Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
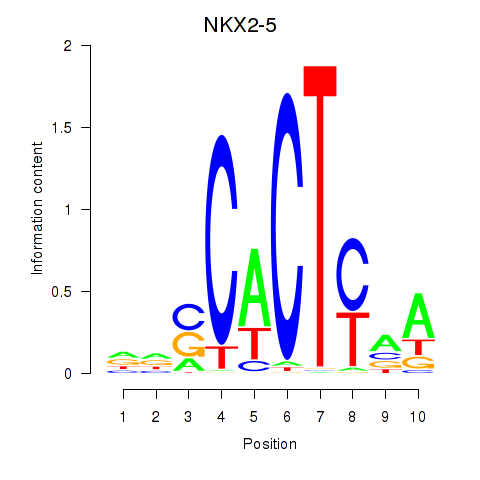
Results for NKX2-5
Z-value: 0.33
Transcription factors associated with NKX2-5
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-5
|
ENSG00000183072.9 | NKX2-5 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX2-5 | hg19_v2_chr5_-_172662303_172662360 | 0.67 | 7.1e-02 | Click! |
Activity profile of NKX2-5 motif
Sorted Z-values of NKX2-5 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-5
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_-_120651020 | 0.48 |
ENST00000522826.1 ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr5_+_75699040 | 0.41 |
ENST00000274364.6 |
IQGAP2 |
IQ motif containing GTPase activating protein 2 |
| chrX_-_142722897 | 0.34 |
ENST00000338017.4 |
SLITRK4 |
SLIT and NTRK-like family, member 4 |
| chr2_-_175499294 | 0.31 |
ENST00000392547.2 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chrX_+_86772787 | 0.29 |
ENST00000373114.4 |
KLHL4 |
kelch-like family member 4 |
| chr2_+_152214098 | 0.28 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr3_-_112356944 | 0.24 |
ENST00000461431.1 |
CCDC80 |
coiled-coil domain containing 80 |
| chrX_+_86772707 | 0.24 |
ENST00000373119.4 |
KLHL4 |
kelch-like family member 4 |
| chr15_-_52944231 | 0.22 |
ENST00000546305.2 |
FAM214A |
family with sequence similarity 214, member A |
| chr14_-_106406090 | 0.21 |
ENST00000390593.2 |
IGHV6-1 |
immunoglobulin heavy variable 6-1 |
| chr4_-_2264015 | 0.20 |
ENST00000337190.2 |
MXD4 |
MAX dimerization protein 4 |
| chr9_+_17579084 | 0.18 |
ENST00000380607.4 |
SH3GL2 |
SH3-domain GRB2-like 2 |
| chr11_+_64879317 | 0.16 |
ENST00000526809.1 ENST00000279263.7 ENST00000524986.1 ENST00000534371.1 ENST00000540748.1 ENST00000525385.1 ENST00000345348.5 ENST00000531321.1 ENST00000529414.1 ENST00000526085.1 ENST00000530750.1 |
TM7SF2 |
transmembrane 7 superfamily member 2 |
| chr16_+_31483451 | 0.15 |
ENST00000565360.1 ENST00000361773.3 |
TGFB1I1 |
transforming growth factor beta 1 induced transcript 1 |
| chr17_+_58755184 | 0.15 |
ENST00000589222.1 ENST00000407086.3 ENST00000390652.5 |
BCAS3 |
breast carcinoma amplified sequence 3 |
| chr10_-_49813090 | 0.15 |
ENST00000249601.4 |
ARHGAP22 |
Rho GTPase activating protein 22 |
| chr14_+_21467414 | 0.14 |
ENST00000554422.1 ENST00000298681.4 |
SLC39A2 |
solute carrier family 39 (zinc transporter), member 2 |
| chr1_+_202431859 | 0.13 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr5_+_140729649 | 0.12 |
ENST00000523390.1 |
PCDHGB1 |
protocadherin gamma subfamily B, 1 |
| chr1_+_154300217 | 0.12 |
ENST00000368489.3 |
ATP8B2 |
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr17_-_56621665 | 0.12 |
ENST00000321691.3 |
C17orf47 |
chromosome 17 open reading frame 47 |
| chr5_+_78407602 | 0.12 |
ENST00000274353.5 ENST00000524080.1 |
BHMT |
betaine--homocysteine S-methyltransferase |
| chr5_-_76788317 | 0.11 |
ENST00000296679.4 |
WDR41 |
WD repeat domain 41 |
| chr1_+_211433275 | 0.11 |
ENST00000367005.4 |
RCOR3 |
REST corepressor 3 |
| chr10_+_76586348 | 0.10 |
ENST00000372724.1 ENST00000287239.4 ENST00000372714.1 |
KAT6B |
K(lysine) acetyltransferase 6B |
| chr1_+_26606608 | 0.10 |
ENST00000319041.6 |
SH3BGRL3 |
SH3 domain binding glutamic acid-rich protein like 3 |
| chr3_-_15374659 | 0.10 |
ENST00000426925.1 |
SH3BP5 |
SH3-domain binding protein 5 (BTK-associated) |
| chr4_+_113970772 | 0.09 |
ENST00000504454.1 ENST00000394537.3 ENST00000357077.4 ENST00000264366.6 |
ANK2 |
ankyrin 2, neuronal |
| chr2_+_201997492 | 0.09 |
ENST00000494258.1 |
CFLAR |
CASP8 and FADD-like apoptosis regulator |
| chr3_+_108321623 | 0.09 |
ENST00000497905.1 ENST00000463306.1 |
DZIP3 |
DAZ interacting zinc finger protein 3 |
| chr17_-_8279977 | 0.08 |
ENST00000396267.1 |
KRBA2 |
KRAB-A domain containing 2 |
| chr9_+_35906176 | 0.08 |
ENST00000354323.2 |
HRCT1 |
histidine rich carboxyl terminus 1 |
| chr20_-_44485835 | 0.08 |
ENST00000457981.1 ENST00000426915.1 ENST00000217455.4 |
ACOT8 |
acyl-CoA thioesterase 8 |
| chr17_-_8059638 | 0.08 |
ENST00000584202.1 ENST00000354903.5 ENST00000577253.1 |
PER1 |
period circadian clock 1 |
| chr17_-_15244894 | 0.08 |
ENST00000338696.2 ENST00000543896.1 ENST00000539245.1 ENST00000539316.1 ENST00000395930.1 |
TEKT3 |
tektin 3 |
| chr7_-_128415844 | 0.08 |
ENST00000249389.2 |
OPN1SW |
opsin 1 (cone pigments), short-wave-sensitive |
| chr1_-_26232951 | 0.07 |
ENST00000426559.2 ENST00000455785.2 |
STMN1 |
stathmin 1 |
| chr17_+_75447326 | 0.07 |
ENST00000591088.1 |
SEPT9 |
septin 9 |
| chr5_+_140810132 | 0.07 |
ENST00000252085.3 |
PCDHGA12 |
protocadherin gamma subfamily A, 12 |
| chr6_+_33378738 | 0.07 |
ENST00000374512.3 ENST00000374516.3 |
PHF1 |
PHD finger protein 1 |
| chr1_+_114472222 | 0.07 |
ENST00000369558.1 ENST00000369561.4 |
HIPK1 |
homeodomain interacting protein kinase 1 |
| chr7_-_44229022 | 0.07 |
ENST00000403799.3 |
GCK |
glucokinase (hexokinase 4) |
| chr5_+_134181625 | 0.07 |
ENST00000394976.3 |
C5orf24 |
chromosome 5 open reading frame 24 |
| chr4_+_2470664 | 0.07 |
ENST00000314289.8 ENST00000541204.1 ENST00000502316.1 ENST00000507247.1 ENST00000509258.1 ENST00000511859.1 |
RNF4 |
ring finger protein 4 |
| chr4_-_76861392 | 0.07 |
ENST00000505594.1 |
NAAA |
N-acylethanolamine acid amidase |
| chr7_-_150675372 | 0.06 |
ENST00000262186.5 |
KCNH2 |
potassium voltage-gated channel, subfamily H (eag-related), member 2 |
| chr10_-_27529486 | 0.06 |
ENST00000375888.1 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chr16_-_30134266 | 0.06 |
ENST00000484663.1 ENST00000478356.1 |
MAPK3 |
mitogen-activated protein kinase 3 |
| chr3_-_47950745 | 0.06 |
ENST00000429422.1 |
MAP4 |
microtubule-associated protein 4 |
| chr9_-_138393580 | 0.06 |
ENST00000371791.1 |
C9orf116 |
chromosome 9 open reading frame 116 |
| chr6_+_31916733 | 0.06 |
ENST00000483004.1 |
CFB |
complement factor B |
| chr12_-_123717643 | 0.06 |
ENST00000541437.1 ENST00000606320.1 |
MPHOSPH9 |
M-phase phosphoprotein 9 |
| chrX_-_13835147 | 0.06 |
ENST00000493677.1 ENST00000355135.2 |
GPM6B |
glycoprotein M6B |
| chr19_-_14945933 | 0.06 |
ENST00000322301.3 |
OR7A5 |
olfactory receptor, family 7, subfamily A, member 5 |
| chr17_+_40811283 | 0.06 |
ENST00000251412.7 |
TUBG2 |
tubulin, gamma 2 |
| chr6_-_112115074 | 0.06 |
ENST00000368667.2 |
FYN |
FYN oncogene related to SRC, FGR, YES |
| chr11_+_108093839 | 0.06 |
ENST00000452508.2 |
ATM |
ataxia telangiectasia mutated |
| chr7_-_28220354 | 0.06 |
ENST00000283928.5 |
JAZF1 |
JAZF zinc finger 1 |
| chr1_-_202858227 | 0.06 |
ENST00000367262.3 |
RABIF |
RAB interacting factor |
| chr2_-_27545431 | 0.05 |
ENST00000233545.2 |
MPV17 |
MpV17 mitochondrial inner membrane protein |
| chr1_-_214638146 | 0.05 |
ENST00000543945.1 |
PTPN14 |
protein tyrosine phosphatase, non-receptor type 14 |
| chr7_+_142457315 | 0.05 |
ENST00000486171.1 ENST00000311737.7 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
| chr17_-_29641084 | 0.05 |
ENST00000544462.1 |
EVI2B |
ecotropic viral integration site 2B |
| chr16_-_30134524 | 0.05 |
ENST00000395202.1 ENST00000395199.3 ENST00000263025.4 ENST00000322266.5 ENST00000403394.1 |
MAPK3 |
mitogen-activated protein kinase 3 |
| chrX_-_48858630 | 0.05 |
ENST00000376425.3 ENST00000376444.3 |
GRIPAP1 |
GRIP1 associated protein 1 |
| chr21_+_17791648 | 0.05 |
ENST00000602892.1 ENST00000418813.2 ENST00000435697.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr21_+_17791838 | 0.05 |
ENST00000453910.1 |
LINC00478 |
long intergenic non-protein coding RNA 478 |
| chr1_-_102312517 | 0.05 |
ENST00000338858.5 |
OLFM3 |
olfactomedin 3 |
| chr20_-_271304 | 0.04 |
ENST00000400269.3 ENST00000360321.2 |
C20orf96 |
chromosome 20 open reading frame 96 |
| chr12_+_123717967 | 0.04 |
ENST00000536130.1 ENST00000546132.1 |
C12orf65 |
chromosome 12 open reading frame 65 |
| chr3_-_39234074 | 0.04 |
ENST00000340369.3 ENST00000421646.1 ENST00000396251.1 |
XIRP1 |
xin actin-binding repeat containing 1 |
| chr11_-_118436707 | 0.04 |
ENST00000264020.2 ENST00000264021.3 |
IFT46 |
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr3_-_107941209 | 0.04 |
ENST00000492106.1 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr20_-_271009 | 0.04 |
ENST00000382369.5 |
C20orf96 |
chromosome 20 open reading frame 96 |
| chr11_+_120973375 | 0.04 |
ENST00000264037.2 |
TECTA |
tectorin alpha |
| chr11_-_19223523 | 0.04 |
ENST00000265968.3 |
CSRP3 |
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr3_-_107941230 | 0.04 |
ENST00000264538.3 |
IFT57 |
intraflagellar transport 57 homolog (Chlamydomonas) |
| chr10_+_111967345 | 0.04 |
ENST00000332674.5 ENST00000453116.1 |
MXI1 |
MAX interactor 1, dimerization protein |
| chr9_+_116263778 | 0.04 |
ENST00000394646.3 |
RGS3 |
regulator of G-protein signaling 3 |
| chr17_+_26646121 | 0.04 |
ENST00000226230.6 |
TMEM97 |
transmembrane protein 97 |
| chr15_+_78799948 | 0.04 |
ENST00000566332.1 |
HYKK |
hydroxylysine kinase |
| chr5_+_169780485 | 0.04 |
ENST00000377360.4 |
KCNIP1 |
Kv channel interacting protein 1 |
| chr12_-_50222187 | 0.04 |
ENST00000335999.6 |
NCKAP5L |
NCK-associated protein 5-like |
| chr1_+_203765437 | 0.04 |
ENST00000550078.1 |
ZBED6 |
zinc finger, BED-type containing 6 |
| chr11_-_118436606 | 0.04 |
ENST00000530872.1 |
IFT46 |
intraflagellar transport 46 homolog (Chlamydomonas) |
| chr16_+_8814563 | 0.04 |
ENST00000425191.2 ENST00000569156.1 |
ABAT |
4-aminobutyrate aminotransferase |
| chr17_-_79623597 | 0.03 |
ENST00000574024.1 ENST00000331056.5 |
PDE6G |
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr12_+_93772402 | 0.03 |
ENST00000546925.1 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr9_+_118950325 | 0.03 |
ENST00000534838.1 |
PAPPA |
pregnancy-associated plasma protein A, pappalysin 1 |
| chr17_+_66521936 | 0.03 |
ENST00000592800.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr9_+_116263639 | 0.03 |
ENST00000343817.5 |
RGS3 |
regulator of G-protein signaling 3 |
| chr17_+_26646175 | 0.03 |
ENST00000583381.1 ENST00000582113.1 ENST00000582384.1 |
TMEM97 |
transmembrane protein 97 |
| chr12_-_122712038 | 0.03 |
ENST00000413918.1 ENST00000443649.3 |
DIABLO |
diablo, IAP-binding mitochondrial protein |
| chr17_+_27055798 | 0.03 |
ENST00000268766.6 |
NEK8 |
NIMA-related kinase 8 |
| chr1_-_23520755 | 0.03 |
ENST00000314113.3 |
HTR1D |
5-hydroxytryptamine (serotonin) receptor 1D, G protein-coupled |
| chr3_-_149095652 | 0.03 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr15_+_78799895 | 0.03 |
ENST00000408962.2 ENST00000388988.4 ENST00000360519.3 |
HYKK |
hydroxylysine kinase |
| chr1_-_149982624 | 0.03 |
ENST00000417191.1 ENST00000369135.4 |
OTUD7B |
OTU domain containing 7B |
| chr8_+_87878640 | 0.03 |
ENST00000518476.1 |
CNBD1 |
cyclic nucleotide binding domain containing 1 |
| chr14_+_81421861 | 0.03 |
ENST00000298171.2 |
TSHR |
thyroid stimulating hormone receptor |
| chr8_-_95220775 | 0.03 |
ENST00000441892.2 ENST00000521491.1 ENST00000027335.3 |
CDH17 |
cadherin 17, LI cadherin (liver-intestine) |
| chr11_-_47616210 | 0.03 |
ENST00000302514.3 |
C1QTNF4 |
C1q and tumor necrosis factor related protein 4 |
| chr14_+_81421355 | 0.03 |
ENST00000541158.2 |
TSHR |
thyroid stimulating hormone receptor |
| chr12_+_93772326 | 0.03 |
ENST00000550056.1 ENST00000549992.1 ENST00000548662.1 ENST00000547014.1 |
NUDT4 |
nudix (nucleoside diphosphate linked moiety X)-type motif 4 |
| chr12_+_111051832 | 0.03 |
ENST00000550703.2 ENST00000551590.1 |
TCTN1 |
tectonic family member 1 |
| chr5_+_122110691 | 0.02 |
ENST00000379516.2 ENST00000505934.1 ENST00000514949.1 |
SNX2 |
sorting nexin 2 |
| chrX_-_154299501 | 0.02 |
ENST00000369476.3 ENST00000369484.3 |
MTCP1 CMC4 |
mature T-cell proliferation 1 C-x(9)-C motif containing 4 |
| chr11_-_116663127 | 0.02 |
ENST00000433069.1 ENST00000542499.1 |
APOA5 |
apolipoprotein A-V |
| chr7_-_27224795 | 0.02 |
ENST00000006015.3 |
HOXA11 |
homeobox A11 |
| chr1_-_22109682 | 0.02 |
ENST00000400301.1 ENST00000532737.1 |
USP48 |
ubiquitin specific peptidase 48 |
| chr19_+_16999654 | 0.02 |
ENST00000248076.3 |
F2RL3 |
coagulation factor II (thrombin) receptor-like 3 |
| chrX_-_30326445 | 0.02 |
ENST00000378963.1 |
NR0B1 |
nuclear receptor subfamily 0, group B, member 1 |
| chr5_+_179105615 | 0.02 |
ENST00000514383.1 |
CANX |
calnexin |
| chr2_-_25194963 | 0.02 |
ENST00000264711.2 |
DNAJC27 |
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr9_-_14313641 | 0.02 |
ENST00000380953.1 |
NFIB |
nuclear factor I/B |
| chr15_+_42696992 | 0.02 |
ENST00000561817.1 |
CAPN3 |
calpain 3, (p94) |
| chr7_-_27224842 | 0.02 |
ENST00000517402.1 |
HOXA11 |
homeobox A11 |
| chrX_-_83442915 | 0.02 |
ENST00000262752.2 ENST00000543399.1 |
RPS6KA6 |
ribosomal protein S6 kinase, 90kDa, polypeptide 6 |
| chr11_-_73689037 | 0.02 |
ENST00000544615.1 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr12_+_111051902 | 0.02 |
ENST00000397655.3 ENST00000471804.2 ENST00000377654.3 ENST00000397659.4 |
TCTN1 |
tectonic family member 1 |
| chr1_-_22109484 | 0.02 |
ENST00000529637.1 |
USP48 |
ubiquitin specific peptidase 48 |
| chr15_-_42840961 | 0.02 |
ENST00000563454.1 ENST00000397130.3 ENST00000570160.1 ENST00000323443.2 |
LRRC57 |
leucine rich repeat containing 57 |
| chr15_+_81589254 | 0.02 |
ENST00000394652.2 |
IL16 |
interleukin 16 |
| chr3_-_187388173 | 0.02 |
ENST00000287641.3 |
SST |
somatostatin |
| chrX_+_71353499 | 0.02 |
ENST00000373677.1 |
NHSL2 |
NHS-like 2 |
| chr4_-_168155417 | 0.02 |
ENST00000511269.1 ENST00000506697.1 ENST00000512042.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr11_+_118754475 | 0.02 |
ENST00000292174.4 |
CXCR5 |
chemokine (C-X-C motif) receptor 5 |
| chr17_+_4699439 | 0.02 |
ENST00000270586.3 |
PSMB6 |
proteasome (prosome, macropain) subunit, beta type, 6 |
| chr9_+_127020202 | 0.02 |
ENST00000373600.3 ENST00000320246.5 |
NEK6 |
NIMA-related kinase 6 |
| chr1_-_160616804 | 0.02 |
ENST00000538290.1 |
SLAMF1 |
signaling lymphocytic activation molecule family member 1 |
| chr16_+_68344877 | 0.02 |
ENST00000566657.1 ENST00000565745.1 ENST00000569571.1 ENST00000569047.3 ENST00000449359.3 |
PRMT7 |
protein arginine methyltransferase 7 |
| chr7_-_81399411 | 0.02 |
ENST00000423064.2 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr17_-_80291818 | 0.01 |
ENST00000269389.3 ENST00000581691.1 |
SECTM1 |
secreted and transmembrane 1 |
| chr20_+_57226284 | 0.01 |
ENST00000458280.1 ENST00000355957.5 ENST00000361770.5 ENST00000312283.8 ENST00000412911.1 ENST00000359617.4 ENST00000371141.4 |
STX16 |
syntaxin 16 |
| chr2_-_85895295 | 0.01 |
ENST00000428225.1 ENST00000519937.2 |
SFTPB |
surfactant protein B |
| chr12_+_123717458 | 0.01 |
ENST00000253233.1 |
C12orf65 |
chromosome 12 open reading frame 65 |
| chr15_+_42697018 | 0.01 |
ENST00000397204.4 |
CAPN3 |
calpain 3, (p94) |
| chr2_+_201936458 | 0.01 |
ENST00000237889.4 |
NDUFB3 |
NADH dehydrogenase (ubiquinone) 1 beta subcomplex, 3, 12kDa |
| chr7_-_81399438 | 0.01 |
ENST00000222390.5 |
HGF |
hepatocyte growth factor (hepapoietin A; scatter factor) |
| chr17_-_40169429 | 0.01 |
ENST00000316603.7 ENST00000588641.1 |
DNAJC7 |
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr11_+_94277017 | 0.01 |
ENST00000358752.2 |
FUT4 |
fucosyltransferase 4 (alpha (1,3) fucosyltransferase, myeloid-specific) |
| chr2_+_136289030 | 0.01 |
ENST00000409478.1 ENST00000264160.4 ENST00000329971.3 ENST00000438014.1 |
R3HDM1 |
R3H domain containing 1 |
| chr8_+_133879193 | 0.01 |
ENST00000377869.1 ENST00000220616.4 |
TG |
thyroglobulin |
| chr10_-_81320151 | 0.01 |
ENST00000372325.2 ENST00000372327.5 ENST00000417041.1 |
SFTPA2 |
surfactant protein A2 |
| chr1_-_115259337 | 0.01 |
ENST00000369535.4 |
NRAS |
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr2_+_219646462 | 0.01 |
ENST00000258415.4 |
CYP27A1 |
cytochrome P450, family 27, subfamily A, polypeptide 1 |
| chr1_-_207143802 | 0.01 |
ENST00000324852.4 ENST00000400962.3 |
FCAMR |
Fc receptor, IgA, IgM, high affinity |
| chr2_+_113403434 | 0.01 |
ENST00000272542.3 |
SLC20A1 |
solute carrier family 20 (phosphate transporter), member 1 |
| chr16_+_33006369 | 0.01 |
ENST00000425181.3 |
IGHV3OR16-10 |
immunoglobulin heavy variable 3/OR16-10 (non-functional) |
| chr17_+_48624450 | 0.01 |
ENST00000006658.6 ENST00000356488.4 ENST00000393244.3 |
SPATA20 |
spermatogenesis associated 20 |
| chr16_+_69458537 | 0.01 |
ENST00000515314.1 ENST00000561792.1 ENST00000568237.1 |
CYB5B |
cytochrome b5 type B (outer mitochondrial membrane) |
| chr17_+_74261413 | 0.01 |
ENST00000587913.1 |
UBALD2 |
UBA-like domain containing 2 |
| chr3_-_113464906 | 0.01 |
ENST00000477813.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr14_+_81421710 | 0.01 |
ENST00000342443.6 |
TSHR |
thyroid stimulating hormone receptor |
| chr15_-_72563585 | 0.01 |
ENST00000287196.9 ENST00000260376.7 |
PARP6 |
poly (ADP-ribose) polymerase family, member 6 |
| chr21_+_44313375 | 0.01 |
ENST00000354250.2 ENST00000340344.4 |
NDUFV3 |
NADH dehydrogenase (ubiquinone) flavoprotein 3, 10kDa |
| chr17_+_46048376 | 0.00 |
ENST00000338399.4 |
CDK5RAP3 |
CDK5 regulatory subunit associated protein 3 |
| chr6_-_36842784 | 0.00 |
ENST00000373699.5 |
PPIL1 |
peptidylprolyl isomerase (cyclophilin)-like 1 |
| chr12_-_53097247 | 0.00 |
ENST00000341809.3 ENST00000537195.1 |
KRT77 |
keratin 77 |
| chr5_-_171615315 | 0.00 |
ENST00000176763.5 |
STK10 |
serine/threonine kinase 10 |
| chr16_+_68344981 | 0.00 |
ENST00000441236.1 ENST00000348497.4 ENST00000339507.5 |
PRMT7 |
protein arginine methyltransferase 7 |
| chr1_-_55352834 | 0.00 |
ENST00000371269.3 |
DHCR24 |
24-dehydrocholesterol reductase |
| chr20_-_56285595 | 0.00 |
ENST00000395816.3 ENST00000347215.4 |
PMEPA1 |
prostate transmembrane protein, androgen induced 1 |
| chr14_+_81421921 | 0.00 |
ENST00000554263.1 ENST00000554435.1 |
TSHR |
thyroid stimulating hormone receptor |
| chr14_+_75179840 | 0.00 |
ENST00000554590.1 ENST00000341162.4 ENST00000534938.2 ENST00000553615.1 |
FCF1 |
FCF1 rRNA-processing protein |
| chr1_+_159796534 | 0.00 |
ENST00000289707.5 |
SLAMF8 |
SLAM family member 8 |
| chr19_-_45873642 | 0.00 |
ENST00000485403.2 ENST00000586856.1 ENST00000586131.1 ENST00000391940.4 ENST00000221481.6 ENST00000391944.3 ENST00000391945.4 |
ERCC2 |
excision repair cross-complementing rodent repair deficiency, complementation group 2 |
| chr12_+_14518598 | 0.00 |
ENST00000261168.4 ENST00000538511.1 ENST00000545723.1 ENST00000543189.1 ENST00000536444.1 |
ATF7IP |
activating transcription factor 7 interacting protein |
| chr15_+_42696954 | 0.00 |
ENST00000337571.4 ENST00000569136.1 |
CAPN3 |
calpain 3, (p94) |
| chr17_-_40169659 | 0.00 |
ENST00000457167.4 |
DNAJC7 |
DnaJ (Hsp40) homolog, subfamily C, member 7 |
| chr2_-_187713891 | 0.00 |
ENST00000295131.2 |
ZSWIM2 |
zinc finger, SWIM-type containing 2 |
| chr22_+_40322595 | 0.00 |
ENST00000420971.1 ENST00000544756.1 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr1_+_154301264 | 0.00 |
ENST00000341822.2 |
ATP8B2 |
ATPase, aminophospholipid transporter, class I, type 8B, member 2 |
| chr10_+_81370689 | 0.00 |
ENST00000372308.3 ENST00000398636.3 ENST00000428376.2 ENST00000372313.5 ENST00000419470.2 ENST00000429958.1 ENST00000439264.1 |
SFTPA1 |
surfactant protein A1 |
| chr15_+_42841008 | 0.00 |
ENST00000260372.3 ENST00000568876.1 ENST00000568846.2 ENST00000562398.1 |
HAUS2 |
HAUS augmin-like complex, subunit 2 |
| chr22_+_40322623 | 0.00 |
ENST00000399090.2 |
GRAP2 |
GRB2-related adaptor protein 2 |
| chr11_-_111175739 | 0.00 |
ENST00000532918.1 |
COLCA1 |
colorectal cancer associated 1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.0 | 0.1 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.0 | 0.5 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.0 | 0.1 | GO:0080154 | regulation of fertilization(GO:0080154) |
| 0.0 | 0.5 | GO:0070493 | thrombin receptor signaling pathway(GO:0070493) |
| 0.0 | 0.1 | GO:1902303 | regulation of heart rate by hormone(GO:0003064) negative regulation of potassium ion export(GO:1902303) |
| 0.0 | 0.2 | GO:0016191 | synaptic vesicle uncoating(GO:0016191) |
| 0.0 | 0.1 | GO:1904882 | signal transduction involved in G2 DNA damage checkpoint(GO:0072425) signal transduction involved in mitotic G2 DNA damage checkpoint(GO:0072434) regulation of microglial cell activation(GO:1903978) telomerase catalytic core complex assembly(GO:1904868) regulation of telomerase catalytic core complex assembly(GO:1904882) positive regulation of telomerase catalytic core complex assembly(GO:1904884) |
| 0.0 | 0.1 | GO:0000189 | MAPK import into nucleus(GO:0000189) regulation of Golgi inheritance(GO:0090170) |
| 0.0 | 0.2 | GO:0030579 | ubiquitin-dependent SMAD protein catabolic process(GO:0030579) |
| 0.0 | 0.1 | GO:0090234 | regulation of kinetochore assembly(GO:0090234) |
| 0.0 | 0.1 | GO:1904588 | cellular response to glycoprotein(GO:1904588) cellular response to thyrotropin-releasing hormone(GO:1905229) |
| 0.0 | 0.1 | GO:0072344 | rescue of stalled ribosome(GO:0072344) |
| 0.0 | 0.1 | GO:0051612 | negative regulation of neurotransmitter uptake(GO:0051581) serotonin uptake(GO:0051610) regulation of serotonin uptake(GO:0051611) negative regulation of serotonin uptake(GO:0051612) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.1 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0071543 | diadenosine polyphosphate catabolic process(GO:0015961) diphosphoinositol polyphosphate metabolic process(GO:0071543) diadenosine pentaphosphate metabolic process(GO:1901906) diadenosine pentaphosphate catabolic process(GO:1901907) diadenosine hexaphosphate metabolic process(GO:1901908) diadenosine hexaphosphate catabolic process(GO:1901909) adenosine 5'-(hexahydrogen pentaphosphate) metabolic process(GO:1901910) adenosine 5'-(hexahydrogen pentaphosphate) catabolic process(GO:1901911) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:1902937 | inward rectifier potassium channel complex(GO:1902937) |
| 0.0 | 0.1 | GO:0000836 | Hrd1p ubiquitin ligase complex(GO:0000836) |
| 0.0 | 0.2 | GO:0005614 | interstitial matrix(GO:0005614) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.1 | 0.2 | GO:0050613 | delta14-sterol reductase activity(GO:0050613) |
| 0.0 | 0.1 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.0 | 0.1 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0003986 | acetyl-CoA hydrolase activity(GO:0003986) |
| 0.0 | 0.2 | GO:0048495 | Roundabout binding(GO:0048495) |
| 0.0 | 0.3 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0032184 | SUMO polymer binding(GO:0032184) |
| 0.0 | 0.1 | GO:0042610 | CD8 receptor binding(GO:0042610) |
| 0.0 | 0.1 | GO:0052848 | endopolyphosphatase activity(GO:0000298) diphosphoinositol-polyphosphate diphosphatase activity(GO:0008486) bis(5'-adenosyl)-hexaphosphatase activity(GO:0034431) bis(5'-adenosyl)-pentaphosphatase activity(GO:0034432) inositol diphosphate tetrakisphosphate diphosphatase activity(GO:0052840) inositol bisdiphosphate tetrakisphosphate diphosphatase activity(GO:0052841) inositol diphosphate pentakisphosphate diphosphatase activity(GO:0052842) inositol-1-diphosphate-2,3,4,5,6-pentakisphosphate diphosphatase activity(GO:0052843) inositol-3-diphosphate-1,2,4,5,6-pentakisphosphate diphosphatase activity(GO:0052844) inositol-5-diphosphate-1,2,3,4,6-pentakisphosphate diphosphatase activity(GO:0052845) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 1-diphosphatase activity(GO:0052846) inositol-1,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052847) inositol-3,5-bisdiphosphate-2,3,4,6-tetrakisphosphate 5-diphosphatase activity(GO:0052848) |