Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for NKX2-2
Z-value: 0.61
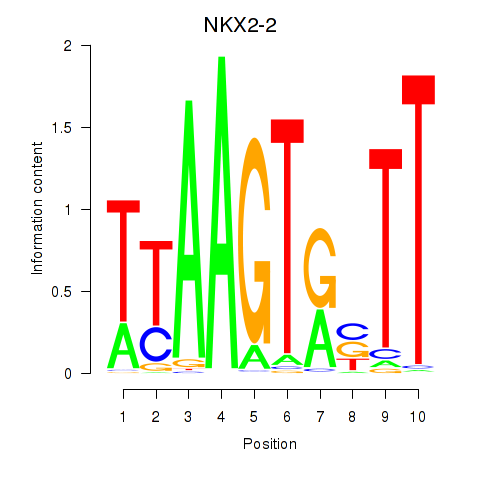
Transcription factors associated with NKX2-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-2
|
ENSG00000125820.5 | NKX2-2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-2 | hg19_v2_chr20_-_21494654_21494678 | 0.19 | 6.5e-01 | Click! |
Activity profile of NKX2-2 motif
Sorted Z-values of NKX2-2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr1_+_78769549 | 0.62 |
ENST00000370758.1 |
PTGFR |
prostaglandin F receptor (FP) |
| chr21_+_25801041 | 0.62 |
ENST00000453784.2 ENST00000423581.1 |
AP000476.1 |
AP000476.1 |
| chr8_+_97597148 | 0.61 |
ENST00000521590.1 |
SDC2 |
syndecan 2 |
| chr4_-_156787425 | 0.60 |
ENST00000537611.2 |
ASIC5 |
acid-sensing (proton-gated) ion channel family member 5 |
| chr5_+_82767583 | 0.56 |
ENST00000512590.2 ENST00000513960.1 ENST00000513984.1 ENST00000502527.2 |
VCAN |
versican |
| chr5_+_82767487 | 0.54 |
ENST00000343200.5 ENST00000342785.4 |
VCAN |
versican |
| chr13_+_102104980 | 0.54 |
ENST00000545560.2 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr8_-_122653630 | 0.53 |
ENST00000303924.4 |
HAS2 |
hyaluronan synthase 2 |
| chr13_+_102104952 | 0.50 |
ENST00000376180.3 |
ITGBL1 |
integrin, beta-like 1 (with EGF-like repeat domains) |
| chr2_-_238323007 | 0.47 |
ENST00000295550.4 |
COL6A3 |
collagen, type VI, alpha 3 |
| chr6_+_151561085 | 0.42 |
ENST00000402676.2 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr2_+_152214098 | 0.42 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr13_-_67802549 | 0.40 |
ENST00000328454.5 ENST00000377865.2 |
PCDH9 |
protocadherin 9 |
| chr17_-_67138015 | 0.39 |
ENST00000284425.2 ENST00000590645.1 |
ABCA6 |
ATP-binding cassette, sub-family A (ABC1), member 6 |
| chr8_+_77593448 | 0.36 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr17_-_29641104 | 0.29 |
ENST00000577894.1 ENST00000330927.4 |
EVI2B |
ecotropic viral integration site 2B |
| chr8_+_77593474 | 0.27 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr4_-_99578789 | 0.27 |
ENST00000511651.1 ENST00000505184.1 |
TSPAN5 |
tetraspanin 5 |
| chr16_+_53241854 | 0.27 |
ENST00000565803.1 |
CHD9 |
chromodomain helicase DNA binding protein 9 |
| chrX_+_100743031 | 0.26 |
ENST00000423738.3 |
ARMCX4 |
armadillo repeat containing, X-linked 4 |
| chr3_+_142838091 | 0.26 |
ENST00000309575.3 |
CHST2 |
carbohydrate (N-acetylglucosamine-6-O) sulfotransferase 2 |
| chr2_-_214014959 | 0.25 |
ENST00000442445.1 ENST00000457361.1 ENST00000342002.2 |
IKZF2 |
IKAROS family zinc finger 2 (Helios) |
| chrM_+_12331 | 0.25 |
ENST00000361567.2 |
MT-ND5 |
mitochondrially encoded NADH dehydrogenase 5 |
| chr5_-_150521192 | 0.24 |
ENST00000523714.1 ENST00000521749.1 |
ANXA6 |
annexin A6 |
| chr9_-_21482312 | 0.23 |
ENST00000448696.3 |
IFNE |
interferon, epsilon |
| chrX_-_70474377 | 0.21 |
ENST00000373978.1 ENST00000373981.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr9_-_20622478 | 0.20 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr17_-_29641084 | 0.20 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr2_+_152266604 | 0.20 |
ENST00000430328.2 |
RIF1 |
RAP1 interacting factor homolog (yeast) |
| chr4_+_114214125 | 0.20 |
ENST00000509550.1 |
ANK2 |
ankyrin 2, neuronal |
| chr12_-_120315074 | 0.20 |
ENST00000261833.7 ENST00000392521.2 |
CIT |
citron (rho-interacting, serine/threonine kinase 21) |
| chr10_+_89420706 | 0.20 |
ENST00000427144.2 |
PAPSS2 |
3'-phosphoadenosine 5'-phosphosulfate synthase 2 |
| chr2_-_145275228 | 0.19 |
ENST00000427902.1 ENST00000409487.3 ENST00000470879.1 ENST00000435831.1 |
ZEB2 |
zinc finger E-box binding homeobox 2 |
| chr13_+_50570019 | 0.19 |
ENST00000442421.1 |
TRIM13 |
tripartite motif containing 13 |
| chr1_+_211433275 | 0.19 |
ENST00000367005.4 |
RCOR3 |
REST corepressor 3 |
| chr1_-_227505289 | 0.19 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr4_-_99578776 | 0.19 |
ENST00000515287.1 |
TSPAN5 |
tetraspanin 5 |
| chr1_+_162602244 | 0.19 |
ENST00000367922.3 ENST00000367921.3 |
DDR2 |
discoidin domain receptor tyrosine kinase 2 |
| chr6_+_151561506 | 0.18 |
ENST00000253332.1 |
AKAP12 |
A kinase (PRKA) anchor protein 12 |
| chr1_+_104293028 | 0.18 |
ENST00000370079.3 |
AMY1C |
amylase, alpha 1C (salivary) |
| chr18_-_70532906 | 0.17 |
ENST00000299430.2 ENST00000397929.1 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr4_+_124320665 | 0.17 |
ENST00000394339.2 |
SPRY1 |
sprouty homolog 1, antagonist of FGF signaling (Drosophila) |
| chr5_-_78281603 | 0.17 |
ENST00000264914.4 |
ARSB |
arylsulfatase B |
| chr15_-_52944231 | 0.16 |
ENST00000546305.2 |
FAM214A |
family with sequence similarity 214, member A |
| chr6_+_28249299 | 0.16 |
ENST00000405948.2 |
PGBD1 |
piggyBac transposable element derived 1 |
| chr6_+_28249332 | 0.16 |
ENST00000259883.3 |
PGBD1 |
piggyBac transposable element derived 1 |
| chr12_-_49582978 | 0.15 |
ENST00000301071.7 |
TUBA1A |
tubulin, alpha 1a |
| chr13_+_111766897 | 0.15 |
ENST00000317133.5 |
ARHGEF7 |
Rho guanine nucleotide exchange factor (GEF) 7 |
| chr1_+_104159999 | 0.15 |
ENST00000414303.2 ENST00000423678.1 |
AMY2A |
amylase, alpha 2A (pancreatic) |
| chr3_+_189507460 | 0.15 |
ENST00000434928.1 |
TP63 |
tumor protein p63 |
| chr12_-_10978957 | 0.15 |
ENST00000240619.2 |
TAS2R10 |
taste receptor, type 2, member 10 |
| chr3_+_63953415 | 0.14 |
ENST00000484332.1 |
ATXN7 |
ataxin 7 |
| chr5_+_140602904 | 0.14 |
ENST00000515856.2 ENST00000239449.4 |
PCDHB14 |
protocadherin beta 14 |
| chr1_-_159880159 | 0.14 |
ENST00000599780.1 |
AL590560.1 |
HCG1995379; Uncharacterized protein |
| chr9_+_37667978 | 0.14 |
ENST00000539465.1 |
FRMPD1 |
FERM and PDZ domain containing 1 |
| chrX_-_70473957 | 0.14 |
ENST00000373984.3 ENST00000314425.5 ENST00000373982.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr4_+_113739244 | 0.14 |
ENST00000503271.1 ENST00000503423.1 ENST00000506722.1 |
ANK2 |
ankyrin 2, neuronal |
| chr14_-_82000140 | 0.14 |
ENST00000555824.1 ENST00000557372.1 ENST00000336735.4 |
SEL1L |
sel-1 suppressor of lin-12-like (C. elegans) |
| chr12_-_102591604 | 0.13 |
ENST00000329406.4 |
PMCH |
pro-melanin-concentrating hormone |
| chr15_-_56209306 | 0.13 |
ENST00000506154.1 ENST00000338963.2 ENST00000508342.1 |
NEDD4 |
neural precursor cell expressed, developmentally down-regulated 4, E3 ubiquitin protein ligase |
| chr17_+_42264322 | 0.13 |
ENST00000446571.3 ENST00000357984.3 ENST00000538716.2 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr4_-_119759795 | 0.13 |
ENST00000419654.2 |
SEC24D |
SEC24 family member D |
| chr12_+_52056548 | 0.13 |
ENST00000545061.1 ENST00000355133.3 |
SCN8A |
sodium channel, voltage gated, type VIII, alpha subunit |
| chr20_+_30102231 | 0.13 |
ENST00000335574.5 ENST00000340852.5 ENST00000398174.3 ENST00000376127.3 ENST00000344042.5 |
HM13 |
histocompatibility (minor) 13 |
| chr7_-_92855762 | 0.12 |
ENST00000453812.2 ENST00000394468.2 |
HEPACAM2 |
HEPACAM family member 2 |
| chr2_-_40739501 | 0.12 |
ENST00000403092.1 ENST00000402441.1 ENST00000448531.1 |
SLC8A1 |
solute carrier family 8 (sodium/calcium exchanger), member 1 |
| chr11_-_124981475 | 0.12 |
ENST00000532156.1 ENST00000532407.1 ENST00000279968.4 ENST00000527766.1 ENST00000529583.1 ENST00000524373.1 ENST00000527271.1 ENST00000526175.1 ENST00000529609.1 ENST00000533273.1 ENST00000531909.1 ENST00000529530.1 |
TMEM218 |
transmembrane protein 218 |
| chr3_-_150920979 | 0.12 |
ENST00000309180.5 ENST00000480322.1 |
GPR171 |
G protein-coupled receptor 171 |
| chr11_+_65266507 | 0.12 |
ENST00000544868.1 |
MALAT1 |
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr9_+_99690592 | 0.12 |
ENST00000354649.3 |
NUTM2G |
NUT family member 2G |
| chr8_-_133772794 | 0.12 |
ENST00000519187.1 ENST00000523829.1 ENST00000356838.3 ENST00000377901.4 ENST00000519304.1 |
TMEM71 |
transmembrane protein 71 |
| chr16_+_22501658 | 0.11 |
ENST00000415833.2 |
NPIPB5 |
nuclear pore complex interacting protein family, member B5 |
| chr17_-_8059638 | 0.11 |
ENST00000584202.1 ENST00000354903.5 ENST00000577253.1 |
PER1 |
period circadian clock 1 |
| chrX_-_70474499 | 0.11 |
ENST00000353904.2 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr13_-_107220455 | 0.11 |
ENST00000400198.3 |
ARGLU1 |
arginine and glutamate rich 1 |
| chrX_-_30326445 | 0.11 |
ENST00000378963.1 |
NR0B1 |
nuclear receptor subfamily 0, group B, member 1 |
| chr10_+_124670121 | 0.11 |
ENST00000368894.1 |
FAM24A |
family with sequence similarity 24, member A |
| chr15_-_60690163 | 0.11 |
ENST00000558998.1 ENST00000560165.1 ENST00000557986.1 ENST00000559780.1 ENST00000559467.1 ENST00000559956.1 ENST00000332680.4 ENST00000396024.3 ENST00000421017.2 ENST00000560466.1 ENST00000558132.1 ENST00000559113.1 ENST00000557906.1 ENST00000558558.1 ENST00000560468.1 ENST00000559370.1 ENST00000558169.1 ENST00000559725.1 ENST00000558985.1 ENST00000451270.2 |
ANXA2 |
annexin A2 |
| chr3_-_122134882 | 0.11 |
ENST00000330689.4 |
WDR5B |
WD repeat domain 5B |
| chr8_-_62559366 | 0.10 |
ENST00000522919.1 |
ASPH |
aspartate beta-hydroxylase |
| chr17_-_2966901 | 0.10 |
ENST00000575751.1 |
OR1D5 |
olfactory receptor, family 1, subfamily D, member 5 |
| chr7_+_150065278 | 0.10 |
ENST00000519397.1 ENST00000479668.1 ENST00000540729.1 |
REPIN1 |
replication initiator 1 |
| chr9_-_139361503 | 0.10 |
ENST00000453963.1 |
SEC16A |
SEC16 homolog A (S. cerevisiae) |
| chr11_+_94277017 | 0.10 |
ENST00000358752.2 |
FUT4 |
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr5_-_58335281 | 0.10 |
ENST00000358923.6 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr4_+_36283213 | 0.10 |
ENST00000357504.3 |
DTHD1 |
death domain containing 1 |
| chr5_+_140800638 | 0.10 |
ENST00000398587.2 ENST00000518882.1 |
PCDHGA11 |
protocadherin gamma subfamily A, 11 |
| chrX_+_47092314 | 0.10 |
ENST00000218348.3 |
USP11 |
ubiquitin specific peptidase 11 |
| chr15_-_56757329 | 0.10 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chr6_-_116833500 | 0.10 |
ENST00000356128.4 |
TRAPPC3L |
trafficking protein particle complex 3-like |
| chr2_+_170655322 | 0.09 |
ENST00000260956.4 ENST00000417292.1 |
SSB |
Sjogren syndrome antigen B (autoantigen La) |
| chr19_-_39881669 | 0.09 |
ENST00000221266.7 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr22_-_24093267 | 0.09 |
ENST00000341976.3 |
ZNF70 |
zinc finger protein 70 |
| chr17_+_48823896 | 0.09 |
ENST00000511974.1 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
| chr11_+_93479588 | 0.09 |
ENST00000526335.1 |
C11orf54 |
chromosome 11 open reading frame 54 |
| chr7_-_102715263 | 0.09 |
ENST00000379305.3 |
FBXL13 |
F-box and leucine-rich repeat protein 13 |
| chr15_+_69365265 | 0.09 |
ENST00000415504.1 |
LINC00277 |
long intergenic non-protein coding RNA 277 |
| chr2_+_152266392 | 0.09 |
ENST00000444746.2 ENST00000453091.2 ENST00000428287.2 ENST00000433166.2 ENST00000420714.3 ENST00000243326.5 ENST00000414861.2 |
RIF1 |
RAP1 interacting factor homolog (yeast) |
| chrX_+_34960907 | 0.09 |
ENST00000329357.5 |
FAM47B |
family with sequence similarity 47, member B |
| chr12_+_109826524 | 0.09 |
ENST00000431443.2 |
MYO1H |
myosin IH |
| chr15_+_65843130 | 0.09 |
ENST00000569894.1 |
PTPLAD1 |
protein tyrosine phosphatase-like A domain containing 1 |
| chr10_+_13203543 | 0.09 |
ENST00000378714.3 ENST00000479669.1 ENST00000484800.2 |
MCM10 |
minichromosome maintenance complex component 10 |
| chr2_+_201994569 | 0.09 |
ENST00000457277.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chrX_+_83116142 | 0.09 |
ENST00000329312.4 |
CYLC1 |
cylicin, basic protein of sperm head cytoskeleton 1 |
| chr1_-_54405773 | 0.09 |
ENST00000371376.1 |
HSPB11 |
heat shock protein family B (small), member 11 |
| chr11_-_77850629 | 0.09 |
ENST00000376156.3 ENST00000525870.1 ENST00000530454.1 ENST00000525755.1 ENST00000527099.1 ENST00000525761.1 ENST00000299626.5 |
ALG8 |
ALG8, alpha-1,3-glucosyltransferase |
| chr5_+_108083517 | 0.09 |
ENST00000281092.4 ENST00000536402.1 |
FER |
fer (fps/fes related) tyrosine kinase |
| chrX_-_19817869 | 0.09 |
ENST00000379698.4 |
SH3KBP1 |
SH3-domain kinase binding protein 1 |
| chr8_+_71485681 | 0.08 |
ENST00000391684.1 |
AC120194.1 |
AC120194.1 |
| chr2_+_189157498 | 0.08 |
ENST00000359135.3 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
| chr6_-_27440460 | 0.08 |
ENST00000377419.1 |
ZNF184 |
zinc finger protein 184 |
| chrX_+_36053908 | 0.08 |
ENST00000378660.2 |
CHDC2 |
calponin homology domain containing 2 |
| chr7_-_120497178 | 0.08 |
ENST00000441017.1 ENST00000424710.1 ENST00000433758.1 |
TSPAN12 |
tetraspanin 12 |
| chr5_-_110848189 | 0.08 |
ENST00000296632.3 ENST00000512160.1 ENST00000509887.1 |
STARD4 |
StAR-related lipid transfer (START) domain containing 4 |
| chr3_-_33686925 | 0.08 |
ENST00000485378.2 ENST00000313350.6 ENST00000487200.1 |
CLASP2 |
cytoplasmic linker associated protein 2 |
| chr19_-_44174305 | 0.08 |
ENST00000601723.1 ENST00000339082.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr9_+_71819927 | 0.08 |
ENST00000535702.1 |
TJP2 |
tight junction protein 2 |
| chr9_-_15510287 | 0.08 |
ENST00000397519.2 |
PSIP1 |
PC4 and SFRS1 interacting protein 1 |
| chr7_-_102715172 | 0.07 |
ENST00000456695.1 ENST00000455112.2 ENST00000440067.1 |
FBXL13 |
F-box and leucine-rich repeat protein 13 |
| chr3_-_69101461 | 0.07 |
ENST00000543976.1 |
TMF1 |
TATA element modulatory factor 1 |
| chr2_+_189157536 | 0.07 |
ENST00000409580.1 ENST00000409637.3 |
GULP1 |
GULP, engulfment adaptor PTB domain containing 1 |
| chr10_-_27529486 | 0.07 |
ENST00000375888.1 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chrX_-_70474910 | 0.07 |
ENST00000373988.1 ENST00000373998.1 |
ZMYM3 |
zinc finger, MYM-type 3 |
| chr20_+_56964169 | 0.07 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr5_-_11588907 | 0.07 |
ENST00000513598.1 ENST00000503622.1 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
| chr20_+_18488137 | 0.07 |
ENST00000450074.1 ENST00000262544.2 ENST00000336714.3 ENST00000377475.3 |
SEC23B |
Sec23 homolog B (S. cerevisiae) |
| chrX_+_115567767 | 0.07 |
ENST00000371900.4 |
SLC6A14 |
solute carrier family 6 (amino acid transporter), member 14 |
| chr9_+_71820057 | 0.07 |
ENST00000539225.1 |
TJP2 |
tight junction protein 2 |
| chr1_-_115238207 | 0.07 |
ENST00000520113.2 ENST00000369538.3 ENST00000353928.6 |
AMPD1 |
adenosine monophosphate deaminase 1 |
| chr14_-_50154921 | 0.07 |
ENST00000553805.2 ENST00000554396.1 ENST00000216367.5 ENST00000539565.2 |
POLE2 |
polymerase (DNA directed), epsilon 2, accessory subunit |
| chr4_+_88720698 | 0.07 |
ENST00000226284.5 |
IBSP |
integrin-binding sialoprotein |
| chr3_+_159943362 | 0.07 |
ENST00000326474.3 |
C3orf80 |
chromosome 3 open reading frame 80 |
| chr20_-_13765526 | 0.07 |
ENST00000202816.1 |
ESF1 |
ESF1, nucleolar pre-rRNA processing protein, homolog (S. cerevisiae) |
| chr5_+_135394840 | 0.07 |
ENST00000503087.1 |
TGFBI |
transforming growth factor, beta-induced, 68kDa |
| chr4_+_56719782 | 0.06 |
ENST00000381295.2 ENST00000346134.7 ENST00000349598.6 |
EXOC1 |
exocyst complex component 1 |
| chr13_-_61989655 | 0.06 |
ENST00000409204.4 |
PCDH20 |
protocadherin 20 |
| chr12_-_71551652 | 0.06 |
ENST00000546561.1 |
TSPAN8 |
tetraspanin 8 |
| chr2_+_201994208 | 0.06 |
ENST00000440180.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr5_+_56205878 | 0.06 |
ENST00000423328.1 |
SETD9 |
SET domain containing 9 |
| chr19_-_14889349 | 0.06 |
ENST00000315576.3 ENST00000392967.2 ENST00000346057.1 ENST00000353876.1 ENST00000353005.1 |
EMR2 |
egf-like module containing, mucin-like, hormone receptor-like 2 |
| chr17_-_43339474 | 0.06 |
ENST00000331780.4 |
SPATA32 |
spermatogenesis associated 32 |
| chr1_+_158901329 | 0.06 |
ENST00000368140.1 ENST00000368138.3 ENST00000392254.2 ENST00000392252.3 ENST00000368135.4 |
PYHIN1 |
pyrin and HIN domain family, member 1 |
| chr1_-_169429690 | 0.06 |
ENST00000445428.1 |
RP1-206D15.3 |
RP1-206D15.3 |
| chr7_+_93535817 | 0.06 |
ENST00000248572.5 |
GNGT1 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 1 |
| chr10_+_63808970 | 0.06 |
ENST00000309334.5 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr2_-_24270217 | 0.06 |
ENST00000295148.4 ENST00000406895.3 |
C2orf44 |
chromosome 2 open reading frame 44 |
| chr1_+_104068312 | 0.06 |
ENST00000524631.1 ENST00000531883.1 ENST00000533099.1 ENST00000527062.1 |
RNPC3 |
RNA-binding region (RNP1, RRM) containing 3 |
| chrM_+_4431 | 0.06 |
ENST00000361453.3 |
MT-ND2 |
mitochondrially encoded NADH dehydrogenase 2 |
| chr12_-_111926342 | 0.06 |
ENST00000389154.3 |
ATXN2 |
ataxin 2 |
| chr11_+_22689648 | 0.06 |
ENST00000278187.3 |
GAS2 |
growth arrest-specific 2 |
| chr2_-_163008903 | 0.06 |
ENST00000418842.2 ENST00000375497.3 |
GCG |
glucagon |
| chr2_-_55276320 | 0.05 |
ENST00000357376.3 |
RTN4 |
reticulon 4 |
| chr12_-_4488872 | 0.05 |
ENST00000237837.1 |
FGF23 |
fibroblast growth factor 23 |
| chr14_+_94547628 | 0.05 |
ENST00000555523.1 |
IFI27L1 |
interferon, alpha-inducible protein 27-like 1 |
| chr14_+_94547675 | 0.05 |
ENST00000393115.3 ENST00000554166.1 ENST00000556381.1 ENST00000553664.1 ENST00000555341.1 ENST00000557218.1 ENST00000554544.1 ENST00000557066.1 |
IFI27L1 |
interferon, alpha-inducible protein 27-like 1 |
| chr5_+_148206156 | 0.05 |
ENST00000305988.4 |
ADRB2 |
adrenoceptor beta 2, surface |
| chr9_-_39239171 | 0.05 |
ENST00000358144.2 |
CNTNAP3 |
contactin associated protein-like 3 |
| chr7_+_32535060 | 0.05 |
ENST00000318709.4 ENST00000409301.1 ENST00000404479.1 |
AVL9 |
AVL9 homolog (S. cerevisiase) |
| chr9_+_37753795 | 0.05 |
ENST00000377753.2 ENST00000537911.1 ENST00000377754.2 ENST00000297994.3 |
TRMT10B |
tRNA methyltransferase 10 homolog B (S. cerevisiae) |
| chr22_+_42086547 | 0.05 |
ENST00000402966.1 |
C22orf46 |
chromosome 22 open reading frame 46 |
| chr16_+_30212378 | 0.05 |
ENST00000569485.1 |
SULT1A3 |
sulfotransferase family, cytosolic, 1A, phenol-preferring, member 3 |
| chr1_-_202936394 | 0.05 |
ENST00000367249.4 |
CYB5R1 |
cytochrome b5 reductase 1 |
| chr6_+_55039050 | 0.05 |
ENST00000370862.3 |
HCRTR2 |
hypocretin (orexin) receptor 2 |
| chr19_-_17366257 | 0.05 |
ENST00000594059.1 |
AC010646.3 |
Uncharacterized protein |
| chr21_+_30968360 | 0.05 |
ENST00000333765.4 |
GRIK1-AS2 |
GRIK1 antisense RNA 2 |
| chr7_-_99149715 | 0.05 |
ENST00000449309.1 |
FAM200A |
family with sequence similarity 200, member A |
| chr5_-_148033726 | 0.05 |
ENST00000354217.2 ENST00000314512.6 ENST00000362016.2 |
HTR4 |
5-hydroxytryptamine (serotonin) receptor 4, G protein-coupled |
| chr19_-_44174330 | 0.05 |
ENST00000340093.3 |
PLAUR |
plasminogen activator, urokinase receptor |
| chr15_+_54305101 | 0.05 |
ENST00000260323.11 ENST00000545554.1 ENST00000537900.1 |
UNC13C |
unc-13 homolog C (C. elegans) |
| chr17_-_15469590 | 0.05 |
ENST00000312127.2 |
CDRT1 |
CMT duplicated region transcript 1; Uncharacterized protein |
| chr9_-_97356075 | 0.05 |
ENST00000375337.3 |
FBP2 |
fructose-1,6-bisphosphatase 2 |
| chr6_-_150067632 | 0.05 |
ENST00000460354.2 ENST00000367404.4 ENST00000543637.1 |
NUP43 |
nucleoporin 43kDa |
| chr1_-_213188772 | 0.04 |
ENST00000544555.1 |
ANGEL2 |
angel homolog 2 (Drosophila) |
| chr4_+_158493642 | 0.04 |
ENST00000507108.1 ENST00000455598.1 ENST00000509450.1 |
RP11-364P22.1 |
RP11-364P22.1 |
| chr17_+_48823975 | 0.04 |
ENST00000513969.1 ENST00000503728.1 |
LUC7L3 |
LUC7-like 3 (S. cerevisiae) |
| chr2_+_113299990 | 0.04 |
ENST00000537335.1 ENST00000417433.2 |
POLR1B |
polymerase (RNA) I polypeptide B, 128kDa |
| chr5_-_139937895 | 0.04 |
ENST00000336283.6 |
SRA1 |
steroid receptor RNA activator 1 |
| chr5_-_98262240 | 0.04 |
ENST00000284049.3 |
CHD1 |
chromodomain helicase DNA binding protein 1 |
| chr9_+_125132803 | 0.04 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr3_+_183770835 | 0.04 |
ENST00000318351.1 |
HTR3C |
5-hydroxytryptamine (serotonin) receptor 3C, ionotropic |
| chr5_-_148033693 | 0.04 |
ENST00000377888.3 ENST00000360693.3 |
HTR4 |
5-hydroxytryptamine (serotonin) receptor 4, G protein-coupled |
| chr6_+_152130240 | 0.04 |
ENST00000427531.2 |
ESR1 |
estrogen receptor 1 |
| chr6_+_158957431 | 0.04 |
ENST00000367090.3 |
TMEM181 |
transmembrane protein 181 |
| chr8_+_11660120 | 0.04 |
ENST00000220584.4 |
FDFT1 |
farnesyl-diphosphate farnesyltransferase 1 |
| chr10_+_13628933 | 0.04 |
ENST00000417658.1 ENST00000320054.4 |
PRPF18 |
pre-mRNA processing factor 18 |
| chrX_-_19988382 | 0.04 |
ENST00000356980.3 ENST00000379687.3 ENST00000379682.4 |
CXorf23 |
chromosome X open reading frame 23 |
| chr13_-_33112899 | 0.04 |
ENST00000267068.3 ENST00000357505.6 ENST00000399396.3 |
N4BP2L2 |
NEDD4 binding protein 2-like 2 |
| chr2_+_79347577 | 0.04 |
ENST00000233735.1 |
REG1A |
regenerating islet-derived 1 alpha |
| chr6_-_26027480 | 0.04 |
ENST00000377364.3 |
HIST1H4B |
histone cluster 1, H4b |
| chr12_-_68696652 | 0.04 |
ENST00000539972.1 |
MDM1 |
Mdm1 nuclear protein homolog (mouse) |
| chr7_+_72742178 | 0.04 |
ENST00000442793.1 ENST00000413573.2 ENST00000252037.4 |
FKBP6 |
FK506 binding protein 6, 36kDa |
| chr1_+_84630053 | 0.04 |
ENST00000394838.2 ENST00000370682.3 ENST00000432111.1 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr5_-_11589131 | 0.04 |
ENST00000511377.1 |
CTNND2 |
catenin (cadherin-associated protein), delta 2 |
| chr20_-_41818373 | 0.04 |
ENST00000373187.1 ENST00000356100.2 ENST00000373184.1 ENST00000373190.1 |
PTPRT |
protein tyrosine phosphatase, receptor type, T |
| chr3_-_69101413 | 0.04 |
ENST00000398559.2 |
TMF1 |
TATA element modulatory factor 1 |
| chr4_+_41937131 | 0.04 |
ENST00000504986.1 ENST00000508448.1 ENST00000513702.1 ENST00000325094.5 |
TMEM33 |
transmembrane protein 33 |
| chr10_-_14504093 | 0.04 |
ENST00000493380.1 ENST00000475141.2 |
FRMD4A |
FERM domain containing 4A |
| chrX_-_39186610 | 0.04 |
ENST00000429281.1 ENST00000448597.1 |
RP11-265P11.2 |
RP11-265P11.2 |
| chr14_-_77495007 | 0.04 |
ENST00000238647.3 |
IRF2BPL |
interferon regulatory factor 2 binding protein-like |
| chr8_+_11666649 | 0.04 |
ENST00000528643.1 ENST00000525777.1 |
FDFT1 |
farnesyl-diphosphate farnesyltransferase 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.5 | GO:0045226 | extracellular polysaccharide biosynthetic process(GO:0045226) extracellular polysaccharide metabolic process(GO:0046379) |
| 0.1 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.6 | GO:0008218 | bioluminescence(GO:0008218) |
| 0.1 | 0.6 | GO:0071798 | response to prostaglandin D(GO:0071798) cellular response to prostaglandin D stimulus(GO:0071799) |
| 0.1 | 0.2 | GO:0044111 | development involved in symbiotic interaction(GO:0044111) |
| 0.1 | 0.2 | GO:0060940 | epithelial to mesenchymal transition involved in cardiac fibroblast development(GO:0060940) |
| 0.0 | 0.2 | GO:0000103 | sulfate assimilation(GO:0000103) |
| 0.0 | 0.3 | GO:2001034 | positive regulation of double-strand break repair via nonhomologous end joining(GO:2001034) |
| 0.0 | 0.7 | GO:0010739 | positive regulation of protein kinase A signaling(GO:0010739) |
| 0.0 | 0.0 | GO:0009405 | pathogenesis(GO:0009405) |
| 0.0 | 1.1 | GO:0030208 | dermatan sulfate biosynthetic process(GO:0030208) |
| 0.0 | 0.1 | GO:0061582 | intestinal epithelial cell migration(GO:0061582) |
| 0.0 | 0.2 | GO:1902748 | positive regulation of lens fiber cell differentiation(GO:1902748) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:0038195 | urokinase plasminogen activator signaling pathway(GO:0038195) |
| 0.0 | 0.1 | GO:0038109 | response to stem cell factor(GO:0036215) cellular response to stem cell factor stimulus(GO:0036216) Kit signaling pathway(GO:0038109) |
| 0.0 | 0.1 | GO:1904253 | positive regulation of bile acid biosynthetic process(GO:0070859) positive regulation of bile acid metabolic process(GO:1904253) |
| 0.0 | 0.1 | GO:2000845 | testosterone secretion(GO:0035936) regulation of testosterone secretion(GO:2000843) positive regulation of testosterone secretion(GO:2000845) |
| 0.0 | 0.2 | GO:0044245 | polysaccharide digestion(GO:0044245) |
| 0.0 | 0.1 | GO:0006696 | ergosterol biosynthetic process(GO:0006696) ergosterol metabolic process(GO:0008204) |
| 0.0 | 0.3 | GO:0090091 | positive regulation of extracellular matrix disassembly(GO:0090091) |
| 0.0 | 0.1 | GO:0042369 | vitamin D catabolic process(GO:0042369) |
| 0.0 | 0.1 | GO:0071048 | nuclear mRNA surveillance of spliceosomal pre-mRNA splicing(GO:0071030) nuclear retention of unspliced pre-mRNA at the site of transcription(GO:0071048) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.2 | GO:1903943 | regulation of hepatocyte apoptotic process(GO:1903943) negative regulation of hepatocyte apoptotic process(GO:1903944) |
| 0.0 | 0.1 | GO:0071847 | TNFSF11-mediated signaling pathway(GO:0071847) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.2 | GO:2000096 | positive regulation of Wnt signaling pathway, planar cell polarity pathway(GO:2000096) |
| 0.0 | 0.1 | GO:0035948 | positive regulation of gluconeogenesis by positive regulation of transcription from RNA polymerase II promoter(GO:0035948) |
| 0.0 | 0.3 | GO:0006044 | N-acetylglucosamine metabolic process(GO:0006044) |
| 0.0 | 0.4 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.0 | GO:0036118 | hyaluranon cable assembly(GO:0036118) regulation of hyaluranon cable assembly(GO:1900104) positive regulation of hyaluranon cable assembly(GO:1900106) |
| 0.0 | 0.1 | GO:0098735 | positive regulation of the force of heart contraction(GO:0098735) |
| 0.0 | 0.0 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.0 | GO:1903371 | regulation of endoplasmic reticulum tubular network organization(GO:1903371) |
| 0.0 | 0.1 | GO:0042780 | tRNA 3'-end processing(GO:0042780) |
| 0.0 | 0.0 | GO:0032203 | telomere formation via telomerase(GO:0032203) |
| 0.0 | 0.0 | GO:0005986 | sucrose biosynthetic process(GO:0005986) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
| 0.0 | 0.1 | GO:0006436 | tryptophanyl-tRNA aminoacylation(GO:0006436) |
| 0.0 | 0.2 | GO:0033141 | positive regulation of peptidyl-serine phosphorylation of STAT protein(GO:0033141) |
| 0.0 | 0.3 | GO:1904380 | endoplasmic reticulum mannose trimming(GO:1904380) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0001940 | male pronucleus(GO:0001940) |
| 0.0 | 0.1 | GO:1990667 | PCSK9-AnxA2 complex(GO:1990667) |
| 0.0 | 0.1 | GO:0000839 | Hrd1p ubiquitin ligase ERAD-L complex(GO:0000839) |
| 0.0 | 0.5 | GO:0098647 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0030008 | TRAPP complex(GO:0030008) |
| 0.0 | 0.1 | GO:0043159 | cytoskeletal calyx(GO:0033150) acrosomal matrix(GO:0043159) |
| 0.0 | 0.1 | GO:0071458 | integral component of cytoplasmic side of endoplasmic reticulum membrane(GO:0071458) |
| 0.0 | 0.1 | GO:0098592 | cytoplasmic side of apical plasma membrane(GO:0098592) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0071020 | post-spliceosomal complex(GO:0071020) |
| 0.0 | 0.1 | GO:0008622 | epsilon DNA polymerase complex(GO:0008622) |
| 0.0 | 0.1 | GO:0000322 | storage vacuole(GO:0000322) |
| 0.0 | 1.6 | GO:0005796 | Golgi lumen(GO:0005796) |
| 0.0 | 0.1 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.0 | GO:0097450 | astrocyte end-foot(GO:0097450) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0004958 | prostaglandin F receptor activity(GO:0004958) |
| 0.1 | 0.5 | GO:0050501 | hyaluronan synthase activity(GO:0050501) |
| 0.1 | 0.6 | GO:0044736 | acid-sensing ion channel activity(GO:0044736) |
| 0.1 | 0.2 | GO:0004781 | adenylylsulfate kinase activity(GO:0004020) sulfate adenylyltransferase activity(GO:0004779) sulfate adenylyltransferase (ATP) activity(GO:0004781) |
| 0.0 | 1.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0030377 | urokinase plasminogen activator receptor activity(GO:0030377) |
| 0.0 | 0.7 | GO:0008179 | adenylate cyclase binding(GO:0008179) |
| 0.0 | 0.1 | GO:0003943 | N-acetylgalactosamine-4-sulfatase activity(GO:0003943) |
| 0.0 | 0.2 | GO:0016160 | amylase activity(GO:0016160) |
| 0.0 | 0.2 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.0 | 0.3 | GO:0001517 | N-acetylglucosamine 6-O-sulfotransferase activity(GO:0001517) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.1 | GO:0030626 | U12 snRNA binding(GO:0030626) |
| 0.0 | 0.1 | GO:0051996 | farnesyl-diphosphate farnesyltransferase activity(GO:0004310) squalene synthase activity(GO:0051996) |
| 0.0 | 0.1 | GO:0033149 | FFAT motif binding(GO:0033149) |
| 0.0 | 0.1 | GO:0000033 | alpha-1,3-mannosyltransferase activity(GO:0000033) |
| 0.0 | 0.1 | GO:0042500 | aspartic endopeptidase activity, intramembrane cleaving(GO:0042500) |
| 0.0 | 0.1 | GO:0009019 | tRNA (guanine-N1-)-methyltransferase activity(GO:0009019) |
| 0.0 | 0.1 | GO:0099580 | ion antiporter activity involved in regulation of postsynaptic membrane potential(GO:0099580) |
| 0.0 | 0.1 | GO:0050815 | phosphoserine binding(GO:0050815) phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0019834 | phospholipase A2 inhibitor activity(GO:0019834) |
| 0.0 | 0.1 | GO:0001594 | trace-amine receptor activity(GO:0001594) |
| 0.0 | 0.2 | GO:0005132 | type I interferon receptor binding(GO:0005132) |
| 0.0 | 0.0 | GO:0015643 | toxic substance binding(GO:0015643) |
| 0.0 | 0.1 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0000386 | second spliceosomal transesterification activity(GO:0000386) |
| 0.0 | 0.0 | GO:0016499 | orexin receptor activity(GO:0016499) |
| 0.0 | 0.0 | GO:0042132 | fructose 1,6-bisphosphate 1-phosphatase activity(GO:0042132) |
| 0.0 | 0.2 | GO:1904264 | ubiquitin protein ligase activity involved in ERAD pathway(GO:1904264) |
| 0.0 | 0.0 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.0 | 0.1 | GO:0004830 | tryptophan-tRNA ligase activity(GO:0004830) |
| 0.0 | 0.1 | GO:0046920 | alpha-(1->3)-fucosyltransferase activity(GO:0046920) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID TOLL ENDOGENOUS PATHWAY | Endogenous TLR signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.6 | REACTOME PROSTANOID LIGAND RECEPTORS | Genes involved in Prostanoid ligand receptors |
| 0.0 | 1.2 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.3 | REACTOME DIGESTION OF DIETARY CARBOHYDRATE | Genes involved in Digestion of dietary carbohydrate |
| 0.0 | 0.5 | REACTOME HYALURONAN METABOLISM | Genes involved in Hyaluronan metabolism |
| 0.0 | 0.6 | REACTOME HS GAG DEGRADATION | Genes involved in HS-GAG degradation |
| 0.0 | 0.4 | REACTOME ABCA TRANSPORTERS IN LIPID HOMEOSTASIS | Genes involved in ABCA transporters in lipid homeostasis |