Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
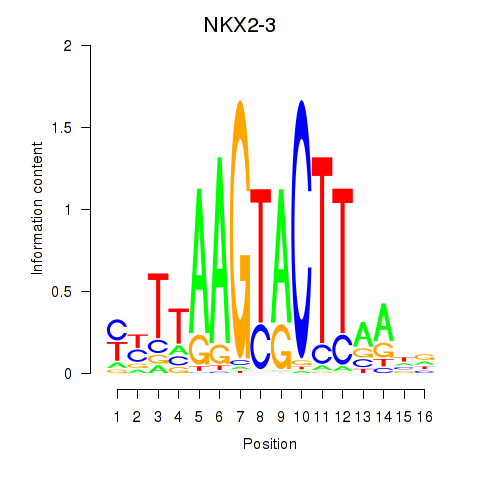
Results for NKX2-3
Z-value: 0.51
Transcription factors associated with NKX2-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-3
|
ENSG00000119919.9 | NKX2-3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-3 | hg19_v2_chr10_+_101292684_101292706 | -0.13 | 7.6e-01 | Click! |
Activity profile of NKX2-3 motif
Sorted Z-values of NKX2-3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_-_100242549 | 0.57 |
ENST00000305046.8 ENST00000394887.3 |
ADH1B |
alcohol dehydrogenase 1B (class I), beta polypeptide |
| chr4_-_140201333 | 0.31 |
ENST00000398955.1 |
MGARP |
mitochondria-localized glutamic acid-rich protein |
| chr19_+_45409011 | 0.29 |
ENST00000252486.4 ENST00000446996.1 ENST00000434152.1 |
APOE |
apolipoprotein E |
| chr9_-_95166841 | 0.26 |
ENST00000262551.4 |
OGN |
osteoglycin |
| chr9_-_95166884 | 0.25 |
ENST00000375561.5 |
OGN |
osteoglycin |
| chr12_-_108714412 | 0.21 |
ENST00000412676.1 ENST00000550573.1 |
CMKLR1 |
chemokine-like receptor 1 |
| chr3_-_114790179 | 0.20 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr6_+_73076432 | 0.18 |
ENST00000414192.2 |
RIMS1 |
regulating synaptic membrane exocytosis 1 |
| chr16_-_66952742 | 0.17 |
ENST00000565235.2 ENST00000568632.1 ENST00000565796.1 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr16_-_66952779 | 0.17 |
ENST00000570262.1 ENST00000394055.3 ENST00000299752.4 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr19_-_1401486 | 0.17 |
ENST00000252288.2 ENST00000447102.3 |
GAMT |
guanidinoacetate N-methyltransferase |
| chr17_-_76870126 | 0.16 |
ENST00000586057.1 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr17_-_76870222 | 0.16 |
ENST00000585421.1 |
TIMP2 |
TIMP metallopeptidase inhibitor 2 |
| chr12_+_71833550 | 0.16 |
ENST00000266674.5 |
LGR5 |
leucine-rich repeat containing G protein-coupled receptor 5 |
| chr2_-_38303218 | 0.16 |
ENST00000407341.1 ENST00000260630.3 |
CYP1B1 |
cytochrome P450, family 1, subfamily B, polypeptide 1 |
| chr5_+_156693091 | 0.15 |
ENST00000318218.6 ENST00000442283.2 ENST00000522463.1 ENST00000521420.1 |
CYFIP2 |
cytoplasmic FMR1 interacting protein 2 |
| chr4_-_57524061 | 0.15 |
ENST00000508121.1 |
HOPX |
HOP homeobox |
| chr9_+_36036430 | 0.14 |
ENST00000377966.3 |
RECK |
reversion-inducing-cysteine-rich protein with kazal motifs |
| chr5_+_149569520 | 0.14 |
ENST00000230671.2 ENST00000524041.1 |
SLC6A7 |
solute carrier family 6 (neurotransmitter transporter), member 7 |
| chr1_+_26147319 | 0.14 |
ENST00000374300.3 |
MTFR1L |
mitochondrial fission regulator 1-like |
| chr14_+_45366518 | 0.13 |
ENST00000557112.1 |
C14orf28 |
chromosome 14 open reading frame 28 |
| chr7_-_150974494 | 0.13 |
ENST00000392811.2 |
SMARCD3 |
SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily d, member 3 |
| chr16_+_32264040 | 0.12 |
ENST00000398664.3 |
TP53TG3D |
TP53 target 3D |
| chr16_-_32688053 | 0.12 |
ENST00000398682.4 |
TP53TG3 |
TP53 target 3 |
| chr8_-_93029865 | 0.12 |
ENST00000422361.2 |
RUNX1T1 |
runt-related transcription factor 1; translocated to, 1 (cyclin D-related) |
| chr10_-_70092671 | 0.12 |
ENST00000358769.2 ENST00000432941.1 ENST00000495025.2 |
PBLD |
phenazine biosynthesis-like protein domain containing |
| chr1_-_79472365 | 0.12 |
ENST00000370742.3 |
ELTD1 |
EGF, latrophilin and seven transmembrane domain containing 1 |
| chr6_+_69942298 | 0.12 |
ENST00000238918.8 |
BAI3 |
brain-specific angiogenesis inhibitor 3 |
| chr15_+_33022885 | 0.11 |
ENST00000322805.4 |
GREM1 |
gremlin 1, DAN family BMP antagonist |
| chr4_-_84035905 | 0.11 |
ENST00000311507.4 |
PLAC8 |
placenta-specific 8 |
| chr11_+_62379194 | 0.11 |
ENST00000525801.1 ENST00000534093.1 |
ROM1 |
retinal outer segment membrane protein 1 |
| chr2_+_161993465 | 0.11 |
ENST00000457476.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr2_-_188419200 | 0.10 |
ENST00000233156.3 ENST00000426055.1 ENST00000453013.1 ENST00000417013.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr17_-_26662464 | 0.10 |
ENST00000579419.1 ENST00000585313.1 ENST00000395418.3 ENST00000578985.1 ENST00000577498.1 ENST00000585089.1 ENST00000357896.3 |
IFT20 |
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr4_-_84035868 | 0.10 |
ENST00000426923.2 ENST00000509973.1 |
PLAC8 |
placenta-specific 8 |
| chr11_+_65266507 | 0.10 |
ENST00000544868.1 |
MALAT1 |
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr3_+_108541545 | 0.10 |
ENST00000295756.6 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr12_+_56546363 | 0.10 |
ENST00000551834.1 ENST00000552568.1 |
MYL6B |
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr9_+_118950325 | 0.09 |
ENST00000534838.1 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
| chr4_-_100212132 | 0.09 |
ENST00000209668.2 |
ADH1A |
alcohol dehydrogenase 1A (class I), alpha polypeptide |
| chr7_+_141463897 | 0.09 |
ENST00000247879.2 |
TAS2R3 |
taste receptor, type 2, member 3 |
| chr12_+_56546223 | 0.09 |
ENST00000550443.1 ENST00000207437.5 |
MYL6B |
myosin, light chain 6B, alkali, smooth muscle and non-muscle |
| chr6_-_150212029 | 0.09 |
ENST00000529948.1 ENST00000357183.4 ENST00000367363.3 |
RAET1E |
retinoic acid early transcript 1E |
| chr12_+_10460549 | 0.09 |
ENST00000543420.1 ENST00000543777.1 |
KLRD1 |
killer cell lectin-like receptor subfamily D, member 1 |
| chr13_-_50018140 | 0.09 |
ENST00000410043.1 ENST00000347776.5 |
CAB39L |
calcium binding protein 39-like |
| chr17_-_8059638 | 0.09 |
ENST00000584202.1 ENST00000354903.5 ENST00000577253.1 |
PER1 |
period circadian clock 1 |
| chr1_-_85870177 | 0.09 |
ENST00000542148.1 |
DDAH1 |
dimethylarginine dimethylaminohydrolase 1 |
| chr20_+_32250079 | 0.09 |
ENST00000375222.3 |
C20orf144 |
chromosome 20 open reading frame 144 |
| chr1_+_38273419 | 0.09 |
ENST00000468084.1 |
C1orf122 |
chromosome 1 open reading frame 122 |
| chr13_-_50018241 | 0.08 |
ENST00000409308.1 |
CAB39L |
calcium binding protein 39-like |
| chr17_-_26662440 | 0.08 |
ENST00000578122.1 |
IFT20 |
intraflagellar transport 20 homolog (Chlamydomonas) |
| chr19_-_18433910 | 0.08 |
ENST00000594828.3 ENST00000593829.1 |
LSM4 |
LSM4 homolog, U6 small nuclear RNA associated (S. cerevisiae) |
| chr3_-_122134882 | 0.08 |
ENST00000330689.4 |
WDR5B |
WD repeat domain 5B |
| chr11_+_63998006 | 0.08 |
ENST00000355040.4 |
DNAJC4 |
DnaJ (Hsp40) homolog, subfamily C, member 4 |
| chr9_-_138391692 | 0.08 |
ENST00000429260.2 |
C9orf116 |
chromosome 9 open reading frame 116 |
| chr7_-_80548667 | 0.08 |
ENST00000265361.3 |
SEMA3C |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C |
| chr1_+_205682497 | 0.08 |
ENST00000598338.1 |
AC119673.1 |
AC119673.1 |
| chr18_-_25616519 | 0.08 |
ENST00000399380.3 |
CDH2 |
cadherin 2, type 1, N-cadherin (neuronal) |
| chrX_-_69509738 | 0.08 |
ENST00000374454.1 ENST00000239666.4 |
PDZD11 |
PDZ domain containing 11 |
| chr6_+_46761118 | 0.07 |
ENST00000230588.4 |
MEP1A |
meprin A, alpha (PABA peptide hydrolase) |
| chr20_-_31071309 | 0.07 |
ENST00000326071.4 |
C20orf112 |
chromosome 20 open reading frame 112 |
| chr11_+_120195992 | 0.07 |
ENST00000314475.2 ENST00000529187.1 |
TMEM136 |
transmembrane protein 136 |
| chr8_+_134125727 | 0.07 |
ENST00000521107.1 |
TG |
thyroglobulin |
| chr15_-_50558223 | 0.07 |
ENST00000267845.3 |
HDC |
histidine decarboxylase |
| chr12_+_111051832 | 0.07 |
ENST00000550703.2 ENST00000551590.1 |
TCTN1 |
tectonic family member 1 |
| chr4_-_104021009 | 0.07 |
ENST00000509245.1 ENST00000296424.4 |
BDH2 |
3-hydroxybutyrate dehydrogenase, type 2 |
| chr2_-_188419078 | 0.07 |
ENST00000437725.1 ENST00000409676.1 ENST00000339091.4 ENST00000420747.1 |
TFPI |
tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) |
| chr13_+_31506818 | 0.07 |
ENST00000380473.3 |
TEX26 |
testis expressed 26 |
| chr8_+_28196157 | 0.07 |
ENST00000522209.1 |
PNOC |
prepronociceptin |
| chr1_-_46642154 | 0.07 |
ENST00000540385.1 |
PIK3R3 |
phosphoinositide-3-kinase, regulatory subunit 3 (gamma) |
| chr19_+_17622415 | 0.07 |
ENST00000252603.2 ENST00000600923.1 |
PGLS |
6-phosphogluconolactonase |
| chr8_+_24298531 | 0.06 |
ENST00000175238.6 |
ADAM7 |
ADAM metallopeptidase domain 7 |
| chr14_+_94547675 | 0.06 |
ENST00000393115.3 ENST00000554166.1 ENST00000556381.1 ENST00000553664.1 ENST00000555341.1 ENST00000557218.1 ENST00000554544.1 ENST00000557066.1 |
IFI27L1 |
interferon, alpha-inducible protein 27-like 1 |
| chr8_+_24298597 | 0.06 |
ENST00000380789.1 |
ADAM7 |
ADAM metallopeptidase domain 7 |
| chr4_-_90758118 | 0.06 |
ENST00000420646.2 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr1_-_89488510 | 0.06 |
ENST00000564665.1 ENST00000370481.4 |
GBP3 |
guanylate binding protein 3 |
| chr3_+_98482175 | 0.06 |
ENST00000485391.1 ENST00000492254.1 |
ST3GAL6 |
ST3 beta-galactoside alpha-2,3-sialyltransferase 6 |
| chr14_+_100531615 | 0.06 |
ENST00000392920.3 |
EVL |
Enah/Vasp-like |
| chr5_+_112312399 | 0.06 |
ENST00000515408.1 ENST00000513585.1 |
DCP2 |
decapping mRNA 2 |
| chr14_+_94547628 | 0.06 |
ENST00000555523.1 |
IFI27L1 |
interferon, alpha-inducible protein 27-like 1 |
| chr5_+_49961727 | 0.06 |
ENST00000505697.2 ENST00000503750.2 ENST00000514342.2 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr2_-_180129484 | 0.06 |
ENST00000428443.3 |
SESTD1 |
SEC14 and spectrin domains 1 |
| chr1_+_202317815 | 0.06 |
ENST00000608999.1 ENST00000336894.4 ENST00000480184.1 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr1_+_54359854 | 0.06 |
ENST00000361921.3 ENST00000322679.6 ENST00000532493.1 ENST00000525202.1 ENST00000524406.1 ENST00000388876.3 |
DIO1 |
deiodinase, iodothyronine, type I |
| chr19_+_53761545 | 0.06 |
ENST00000341702.3 |
VN1R2 |
vomeronasal 1 receptor 2 |
| chr3_+_108541608 | 0.06 |
ENST00000426646.1 |
TRAT1 |
T cell receptor associated transmembrane adaptor 1 |
| chr1_+_202431859 | 0.06 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr5_+_179105615 | 0.06 |
ENST00000514383.1 |
CANX |
calnexin |
| chrX_+_37865804 | 0.06 |
ENST00000297875.2 ENST00000357972.5 |
SYTL5 |
synaptotagmin-like 5 |
| chr5_-_180688105 | 0.06 |
ENST00000327767.4 |
TRIM52 |
tripartite motif containing 52 |
| chr15_+_85147127 | 0.06 |
ENST00000541040.1 ENST00000538076.1 ENST00000485222.2 |
ZSCAN2 |
zinc finger and SCAN domain containing 2 |
| chr20_-_33735070 | 0.06 |
ENST00000374491.3 ENST00000542871.1 ENST00000374492.3 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr14_-_65769392 | 0.06 |
ENST00000555736.1 |
CTD-2509G16.5 |
CTD-2509G16.5 |
| chr4_+_76439665 | 0.06 |
ENST00000508105.1 ENST00000311638.3 ENST00000380837.3 ENST00000507556.1 ENST00000504190.1 ENST00000507885.1 ENST00000502620.1 ENST00000514480.1 |
THAP6 |
THAP domain containing 6 |
| chr15_+_43477580 | 0.06 |
ENST00000356633.5 |
CCNDBP1 |
cyclin D-type binding-protein 1 |
| chr12_+_111051902 | 0.06 |
ENST00000397655.3 ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1 |
tectonic family member 1 |
| chr11_+_12399071 | 0.05 |
ENST00000539723.1 ENST00000550549.1 |
PARVA |
parvin, alpha |
| chr7_-_127255724 | 0.05 |
ENST00000463946.1 ENST00000341640.2 |
PAX4 |
paired box 4 |
| chr9_+_116263778 | 0.05 |
ENST00000394646.3 |
RGS3 |
regulator of G-protein signaling 3 |
| chr7_+_134430212 | 0.05 |
ENST00000436461.2 |
CALD1 |
caldesmon 1 |
| chr12_-_42719885 | 0.05 |
ENST00000552673.1 ENST00000266529.3 ENST00000552235.1 |
ZCRB1 |
zinc finger CCHC-type and RNA binding motif 1 |
| chr1_+_109632425 | 0.05 |
ENST00000338272.8 |
TMEM167B |
transmembrane protein 167B |
| chr10_-_49813090 | 0.05 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr14_-_45252031 | 0.05 |
ENST00000556405.1 |
RP11-398E10.1 |
RP11-398E10.1 |
| chr19_-_10341948 | 0.05 |
ENST00000590320.1 ENST00000592342.1 ENST00000588952.1 |
S1PR2 DNMT1 |
sphingosine-1-phosphate receptor 2 DNA (cytosine-5-)-methyltransferase 1 |
| chr16_-_31076332 | 0.05 |
ENST00000539836.3 ENST00000535577.1 ENST00000442862.2 |
ZNF668 |
zinc finger protein 668 |
| chr3_+_57094469 | 0.05 |
ENST00000334325.1 |
SPATA12 |
spermatogenesis associated 12 |
| chr15_+_43477455 | 0.05 |
ENST00000300213.4 |
CCNDBP1 |
cyclin D-type binding-protein 1 |
| chr2_+_161993412 | 0.05 |
ENST00000259075.2 ENST00000432002.1 |
TANK |
TRAF family member-associated NFKB activator |
| chr3_-_3221358 | 0.05 |
ENST00000424814.1 ENST00000450014.1 ENST00000231948.4 ENST00000432408.2 |
CRBN |
cereblon |
| chr20_+_57427765 | 0.05 |
ENST00000371100.4 |
GNAS |
GNAS complex locus |
| chr1_-_232598163 | 0.05 |
ENST00000308942.4 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr15_-_78369994 | 0.05 |
ENST00000300584.3 ENST00000409931.3 |
TBC1D2B |
TBC1 domain family, member 2B |
| chr12_-_104531945 | 0.05 |
ENST00000551446.1 |
NFYB |
nuclear transcription factor Y, beta |
| chr1_-_19746236 | 0.05 |
ENST00000375144.1 |
CAPZB |
capping protein (actin filament) muscle Z-line, beta |
| chr16_+_3184924 | 0.05 |
ENST00000574902.1 ENST00000396878.3 |
ZNF213 |
zinc finger protein 213 |
| chr17_-_2966901 | 0.05 |
ENST00000575751.1 |
OR1D5 |
olfactory receptor, family 1, subfamily D, member 5 |
| chr4_-_90758227 | 0.05 |
ENST00000506691.1 ENST00000394986.1 ENST00000506244.1 ENST00000394989.2 ENST00000394991.3 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr10_+_35464513 | 0.05 |
ENST00000494479.1 ENST00000463314.1 ENST00000342105.3 ENST00000495301.1 ENST00000463960.1 |
CREM |
cAMP responsive element modulator |
| chr5_-_70320941 | 0.05 |
ENST00000523981.1 |
NAIP |
NLR family, apoptosis inhibitory protein |
| chr19_+_15838834 | 0.05 |
ENST00000305899.3 |
OR10H2 |
olfactory receptor, family 10, subfamily H, member 2 |
| chr9_+_125376948 | 0.05 |
ENST00000297913.2 |
OR1Q1 |
olfactory receptor, family 1, subfamily Q, member 1 |
| chr9_+_75229616 | 0.05 |
ENST00000340019.3 |
TMC1 |
transmembrane channel-like 1 |
| chrX_-_21776281 | 0.05 |
ENST00000379494.3 |
SMPX |
small muscle protein, X-linked |
| chr19_-_12946236 | 0.05 |
ENST00000589272.1 ENST00000393233.2 |
RTBDN |
retbindin |
| chr16_-_31076273 | 0.05 |
ENST00000426488.2 |
ZNF668 |
zinc finger protein 668 |
| chr19_+_36142147 | 0.05 |
ENST00000590618.1 |
COX6B1 |
cytochrome c oxidase subunit VIb polypeptide 1 (ubiquitous) |
| chr12_+_123465033 | 0.05 |
ENST00000454885.2 |
ARL6IP4 |
ADP-ribosylation-like factor 6 interacting protein 4 |
| chr8_+_11961898 | 0.05 |
ENST00000400085.3 |
ZNF705D |
zinc finger protein 705D |
| chr9_-_115819039 | 0.04 |
ENST00000555206.1 |
ZFP37 |
ZFP37 zinc finger protein |
| chr10_-_112678692 | 0.04 |
ENST00000605742.1 |
BBIP1 |
BBSome interacting protein 1 |
| chr6_+_32006042 | 0.04 |
ENST00000418967.2 |
CYP21A2 |
cytochrome P450, family 21, subfamily A, polypeptide 2 |
| chr4_-_52883786 | 0.04 |
ENST00000343457.3 |
LRRC66 |
leucine rich repeat containing 66 |
| chr1_+_32666188 | 0.04 |
ENST00000421922.2 |
CCDC28B |
coiled-coil domain containing 28B |
| chrX_+_153029633 | 0.04 |
ENST00000538966.1 ENST00000361971.5 ENST00000538776.1 ENST00000538543.1 |
PLXNB3 |
plexin B3 |
| chr9_-_116840728 | 0.04 |
ENST00000265132.3 |
AMBP |
alpha-1-microglobulin/bikunin precursor |
| chr20_-_30539773 | 0.04 |
ENST00000202017.4 |
PDRG1 |
p53 and DNA-damage regulated 1 |
| chr3_+_10157276 | 0.04 |
ENST00000530758.1 ENST00000256463.6 |
BRK1 |
BRICK1, SCAR/WAVE actin-nucleating complex subunit |
| chr5_+_150051149 | 0.04 |
ENST00000523553.1 |
MYOZ3 |
myozenin 3 |
| chr7_-_29186008 | 0.04 |
ENST00000396276.3 ENST00000265394.5 |
CPVL |
carboxypeptidase, vitellogenic-like |
| chr8_-_49833978 | 0.04 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr8_-_49834299 | 0.04 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chrX_-_118284542 | 0.04 |
ENST00000402510.2 |
KIAA1210 |
KIAA1210 |
| chr5_+_169780485 | 0.04 |
ENST00000377360.4 |
KCNIP1 |
Kv channel interacting protein 1 |
| chr20_+_60962143 | 0.04 |
ENST00000343986.4 |
RPS21 |
ribosomal protein S21 |
| chr12_+_123464607 | 0.04 |
ENST00000543566.1 ENST00000315580.5 ENST00000542099.1 ENST00000392435.2 ENST00000413381.2 ENST00000426960.2 ENST00000453766.2 |
ARL6IP4 |
ADP-ribosylation-like factor 6 interacting protein 4 |
| chr7_-_2883928 | 0.04 |
ENST00000275364.3 |
GNA12 |
guanine nucleotide binding protein (G protein) alpha 12 |
| chr7_+_117251671 | 0.04 |
ENST00000468795.1 |
CFTR |
cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub-family C, member 7) |
| chr18_-_70532906 | 0.04 |
ENST00000299430.2 ENST00000397929.1 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr20_+_33104199 | 0.04 |
ENST00000357156.2 ENST00000417166.2 ENST00000300469.9 ENST00000374846.3 |
DYNLRB1 |
dynein, light chain, roadblock-type 1 |
| chr2_-_492655 | 0.04 |
ENST00000427398.1 |
AC093326.1 |
AC093326.1 |
| chr7_-_86849883 | 0.04 |
ENST00000433078.1 |
TMEM243 |
transmembrane protein 243, mitochondrial |
| chr15_+_67841330 | 0.04 |
ENST00000354498.5 |
MAP2K5 |
mitogen-activated protein kinase kinase 5 |
| chr16_+_78133293 | 0.04 |
ENST00000566780.1 |
WWOX |
WW domain containing oxidoreductase |
| chr17_-_29641084 | 0.04 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr11_+_98891797 | 0.04 |
ENST00000527185.1 ENST00000528682.1 ENST00000524871.1 |
CNTN5 |
contactin 5 |
| chr17_+_36283971 | 0.04 |
ENST00000327454.6 ENST00000378174.5 |
TBC1D3F |
TBC1 domain family, member 3F |
| chr12_-_104532062 | 0.04 |
ENST00000240055.3 |
NFYB |
nuclear transcription factor Y, beta |
| chr16_+_2031633 | 0.04 |
ENST00000598236.1 |
AC005606.1 |
LOC283868 protein; Uncharacterized protein |
| chr3_-_52002403 | 0.04 |
ENST00000490063.1 ENST00000468324.1 ENST00000497653.1 ENST00000484633.1 |
PCBP4 |
poly(rC) binding protein 4 |
| chr8_-_74791051 | 0.04 |
ENST00000453587.2 ENST00000602969.1 ENST00000602593.1 ENST00000419880.3 ENST00000517608.1 |
UBE2W |
ubiquitin-conjugating enzyme E2W (putative) |
| chr20_-_31071239 | 0.03 |
ENST00000359676.5 |
C20orf112 |
chromosome 20 open reading frame 112 |
| chrX_-_134049233 | 0.03 |
ENST00000370779.4 |
MOSPD1 |
motile sperm domain containing 1 |
| chr2_-_178128528 | 0.03 |
ENST00000397063.4 ENST00000421929.1 |
NFE2L2 |
nuclear factor, erythroid 2-like 2 |
| chr17_-_58096336 | 0.03 |
ENST00000587125.1 ENST00000407042.3 |
TBC1D3P1-DHX40P1 |
TBC1D3P1-DHX40P1 readthrough transcribed pseudogene |
| chr12_+_10460417 | 0.03 |
ENST00000381908.3 ENST00000336164.4 ENST00000350274.5 |
KLRD1 |
killer cell lectin-like receptor subfamily D, member 1 |
| chr7_-_38948774 | 0.03 |
ENST00000395969.2 ENST00000414632.1 ENST00000310301.4 |
VPS41 |
vacuolar protein sorting 41 homolog (S. cerevisiae) |
| chr17_-_34808047 | 0.03 |
ENST00000592614.1 ENST00000591542.1 ENST00000330458.7 ENST00000341264.6 ENST00000592987.1 ENST00000400684.4 |
TBC1D3G TBC1D3H |
TBC1 domain family, member 3G TBC1 domain family, member 3H |
| chr19_-_12946215 | 0.03 |
ENST00000591512.1 ENST00000587549.1 ENST00000322912.5 |
RTBDN |
retbindin |
| chr10_-_13523073 | 0.03 |
ENST00000440282.1 |
BEND7 |
BEN domain containing 7 |
| chr3_+_183892635 | 0.03 |
ENST00000427072.1 ENST00000411763.2 ENST00000292807.5 ENST00000448139.1 ENST00000455925.1 |
AP2M1 |
adaptor-related protein complex 2, mu 1 subunit |
| chr17_+_45286387 | 0.03 |
ENST00000572316.1 ENST00000354968.1 ENST00000576874.1 ENST00000536623.2 |
MYL4 |
myosin, light chain 4, alkali; atrial, embryonic |
| chr17_+_42264322 | 0.03 |
ENST00000446571.3 ENST00000357984.3 ENST00000538716.2 |
TMUB2 |
transmembrane and ubiquitin-like domain containing 2 |
| chr5_-_115177247 | 0.03 |
ENST00000500945.2 |
ATG12 |
autophagy related 12 |
| chr19_-_46288917 | 0.03 |
ENST00000537879.1 ENST00000596586.1 ENST00000595946.1 |
DMWD AC011530.4 |
dystrophia myotonica, WD repeat containing Uncharacterized protein |
| chr14_+_102276132 | 0.03 |
ENST00000350249.3 ENST00000557621.1 ENST00000556946.1 |
PPP2R5C |
protein phosphatase 2, regulatory subunit B', gamma |
| chr17_+_66521936 | 0.03 |
ENST00000592800.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr22_+_21213771 | 0.03 |
ENST00000439214.1 |
SNAP29 |
synaptosomal-associated protein, 29kDa |
| chr17_-_36348610 | 0.03 |
ENST00000339023.4 ENST00000354664.4 |
TBC1D3 |
TBC1 domain family, member 3 |
| chr1_-_165668100 | 0.03 |
ENST00000354775.4 |
ALDH9A1 |
aldehyde dehydrogenase 9 family, member A1 |
| chr21_+_17442799 | 0.03 |
ENST00000602580.1 ENST00000458468.1 ENST00000602935.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr14_+_36295504 | 0.03 |
ENST00000216807.7 |
BRMS1L |
breast cancer metastasis-suppressor 1-like |
| chr19_+_41882598 | 0.03 |
ENST00000447302.2 ENST00000544232.1 ENST00000542945.1 ENST00000540732.1 |
TMEM91 CTC-435M10.3 |
transmembrane protein 91 2-oxoisovalerate dehydrogenase subunit alpha, mitochondrial; Uncharacterized protein |
| chr6_-_33285505 | 0.03 |
ENST00000431845.2 |
ZBTB22 |
zinc finger and BTB domain containing 22 |
| chr9_-_99382065 | 0.03 |
ENST00000265659.2 ENST00000375241.1 ENST00000375236.1 |
CDC14B |
cell division cycle 14B |
| chr6_-_26027480 | 0.03 |
ENST00000377364.3 |
HIST1H4B |
histone cluster 1, H4b |
| chr10_-_76868931 | 0.03 |
ENST00000372700.3 ENST00000473072.2 ENST00000491677.2 ENST00000607131.1 ENST00000372702.3 |
DUSP13 |
dual specificity phosphatase 13 |
| chr10_+_124670121 | 0.03 |
ENST00000368894.1 |
FAM24A |
family with sequence similarity 24, member A |
| chr20_-_32262165 | 0.03 |
ENST00000606690.1 ENST00000246190.6 ENST00000439478.1 ENST00000375238.4 |
NECAB3 |
N-terminal EF-hand calcium binding protein 3 |
| chr11_-_62342375 | 0.03 |
ENST00000378019.3 |
EEF1G |
eukaryotic translation elongation factor 1 gamma |
| chr4_-_90757364 | 0.03 |
ENST00000508895.1 |
SNCA |
synuclein, alpha (non A4 component of amyloid precursor) |
| chr1_-_162381907 | 0.03 |
ENST00000367929.2 ENST00000359567.3 |
SH2D1B |
SH2 domain containing 1B |
| chr20_+_55043647 | 0.03 |
ENST00000023939.4 ENST00000395881.3 ENST00000357348.5 ENST00000449062.1 ENST00000435342.2 |
RTFDC1 |
replication termination factor 2 domain containing 1 |
| chr20_+_56964169 | 0.03 |
ENST00000475243.1 |
VAPB |
VAMP (vesicle-associated membrane protein)-associated protein B and C |
| chr12_-_50290839 | 0.03 |
ENST00000552863.1 |
FAIM2 |
Fas apoptotic inhibitory molecule 2 |
| chr16_-_50913188 | 0.03 |
ENST00000569986.1 |
CTD-2034I21.1 |
CTD-2034I21.1 |
| chr13_-_46679144 | 0.03 |
ENST00000181383.4 |
CPB2 |
carboxypeptidase B2 (plasma) |
| chr2_-_201753980 | 0.03 |
ENST00000443398.1 ENST00000286175.8 ENST00000409449.1 |
PPIL3 |
peptidylprolyl isomerase (cyclophilin)-like 3 |
| chr10_-_7513904 | 0.03 |
ENST00000420395.1 |
RP5-1031D4.2 |
RP5-1031D4.2 |
| chr6_-_32731243 | 0.02 |
ENST00000427449.1 ENST00000411527.1 |
HLA-DQB2 |
major histocompatibility complex, class II, DQ beta 2 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:2000646 | regulation of phospholipid efflux(GO:1902994) positive regulation of phospholipid efflux(GO:1902995) positive regulation of receptor catabolic process(GO:2000646) |
| 0.1 | 0.3 | GO:0032487 | regulation of Rap protein signal transduction(GO:0032487) |
| 0.1 | 0.3 | GO:0097211 | response to gonadotropin-releasing hormone(GO:0097210) cellular response to gonadotropin-releasing hormone(GO:0097211) |
| 0.0 | 0.7 | GO:0006069 | ethanol oxidation(GO:0006069) |
| 0.0 | 0.2 | GO:0061304 | retinal blood vessel morphogenesis(GO:0061304) |
| 0.0 | 0.1 | GO:1900158 | negative regulation of osteoclast proliferation(GO:0090291) negative regulation of bone mineralization involved in bone maturation(GO:1900158) |
| 0.0 | 0.1 | GO:0060392 | negative regulation of SMAD protein import into nucleus(GO:0060392) |
| 0.0 | 0.1 | GO:0051620 | negative regulation of dopamine uptake involved in synaptic transmission(GO:0051585) norepinephrine uptake(GO:0051620) regulation of norepinephrine uptake(GO:0051621) negative regulation of norepinephrine uptake(GO:0051622) negative regulation of catecholamine uptake involved in synaptic transmission(GO:0051945) regulation of glutathione peroxidase activity(GO:1903282) positive regulation of glutathione peroxidase activity(GO:1903284) positive regulation of hydrogen peroxide catabolic process(GO:1903285) positive regulation of peroxidase activity(GO:2000470) |
| 0.0 | 0.1 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.0 | 0.5 | GO:0042340 | keratan sulfate catabolic process(GO:0042340) |
| 0.0 | 0.1 | GO:0003219 | cardiac right ventricle formation(GO:0003219) |
| 0.0 | 0.2 | GO:2000158 | positive regulation of ubiquitin-specific protease activity(GO:2000158) |
| 0.0 | 0.1 | GO:0001694 | histamine biosynthetic process(GO:0001694) |
| 0.0 | 0.1 | GO:0009051 | pentose-phosphate shunt, oxidative branch(GO:0009051) |
| 0.0 | 0.2 | GO:0007598 | blood coagulation, extrinsic pathway(GO:0007598) |
| 0.0 | 0.2 | GO:0001920 | negative regulation of receptor recycling(GO:0001920) |
| 0.0 | 0.1 | GO:0036511 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:0021523 | somatic motor neuron differentiation(GO:0021523) protein localization to ciliary transition zone(GO:1904491) |
| 0.0 | 0.1 | GO:0035524 | proline transmembrane transport(GO:0035524) |
| 0.0 | 0.2 | GO:0035845 | photoreceptor cell outer segment organization(GO:0035845) |
| 0.0 | 0.2 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:1904684 | negative regulation of metalloendopeptidase activity(GO:1904684) |
| 0.0 | 0.2 | GO:0051388 | positive regulation of neurotrophin TRK receptor signaling pathway(GO:0051388) |
| 0.0 | 0.2 | GO:2001013 | epithelial cell proliferation involved in renal tubule morphogenesis(GO:2001013) |
| 0.0 | 0.1 | GO:0003350 | pulmonary myocardium development(GO:0003350) |
| 0.0 | 0.0 | GO:1902161 | positive regulation of cyclic nucleotide-gated ion channel activity(GO:1902161) |
| 0.0 | 0.1 | GO:2000809 | positive regulation of synaptic vesicle clustering(GO:2000809) |
| 0.0 | 0.0 | GO:0003331 | regulation of extracellular matrix constituent secretion(GO:0003330) positive regulation of extracellular matrix constituent secretion(GO:0003331) |
| 0.0 | 0.1 | GO:0090309 | positive regulation of methylation-dependent chromatin silencing(GO:0090309) |
| 0.0 | 0.3 | GO:0016339 | calcium-dependent cell-cell adhesion via plasma membrane cell adhesion molecules(GO:0016339) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.1 | 0.2 | GO:1902636 | kinociliary basal body(GO:1902636) |
| 0.0 | 0.2 | GO:0016461 | unconventional myosin complex(GO:0016461) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0004024 | alcohol dehydrogenase activity, zinc-dependent(GO:0004024) |
| 0.1 | 0.3 | GO:0070326 | very-low-density lipoprotein particle receptor binding(GO:0070326) |
| 0.0 | 0.1 | GO:0005298 | proline:sodium symporter activity(GO:0005298) |
| 0.0 | 0.2 | GO:0002046 | opsin binding(GO:0002046) |
| 0.0 | 0.1 | GO:0060961 | phospholipase D inhibitor activity(GO:0060961) |
| 0.0 | 0.2 | GO:0035800 | deubiquitinase activator activity(GO:0035800) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0003858 | 3-hydroxybutyrate dehydrogenase activity(GO:0003858) |
| 0.0 | 0.1 | GO:0017057 | 6-phosphogluconolactonase activity(GO:0017057) |
| 0.0 | 0.1 | GO:0016503 | pheromone receptor activity(GO:0016503) |
| 0.0 | 0.5 | GO:0008191 | metalloendopeptidase inhibitor activity(GO:0008191) |
| 0.0 | 0.1 | GO:0052798 | beta-galactoside alpha-2,3-sialyltransferase activity(GO:0052798) |
| 0.0 | 0.0 | GO:0005260 | channel-conductance-controlling ATPase activity(GO:0005260) |
| 0.0 | 0.1 | GO:1902444 | riboflavin binding(GO:1902444) |
| 0.0 | 0.1 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0016015 | morphogen activity(GO:0016015) |
| 0.0 | 0.0 | GO:0019862 | IgA binding(GO:0019862) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | REACTOME ETHANOL OXIDATION | Genes involved in Ethanol oxidation |
| 0.0 | 0.5 | REACTOME KERATAN SULFATE DEGRADATION | Genes involved in Keratan sulfate degradation |
| 0.0 | 0.3 | REACTOME HDL MEDIATED LIPID TRANSPORT | Genes involved in HDL-mediated lipid transport |