.1 ENST00000381488.6 ENST00000412477.3 ENST00000571428.1 ENST00000575877.1 |
ARRB2 |
arrestin, beta 2 |
| chr14_+_74815116 | 0.06 |
ENST00000256362.4 |
VRTN |
vertebrae development associated |
| chr6_-_137113604 | 0.05 |
ENST00000359015.4 |
MAP3K5 |
mitogen-activated protein kinase kinase kinase 5 |
| chr19_-_36231437 | 0.05 |
ENST00000591748.1 |
IGFLR1 |
IGF-like family receptor 1 |
| chr17_+_66521936 | 0.05 |
ENST00000592800.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr18_-_24283586 | 0.05 |
ENST00000579458.1 |
U3 |
Small nucleolar RNA U3 |
| chrX_-_134478012 | 0.05 |
ENST00000370766.3 |
ZNF75D |
zinc finger protein 75D |
| chrX_-_70473957 | 0.05 |
ENST00000373984.3 ENST00000314425.5 ENST00000373982.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr12_-_10978957 | 0.05 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr3_+_187930719 | 0.05 |
ENST00000312675.4 |
LPP |
LIM domain containing preferred translocation partner in lipoma |
| chr17_+_76210267 | 0.05 |
ENST00000301633.4 ENST00000350051.3 ENST00000374948.2 ENST00000590449.1 |
BIRC5 |
baculoviral IAP repeat containing 5 |
| chr21_+_45773515 | 0.05 |
ENST00000397932.2 ENST00000300481.9 |
TRPM2 |
transient receptor potential cation channel, subfamily M, member 2 |
| chr9_-_72374848 | 0.05 |
ENST00000377200.5 ENST00000340434.4 ENST00000472967.2 |
PTAR1 |
protein prenyltransferase alpha subunit repeat containing 1 |
| chrX_+_153170189 | 0.05 |
ENST00000358927.2 |
AVPR2 |
arginine vasopressin receptor 2 |
| chrX_-_70474377 | 0.05 |
ENST00000373978.1 ENST00000373981.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr16_-_67493110 | 0.05 |
ENST00000602876.1 |
ATP6V0D1 |
ATPase, H+ transporting, lysosomal 38kDa, V0 subunit d1 |
| chr2_+_152266604 | 0.05 |
ENST00000430328.2 |
RIF1 |
RAP1 interacting factor homolog (yeast) |
| chr1_-_15735925 | 0.04 |
ENST00000427824.1 |
RP3-467K16.4 |
RP3-467K16.4 |
| chr19_+_39903185 | 0.04 |
ENST00000409794.3 |
PLEKHG2 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr8_+_94752349 | 0.04 |
ENST00000391680.1 |
RBM12B-AS1 |
RBM12B antisense RNA 1 |
| chr11_+_60223225 | 0.04 |
ENST00000524807.1 ENST00000345732.4 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chrX_-_70474499 | 0.04 |
ENST00000353904.2 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr12_+_93772326 | 0.04 |
ENST00000550056.1 ENST00000549992.1 ENST00000548662.1 ENST00000547014.1 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr5_-_172662303 | 0.04 |
ENST00000517440.1 ENST00000329198.4 |
NKX2-5 |
NK2 homeobox 5 |
| chr17_+_26646175 | 0.04 |
ENST00000583381.1 ENST00000582113.1 ENST00000582384.1 |
TMEM97 |
transmembrane protein 97 |
| chr1_-_153599426 | 0.04 |
ENST00000392622.1 |
S100A13 |
S100 calcium binding protein A13 |
| chr15_-_43559055 | 0.04 |
ENST00000220420.5 ENST00000349114.4 |
TGM5 |
transglutaminase 5 |
| chr2_-_152118352 | 0.04 |
ENST00000331426.5 |
RBM43 |
RNA binding motif protein 43 |
| chr11_-_88796803 | 0.04 |
ENST00000418177.2 ENST00000455756.2 |
GRM5 |
glutamate receptor, metabotropic 5 |
| chr1_-_115259337 | 0.04 |
ENST00000369535.4 |
NRAS |
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr14_+_74353320 | 0.04 |
ENST00000540593.1 ENST00000555730.1 |
ZNF410 |
zinc finger protein 410 |
| chr1_-_38397384 | 0.04 |
ENST00000373027.1 |
INPP5B |
inositol polyphosphate-5-phosphatase, 75kDa |
| chr16_-_29757272 | 0.04 |
ENST00000329410.3 |
C16orf54 |
chromosome 16 open reading frame 54 |
| chr17_-_76778339 | 0.04 |
ENST00000591455.1 ENST00000446868.3 ENST00000361101.4 ENST00000589296.1 |
CYTH1 |
cytohesin 1 |
| chr15_-_66790146 | 0.04 |
ENST00000316634.5 |
SNAPC5 |
small nuclear RNA activating complex, polypeptide 5, 19kDa |
| chrX_+_22056165 | 0.04 |
ENST00000535894.1 |
PHEX |
phosphate regulating endopeptidase homolog, X-linked |
| chr12_+_56401268 | 0.04 |
ENST00000262032.5 |
IKZF4 |
IKAROS family zinc finger 4 (Eos) |
| chr2_+_99797636 | 0.04 |
ENST00000409145.1 |
MRPL30 |
mitochondrial ribosomal protein L30 |
| chr17_+_26646121 | 0.04 |
ENST00000226230.6 |
TMEM97 |
transmembrane protein 97 |
| chr9_-_14313641 | 0.04 |
ENST00000380953.1 |
NFIB |
nuclear factor I/B |
| chrX_+_153170455 | 0.04 |
ENST00000430697.1 ENST00000337474.5 ENST00000370049.1 |
AVPR2 |
arginine vasopressin receptor 2 |
| chr19_-_10679644 | 0.04 |
ENST00000393599.2 |
CDKN2D |
cyclin-dependent kinase inhibitor 2D (p19, inhibits CDK4) |
| chr2_+_99797542 | 0.03 |
ENST00000338148.3 ENST00000512183.2 |
MRPL30 C2orf15 |
mitochondrial ribosomal protein L30 chromosome 2 open reading frame 15 |
| chr2_+_119699742 | 0.03 |
ENST00000327097.4 |
MARCO |
macrophage receptor with collagenous structure |
| chr8_-_62602327 | 0.03 |
ENST00000445642.3 ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH |
aspartate beta-hydroxylase |
| chr16_-_67190152 | 0.03 |
ENST00000486556.1 |
TRADD |
TNFRSF1A-associated via death domain |
| chr1_+_207277590 | 0.03 |
ENST00000367070.3 |
C4BPA |
complement component 4 binding protein, alpha |
| chr7_+_127881325 | 0.03 |
ENST00000308868.4 |
LEP |
leptin |
| chr1_-_182369751 | 0.03 |
ENST00000367565.1 |
TEDDM1 |
transmembrane epididymal protein 1 |
| chr12_-_719573 | 0.03 |
ENST00000397265.3 |
NINJ2 |
ninjurin 2 |
| chr1_+_206643806 | 0.03 |
ENST00000537984.1 |
IKBKE |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase epsilon |
| chr16_+_66638003 | 0.03 |
ENST00000562357.1 ENST00000360086.4 ENST00000562707.1 ENST00000361909.4 ENST00000460097.1 ENST00000565666.1 |
CMTM3 |
CKLF-like MARVEL transmembrane domain containing 3 |
| chr5_+_140079919 | 0.03 |
ENST00000274712.3 |
ZMAT2 |
zinc finger, matrin-type 2 |
| chr1_-_175712665 | 0.03 |
ENST00000263525.2 |
TNR |
tenascin R |
| chr7_-_120497178 | 0.03 |
ENST00000441017.1 ENST00000424710.1 ENST00000433758.1 |
TSPAN12 |
tetraspanin 12 |
| chr21_+_45432174 | 0.03 |
ENST00000380221.3 ENST00000291574.4 |
TRAPPC10 |
trafficking protein particle complex 10 |
| chr11_+_60223312 | 0.03 |
ENST00000532491.1 ENST00000532073.1 ENST00000534668.1 ENST00000528313.1 ENST00000533306.1 |
MS4A1 |
membrane-spanning 4-domains, subfamily A, member 1 |
| chr19_-_39390350 | 0.03 |
ENST00000447739.1 ENST00000358931.5 ENST00000407552.1 |
SIRT2 |
sirtuin 2 |
| chr5_-_134783038 | 0.03 |
ENST00000503143.2 |
C5orf20 |
chromosome 5 open reading frame 20 |
| chr16_+_1359138 | 0.03 |
ENST00000325437.5 |
UBE2I |
ubiquitin-conjugating enzyme E2I |
| chr2_-_176032843 | 0.03 |
ENST00000392544.1 ENST00000409499.1 ENST00000426833.3 ENST00000392543.2 ENST00000538946.1 ENST00000487334.2 ENST00000409833.1 ENST00000409635.1 ENST00000264110.2 ENST00000345739.5 |
ATF2 |
activating transcription factor 2 |
| chr2_-_225266743 | 0.03 |
ENST00000409685.3 |
FAM124B |
family with sequence similarity 124B |
| chr11_-_105948040 | 0.03 |
ENST00000534815.1 |
KBTBD3 |
kelch repeat and BTB (POZ) domain containing 3 |
| chr1_-_10003372 | 0.03 |
ENST00000377223.1 ENST00000541052.1 ENST00000377213.1 |
LZIC |
leucine zipper and CTNNBIP1 domain containing |
| chr2_-_225266711 | 0.03 |
ENST00000389874.3 |
FAM124B |
family with sequence similarity 124B |
| chr1_-_68698222 | 0.03 |
ENST00000370976.3 ENST00000354777.2 ENST00000262348.4 ENST00000540432.1 |
WLS |
wntless Wnt ligand secretion mediator |
| chr1_+_55464600 | 0.03 |
ENST00000371265.4 |
BSND |
Bartter syndrome, infantile, with sensorineural deafness (Barttin) |
| chr17_-_46035187 | 0.03 |
ENST00000300557.2 |
PRR15L |
proline rich 15-like |
| chr6_+_42883727 | 0.03 |
ENST00000304672.1 ENST00000441198.1 ENST00000446507.1 |
PTCRA |
pre T-cell antigen receptor alpha |
| chr22_+_24891210 | 0.03 |
ENST00000382760.2 |
UPB1 |
ureidopropionase, beta |
| chr3_-_113464906 | 0.03 |
ENST00000477813.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr12_-_102591604 | 0.03 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr19_+_48972459 | 0.03 |
ENST00000427476.1 |
CYTH2 |
cytohesin 2 |
| chr11_+_105948216 | 0.03 |
ENST00000278618.4 |
AASDHPPT |
aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase |
| chr10_-_81320151 | 0.02 |
ENST00000372325.2 ENST00000372327.5 ENST00000417041.1 |
SFTPA2 |
surfactant protein A2 |
| chr20_-_35724388 | 0.02 |
ENST00000344359.3 ENST00000373664.3 |
RBL1 |
retinoblastoma-like 1 (p107) |
| chrX_+_134478706 | 0.02 |
ENST00000370761.3 ENST00000339249.4 ENST00000370760.3 |
ZNF449 |
zinc finger protein 449 |
| chr2_-_157189180 | 0.02 |
ENST00000539077.1 ENST00000424077.1 ENST00000426264.1 ENST00000339562.4 ENST00000421709.1 |
NR4A2 |
nuclear receptor subfamily 4, group A, member 2 |
| chr11_-_73472096 | 0.02 |
ENST00000541588.1 ENST00000336083.3 ENST00000540771.1 ENST00000310653.6 |
RAB6A |
RAB6A, member RAS oncogene family |
| chr3_-_114790179 | 0.02 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr16_+_640201 | 0.02 |
ENST00000563109.1 |
RAB40C |
RAB40C, member RAS oncogene family |
| chr16_+_71929397 | 0.02 |
ENST00000537613.1 ENST00000424485.2 ENST00000606369.1 ENST00000329908.8 ENST00000538850.1 ENST00000541918.1 ENST00000534994.1 ENST00000378798.5 ENST00000539186.1 |
IST1 |
increased sodium tolerance 1 homolog (yeast) |
| chr1_-_160232197 | 0.02 |
ENST00000419626.1 ENST00000610139.1 ENST00000475733.1 ENST00000407642.2 ENST00000368073.3 ENST00000326837.2 |
DCAF8 |
DDB1 and CUL4 associated factor 8 |
| chr6_-_71012773 | 0.02 |
ENST00000370496.3 ENST00000357250.6 |
COL9A1 |
collagen, type IX, alpha 1 |
| chr14_+_81421861 | 0.02 |
ENST00000298171.2 |
TSHR |
thyroid stimulating hormone receptor |
| chr2_+_119699864 | 0.02 |
ENST00000541757.1 ENST00000412481.1 |
MARCO |
macrophage receptor with collagenous structure |
| chr9_-_97356075 | 0.02 |
ENST00000375337.3 |
FBP2 |
fructose-1,6-bisphosphatase 2 |
| chr16_+_66638616 | 0.02 |
ENST00000564060.1 ENST00000565922.1 |
CMTM3 |
CKLF-like MARVEL transmembrane domain containing 3 |
| chr17_-_40169429 | 0.02 |
ENST00000316603.7 ENST00000588641.1 |
DNAJC7 |
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr1_+_32930647 | 0.02 |
ENST00000609129.1 |
ZBTB8B |
zinc finger and BTB domain containing 8B |
| chr6_-_42858534 | 0.02 |
ENST00000408925.2 |
C6orf226 |
chromosome 6 open reading frame 226 |
| chr4_-_155471528 | 0.02 |
ENST00000302078.5 ENST00000499023.2 |
PLRG1 |
pleiotropic regulator 1 |
| chr14_+_81421355 | 0.02 |
ENST00000541158.2 |
TSHR |
thyroid stimulating hormone receptor |
| chr1_+_24019099 | 0.02 |
ENST00000443624.1 ENST00000458455.1 |
RPL11 |
ribosomal protein L11 |
| chr12_-_5352315 | 0.02 |
ENST00000536518.1 |
RP11-319E16.1 |
RP11-319E16.1 |
| chr1_+_150266256 | 0.02 |
ENST00000309092.7 ENST00000369084.5 |
MRPS21 |
mitochondrial ribosomal protein S21 |
| chr1_+_66999268 | 0.02 |
ENST00000371039.1 ENST00000424320.1 |
SGIP1 |
SH3-domain GRB2-like (endophilin) interacting protein 1 |
| chr6_+_31554779 | 0.02 |
ENST00000376090.2 |
LST1 |
leukocyte specific transcript 1 |
| chr16_+_640055 | 0.02 |
ENST00000568586.1 ENST00000538492.1 ENST00000248139.3 |
RAB40C |
RAB40C, member RAS oncogene family |
| chr21_-_36421401 | 0.02 |
ENST00000486278.2 |
RUNX1 |
runt-related transcription factor 1 |
| chr7_-_27224795 | 0.02 |
ENST00000006015.3 |
HOXA11 |
homeobox A11 |
| chr6_+_43968306 | 0.02 |
ENST00000442114.2 ENST00000336600.5 ENST00000439969.2 |
C6orf223 |
chromosome 6 open reading frame 223 |
| chrX_-_83442915 | 0.02 |
ENST00000262752.2 ENST00000543399.1 |
RPS6KA6 |
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
| chr10_+_63808970 | 0.02 |
ENST00000309334.5 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr11_-_105948129 | 0.02 |
ENST00000526793.1 |
KBTBD3 |
kelch repeat and BTB (POZ) domain containing 3 |
| chr18_+_54814288 | 0.02 |
ENST00000585477.1 |
BOD1L2 |
biorientation of chromosomes in cell division 1-like 2 |
| chr19_+_40477062 | 0.02 |
ENST00000455878.2 |
PSMC4 |
proteasome (prosome, macropain) 26S subunit, ATPase, 4 |
| chr7_-_27224842 | 0.02 |
ENST00000517402.1 |
HOXA11 |
homeobox A11 |
| chr19_+_48958766 | 0.02 |
ENST00000342291.2 |
KCNJ14 |
potassium inwardly-rectifying channel, subfamily J, member 14 |
| chr15_+_40453204 | 0.02 |
ENST00000287598.6 ENST00000412359.3 |
BUB1B |
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr17_+_73642486 | 0.02 |
ENST00000579469.1 |
SMIM6 |
small integral membrane protein 6 |
| chr19_-_51336443 | 0.02 |
ENST00000598673.1 |
KLK15 |
kallikrein-related peptidase 15 |
| chr8_-_141810634 | 0.02 |
ENST00000521986.1 ENST00000523539.1 ENST00000538769.1 |
PTK2 |
protein tyrosine kinase 2 |
| chr18_-_58040000 | 0.02 |
ENST00000299766.3 |
MC4R |
melanocortin 4 receptor |
| chr1_-_41487383 | 0.01 |
ENST00000302946.8 ENST00000372613.2 ENST00000439569.2 ENST00000397197.2 |
SLFNL1 |
schlafen-like 1 |
| chr6_+_31674639 | 0.01 |
ENST00000556581.1 ENST00000375832.4 ENST00000503322.1 |
LY6G6F MEGT1 |
lymphocyte antigen 6 complex, locus G6F HCG43720, isoform CRA_a; Lymphocyte antigen 6 complex locus protein G6f; Megakaryocyte-enhanced gene transcript 1 protein; Uncharacterized protein |
| chr17_-_8198636 | 0.01 |
ENST00000577745.1 ENST00000579192.1 ENST00000396278.1 |
SLC25A35 |
solute carrier family 25, member 35 |
| chr12_+_6961279 | 0.01 |
ENST00000229268.8 ENST00000389231.5 ENST00000542087.1 |
USP5 |
ubiquitin specific peptidase 5 (isopeptidase T) |
| chr1_-_41487415 | 0.01 |
ENST00000372611.1 |
SLFNL1 |
schlafen-like 1 |
| chr17_+_73642315 | 0.01 |
ENST00000556126.2 |
SMIM6 |
small integral membrane protein 6 |
| chr10_-_103454876 | 0.01 |
ENST00000331272.7 |
FBXW4 |
F-box and WD repeat domain containing 4 |
| chr17_-_47492164 | 0.01 |
ENST00000512041.2 ENST00000446735.1 ENST00000504124.1 |
PHB |
prohibitin |
| chr3_+_19189946 | 0.01 |
ENST00000328405.2 |
KCNH8 |
potassium voltage-gated channel, subfamily H (eag-related), member 8 |
| chr20_+_59654146 | 0.01 |
ENST00000441660.1 |
RP5-827L5.1 |
RP5-827L5.1 |
| chr16_+_66638567 | 0.01 |
ENST00000567572.1 |
CMTM3 |
CKLF-like MARVEL transmembrane domain containing 3 |
| chr5_-_137548997 | 0.01 |
ENST00000505120.1 ENST00000394886.2 ENST00000394884.3 |
CDC23 |
cell division cycle 23 |
| chr16_-_21170762 | 0.01 |
ENST00000261383.3 ENST00000415178.1 |
DNAH3 |
dynein, axonemal, heavy chain 3 |
| chr14_+_81421921 | 0.01 |
ENST00000554263.1 ENST00000554435.1 |
TSHR |
thyroid stimulating hormone receptor |
| chr8_-_144679296 | 0.01 |
ENST00000317198.6 |
EEF1D |
eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) |
| chr2_-_85895295 | 0.01 |
ENST00000428225.1 ENST00000519937.2 |
SFTPB |
surfactant protein B |
| chr16_+_28303804 | 0.01 |
ENST00000341901.4 |
SBK1 |
SH3 domain binding kinase 1 |
| chr2_-_180610767 | 0.01 |
ENST00000409343.1 |
ZNF385B |
zinc finger protein 385B |
| chr20_+_30407151 | 0.01 |
ENST00000375985.4 |
MYLK2 |
myosin light chain kinase 2 |
| chr2_+_220379052 | 0.01 |
ENST00000347842.3 ENST00000358078.4 |
ASIC4 |
acid-sensing (proton-gated) ion channel family member 4 |
| chr15_-_44955842 | 0.01 |
ENST00000427534.2 ENST00000559193.1 ENST00000261866.7 ENST00000535302.2 ENST00000558319.1 |
SPG11 |
spastic paraplegia 11 (autosomal recessive) |
| chr8_-_67341208 | 0.01 |
ENST00000499642.1 |
RP11-346I3.4 |
RP11-346I3.4 |
| chr14_+_81421710 | 0.01 |
ENST00000342443.6 |
TSHR |
thyroid stimulating hormone receptor |
| chr20_+_30407105 | 0.01 |
ENST00000375994.2 |
MYLK2 |
myosin light chain kinase 2 |
| chr1_-_17380630 | 0.01 |
ENST00000375499.3 |
SDHB |
succinate dehydrogenase complex, subunit B, iron sulfur (Ip) |
| chrX_-_30326445 | 0.01 |
ENST00000378963.1 |
NR0B1 |
nuclear receptor subfamily 0, group B, member 1 |
| chr2_+_220299547 | 0.01 |
ENST00000312358.7 |
SPEG |
SPEG complex locus |
| chr14_-_106068065 | 0.01 |
ENST00000390541.2 |
IGHE |
immunoglobulin heavy constant epsilon |
| chr13_+_95364963 | 0.01 |
ENST00000438290.2 |
SOX21-AS1 |
SOX21 antisense RNA 1 (head to head) |
| chr10_+_82300575 | 0.01 |
ENST00000313455.4 |
SH2D4B |
SH2 domain containing 4B |
| chr2_+_101591314 | 0.01 |
ENST00000450763.1 |
NPAS2 |
neuronal PAS domain protein 2 |
| chr5_-_58571935 | 0.01 |
ENST00000503258.1 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr2_+_136289030 | 0.00 |
ENST00000409478.1 ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1 |
R3H domain containing 1 |
| chr17_-_47492236 | 0.00 |
ENST00000434917.2 ENST00000300408.3 ENST00000511832.1 ENST00000419140.2 |
PHB |
prohibitin |
| chr6_-_55739542 | 0.00 |
ENST00000446683.2 |
BMP5 |
bone morphogenetic protein 5 |
| chr3_-_52029958 | 0.00 |
ENST00000294189.6 |
RPL29 |
ribosomal protein L29 |
| chr5_-_171615315 | 0.00 |
ENST00000176763.5 |
STK10 |
serine/threonine kinase 10 |
| chr17_-_34195862 | 0.00 |
ENST00000592980.1 ENST00000587626.1 |
C17orf66 |
chromosome 17 open reading frame 66 |
| chr12_-_21810726 | 0.00 |
ENST00000396076.1 |
LDHB |
lactate dehydrogenase B |
| chr15_+_57511609 | 0.00 |
ENST00000543579.1 ENST00000537840.1 ENST00000343827.3 |
TCF12 |
transcription factor 12 |
| chr18_+_29171689 | 0.00 |
ENST00000237014.3 |
TTR |
transthyretin |
| chr14_-_58893832 | 0.00 |
ENST00000556007.2 |
TIMM9 |
translocase of inner mitochondrial membrane 9 homolog (yeast) |
| chr15_+_69452959 | 0.00 |
ENST00000261858.2 |
GLCE |
glucuronic acid epimerase |
| chrX_+_78003204 | 0.00 |
ENST00000435339.3 ENST00000514744.1 |
LPAR4 |
lysophosphatidic acid receptor 4 |
| chr9_-_128246769 | 0.00 |
ENST00000444226.1 |
MAPKAP1 |
mitogen-activated protein kinase associated protein 1 |
| chr16_+_57126482 | 0.00 |
ENST00000537605.1 ENST00000535318.2 |
CPNE2 |
copine II |
| chr17_-_79623597 | 0.00 |
ENST00000574024.1 ENST00000331056.5 |
PDE6G |
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr16_-_28223166 | 0.00 |
ENST00000304658.5 |
XPO6 |
exportin 6 |
| chr12_-_21810765 | 0.00 |
ENST00000450584.1 ENST00000350669.1 |
LDHB |
lactate dehydrogenase B |
| chr16_+_57126428 | 0.00 |
ENST00000290776.8 |
CPNE2 |
copine II |
| chr3_-_52029830 | 0.00 |
ENST00000492277.1 ENST00000475248.1 ENST00000479017.1 ENST00000495383.1 ENST00000466397.1 ENST00000481629.1 |
RPL29 |
ribosomal protein L29 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:1903465 | vacuolar phosphate transport(GO:0007037) positive regulation of mitotic cell cycle DNA replication(GO:1903465) positive regulation of parathyroid hormone secretion(GO:2000830) |
| 0.0 | 0.1 | GO:0006500 | N-terminal protein palmitoylation(GO:0006500) |
| 0.0 | 0.1 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.0 | 0.1 | GO:0007621 | negative regulation of female receptivity(GO:0007621) |
| 0.0 | 0.1 | GO:0010966 | regulation of phosphate transport(GO:0010966) |
| 0.0 | 0.1 | GO:1901909 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
| 0.0 | 0.1 | GO:0001992 | regulation of systemic arterial blood pressure by vasopressin(GO:0001992) |
| 0.0 | 0.1 | GO:0006848 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.1 | GO:1904588 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.0 | 0.0 | GO:0003162 | atrioventricular node development(GO:0003162) right ventricular cardiac muscle tissue morphogenesis(GO:0003221) septum secundum development(GO:0003285) |
| 0.0 | 0.0 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.2 | GO:0048172 | regulation of short-term neuronal synaptic plasticity(GO:0048172) |
| 0.0 | 0.1 | GO:0002032 | desensitization of G-protein coupled receptor protein signaling pathway by arrestin(GO:0002032) |
| 0.0 | 0.1 | GO:0072592 | oxygen metabolic process(GO:0072592) |
| 0.0 | 0.0 | GO:2000366 | negative regulation of glucagon secretion(GO:0070093) regulation of STAT protein import into nucleus(GO:2000364) positive regulation of STAT protein import into nucleus(GO:2000366) |
| 0.0 | 0.0 | GO:2000777 | positive regulation of oocyte maturation(GO:1900195) positive regulation of proteasomal ubiquitin-dependent protein catabolic process involved in cellular response to hypoxia(GO:2000777) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0032133 | chromosome passenger complex(GO:0032133) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0031751 | D4 dopamine receptor binding(GO:0031751) |
| 0.0 | 0.2 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.0 | 0.2 | GO:0035005 | 1-phosphatidylinositol-4-phosphate 3-kinase activity(GO:0035005) |
| 0.0 | 0.1 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.1 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.0 | 0.1 | GO:0004473 | malate dehydrogenase (decarboxylating) (NAD+) activity(GO:0004471) malate dehydrogenase (decarboxylating) (NADP+) activity(GO:0004473) |
| 0.0 | 0.2 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.0 | 0.1 | GO:0052846 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |
| 0.0 | 0.1 | GO:0004803 | transposase activity(GO:0004803) |
| 0.0 | 0.3 | GO:0050786 | RAGE receptor binding(GO:0050786) |
| 0.0 | 0.0 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.0 | GO:0036435 | K48-linked polyubiquitin binding(GO:0036435) |
| 0.0 | 0.0 | GO:0034739 | histone deacetylase activity (H4-K16 specific)(GO:0034739) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | REACTOME HORMONE LIGAND BINDING RECEPTORS | Genes involved in Hormone ligand-binding receptors |