Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
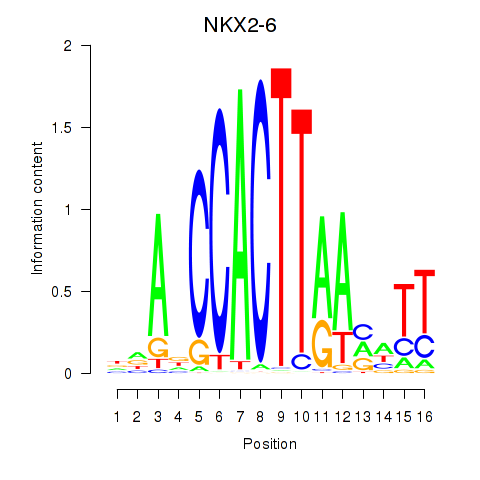
Results for NKX2-6
Z-value: 1.04
Transcription factors associated with NKX2-6
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX2-6
|
ENSG00000180053.6 | NKX2-6 |
Activity profile of NKX2-6 motif
Sorted Z-values of NKX2-6 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX2-6
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr14_-_60337684 | 1.85 |
ENST00000267484.5 |
RTN1 |
reticulon 1 |
| chr12_+_56075330 | 1.81 |
ENST00000394252.3 |
METTL7B |
methyltransferase like 7B |
| chr12_-_56236690 | 1.47 |
ENST00000322569.4 |
MMP19 |
matrix metallopeptidase 19 |
| chr4_+_70916119 | 1.43 |
ENST00000246896.3 ENST00000511674.1 |
HTN1 |
histatin 1 |
| chr8_-_120651020 | 1.01 |
ENST00000522826.1 ENST00000520066.1 ENST00000259486.6 ENST00000075322.6 |
ENPP2 |
ectonucleotide pyrophosphatase/phosphodiesterase 2 |
| chr22_+_31160239 | 0.76 |
ENST00000445781.1 ENST00000401475.1 |
OSBP2 |
oxysterol binding protein 2 |
| chr5_-_111091948 | 0.76 |
ENST00000447165.2 |
NREP |
neuronal regeneration related protein |
| chr11_+_65265141 | 0.68 |
ENST00000534336.1 |
MALAT1 |
metastasis associated lung adenocarcinoma transcript 1 (non-protein coding) |
| chr1_-_227505289 | 0.62 |
ENST00000366765.3 |
CDC42BPA |
CDC42 binding protein kinase alpha (DMPK-like) |
| chr2_-_163100045 | 0.51 |
ENST00000188790.4 |
FAP |
fibroblast activation protein, alpha |
| chr10_-_27529486 | 0.51 |
ENST00000375888.1 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chr2_-_163099885 | 0.50 |
ENST00000443424.1 |
FAP |
fibroblast activation protein, alpha |
| chr5_+_140723601 | 0.49 |
ENST00000253812.6 |
PCDHGA3 |
protocadherin gamma subfamily A, 3 |
| chr8_+_27629459 | 0.45 |
ENST00000523566.1 |
ESCO2 |
establishment of sister chromatid cohesion N-acetyltransferase 2 |
| chr12_-_39300071 | 0.44 |
ENST00000550863.1 |
CPNE8 |
copine VIII |
| chr9_+_96846740 | 0.44 |
ENST00000288976.3 |
PTPDC1 |
protein tyrosine phosphatase domain containing 1 |
| chr5_-_139937895 | 0.44 |
ENST00000336283.6 |
SRA1 |
steroid receptor RNA activator 1 |
| chr10_-_13342097 | 0.43 |
ENST00000263038.4 |
PHYH |
phytanoyl-CoA 2-hydroxylase |
| chr18_+_7754957 | 0.43 |
ENST00000400053.4 |
PTPRM |
protein tyrosine phosphatase, receptor type, M |
| chr2_-_175462934 | 0.40 |
ENST00000392546.2 ENST00000436221.1 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr3_-_114790179 | 0.38 |
ENST00000462705.1 |
ZBTB20 |
zinc finger and BTB domain containing 20 |
| chr11_-_118901559 | 0.37 |
ENST00000330775.7 ENST00000545985.1 ENST00000357590.5 ENST00000538950.1 |
SLC37A4 |
solute carrier family 37 (glucose-6-phosphate transporter), member 4 |
| chr12_-_89920030 | 0.37 |
ENST00000413530.1 ENST00000547474.1 |
GALNT4 POC1B-GALNT4 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr12_+_60083118 | 0.36 |
ENST00000261187.4 ENST00000543448.1 |
SLC16A7 |
solute carrier family 16 (monocarboxylate transporter), member 7 |
| chr10_-_101841588 | 0.34 |
ENST00000370418.3 |
CPN1 |
carboxypeptidase N, polypeptide 1 |
| chr17_-_7216939 | 0.33 |
ENST00000573684.1 |
GPS2 |
G protein pathway suppressor 2 |
| chr4_+_113739244 | 0.32 |
ENST00000503271.1 ENST00000503423.1 ENST00000506722.1 |
ANK2 |
ankyrin 2, neuronal |
| chr12_+_44152740 | 0.32 |
ENST00000440781.2 ENST00000431837.1 ENST00000550616.1 ENST00000448290.2 ENST00000551736.1 |
IRAK4 |
interleukin-1 receptor-associated kinase 4 |
| chr14_+_52164820 | 0.32 |
ENST00000554167.1 |
FRMD6 |
FERM domain containing 6 |
| chr1_+_144989309 | 0.32 |
ENST00000596396.1 |
AL590452.1 |
Uncharacterized protein |
| chr4_-_1685718 | 0.31 |
ENST00000472884.2 ENST00000489363.1 ENST00000308132.6 ENST00000463238.1 |
FAM53A |
family with sequence similarity 53, member A |
| chr11_-_62521614 | 0.31 |
ENST00000527994.1 ENST00000394807.3 |
ZBTB3 |
zinc finger and BTB domain containing 3 |
| chr15_-_56757329 | 0.30 |
ENST00000260453.3 |
MNS1 |
meiosis-specific nuclear structural 1 |
| chr5_+_78407602 | 0.30 |
ENST00000274353.5 ENST00000524080.1 |
BHMT |
betaine--homocysteine S-methyltransferase |
| chr11_+_111126707 | 0.29 |
ENST00000280325.4 |
C11orf53 |
chromosome 11 open reading frame 53 |
| chr6_-_15586238 | 0.29 |
ENST00000462989.2 |
DTNBP1 |
dystrobrevin binding protein 1 |
| chr13_+_115000556 | 0.28 |
ENST00000252458.6 |
CDC16 |
cell division cycle 16 |
| chr22_-_24096562 | 0.28 |
ENST00000398465.3 |
VPREB3 |
pre-B lymphocyte 3 |
| chr11_-_64527425 | 0.27 |
ENST00000377432.3 |
PYGM |
phosphorylase, glycogen, muscle |
| chr4_+_17579110 | 0.27 |
ENST00000606142.1 |
LAP3 |
leucine aminopeptidase 3 |
| chr7_+_142498725 | 0.27 |
ENST00000466254.1 |
TRBC2 |
T cell receptor beta constant 2 |
| chr19_+_39903185 | 0.27 |
ENST00000409794.3 |
PLEKHG2 |
pleckstrin homology domain containing, family G (with RhoGef domain) member 2 |
| chr13_+_115000521 | 0.26 |
ENST00000252457.5 ENST00000375308.1 |
CDC16 |
cell division cycle 16 |
| chr4_-_156875003 | 0.26 |
ENST00000433477.3 |
CTSO |
cathepsin O |
| chr3_-_42003479 | 0.26 |
ENST00000420927.1 |
ULK4 |
unc-51 like kinase 4 |
| chr9_+_35906176 | 0.25 |
ENST00000354323.2 |
HRCT1 |
histidine rich carboxyl terminus 1 |
| chr1_-_48866517 | 0.24 |
ENST00000371841.1 |
SPATA6 |
spermatogenesis associated 6 |
| chr3_-_127541194 | 0.24 |
ENST00000453507.2 |
MGLL |
monoglyceride lipase |
| chr9_-_134406565 | 0.24 |
ENST00000372210.3 ENST00000372211.3 |
UCK1 |
uridine-cytidine kinase 1 |
| chr20_-_271009 | 0.24 |
ENST00000382369.5 |
C20orf96 |
chromosome 20 open reading frame 96 |
| chr11_-_65629497 | 0.24 |
ENST00000532134.1 |
CFL1 |
cofilin 1 (non-muscle) |
| chr17_+_66521936 | 0.23 |
ENST00000592800.1 |
PRKAR1A |
protein kinase, cAMP-dependent, regulatory, type I, alpha |
| chr11_-_105892937 | 0.23 |
ENST00000301919.4 ENST00000534458.1 ENST00000530108.1 ENST00000530788.1 |
MSANTD4 |
Myb/SANT-like DNA-binding domain containing 4 with coiled-coils |
| chr17_+_58755184 | 0.23 |
ENST00000589222.1 ENST00000407086.3 ENST00000390652.5 |
BCAS3 |
breast carcinoma amplified sequence 3 |
| chr13_+_44453969 | 0.23 |
ENST00000325686.6 |
LACC1 |
laccase (multicopper oxidoreductase) domain containing 1 |
| chr17_-_41116454 | 0.22 |
ENST00000427569.2 ENST00000430739.1 |
AARSD1 |
alanyl-tRNA synthetase domain containing 1 |
| chr9_-_134406611 | 0.22 |
ENST00000372208.3 ENST00000372215.4 |
UCK1 |
uridine-cytidine kinase 1 |
| chr9_+_125133315 | 0.21 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr3_-_39234074 | 0.21 |
ENST00000340369.3 ENST00000421646.1 ENST00000396251.1 |
XIRP1 |
xin actin-binding repeat containing 1 |
| chr17_+_33914460 | 0.21 |
ENST00000537622.2 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr6_+_126102292 | 0.20 |
ENST00000368357.3 |
NCOA7 |
nuclear receptor coactivator 7 |
| chr15_-_20170354 | 0.20 |
ENST00000338912.5 |
IGHV1OR15-9 |
immunoglobulin heavy variable 1/OR15-9 (non-functional) |
| chr11_+_65627865 | 0.20 |
ENST00000308110.4 |
MUS81 |
MUS81 structure-specific endonuclease subunit |
| chr22_-_24096630 | 0.20 |
ENST00000248948.3 |
VPREB3 |
pre-B lymphocyte 3 |
| chrX_-_153637612 | 0.20 |
ENST00000369807.1 ENST00000369808.3 |
DNASE1L1 |
deoxyribonuclease I-like 1 |
| chr1_+_202431859 | 0.20 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr2_-_175462456 | 0.19 |
ENST00000409891.1 ENST00000410117.1 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr10_+_124670121 | 0.19 |
ENST00000368894.1 |
FAM24A |
family with sequence similarity 24, member A |
| chr19_-_49552006 | 0.19 |
ENST00000391869.3 |
CGB1 |
chorionic gonadotropin, beta polypeptide 1 |
| chr1_+_64936428 | 0.19 |
ENST00000371073.2 ENST00000290039.5 |
CACHD1 |
cache domain containing 1 |
| chr9_-_137809718 | 0.18 |
ENST00000371806.3 |
FCN1 |
ficolin (collagen/fibrinogen domain containing) 1 |
| chr3_-_160167301 | 0.18 |
ENST00000494486.1 |
TRIM59 |
tripartite motif containing 59 |
| chr14_+_50779071 | 0.18 |
ENST00000426751.2 |
ATP5S |
ATP synthase, H+ transporting, mitochondrial Fo complex, subunit s (factor B) |
| chr3_-_81811312 | 0.18 |
ENST00000429644.2 |
GBE1 |
glucan (1,4-alpha-), branching enzyme 1 |
| chr2_-_27545921 | 0.18 |
ENST00000402310.1 ENST00000405983.1 ENST00000403262.2 ENST00000428910.1 ENST00000402722.1 ENST00000399052.4 ENST00000380044.1 ENST00000405076.1 |
MPV17 |
MpV17 mitochondrial inner membrane protein |
| chrX_-_47518527 | 0.17 |
ENST00000333119.3 |
UXT |
ubiquitously-expressed, prefoldin-like chaperone |
| chr12_-_44152551 | 0.17 |
ENST00000416848.2 ENST00000550784.1 ENST00000547156.1 ENST00000549868.1 ENST00000553166.1 ENST00000551923.1 ENST00000431332.3 ENST00000344862.5 |
PUS7L |
pseudouridylate synthase 7 homolog (S. cerevisiae)-like |
| chrX_-_47518498 | 0.17 |
ENST00000335890.2 |
UXT |
ubiquitously-expressed, prefoldin-like chaperone |
| chr16_+_58783542 | 0.17 |
ENST00000500117.1 ENST00000565722.1 |
RP11-410D17.2 |
RP11-410D17.2 |
| chr1_+_158969752 | 0.17 |
ENST00000566111.1 |
IFI16 |
interferon, gamma-inducible protein 16 |
| chr16_-_20367584 | 0.17 |
ENST00000570689.1 |
UMOD |
uromodulin |
| chr12_+_129028500 | 0.17 |
ENST00000315208.8 |
TMEM132C |
transmembrane protein 132C |
| chr14_+_31494672 | 0.16 |
ENST00000542754.2 ENST00000313566.6 |
AP4S1 |
adaptor-related protein complex 4, sigma 1 subunit |
| chr22_+_44577724 | 0.16 |
ENST00000466375.2 |
PARVG |
parvin, gamma |
| chr5_-_140070897 | 0.16 |
ENST00000448240.1 ENST00000438307.2 ENST00000415192.2 ENST00000457527.2 ENST00000307633.3 ENST00000507746.1 ENST00000431330.2 |
HARS |
histidyl-tRNA synthetase |
| chr5_-_133510456 | 0.16 |
ENST00000520417.1 |
SKP1 |
S-phase kinase-associated protein 1 |
| chr9_+_134065506 | 0.15 |
ENST00000483497.2 |
NUP214 |
nucleoporin 214kDa |
| chr17_+_33914276 | 0.15 |
ENST00000592545.1 ENST00000538556.1 ENST00000312678.8 ENST00000589344.1 |
AP2B1 |
adaptor-related protein complex 2, beta 1 subunit |
| chr19_+_50433476 | 0.15 |
ENST00000596658.1 |
ATF5 |
activating transcription factor 5 |
| chr5_+_140071178 | 0.15 |
ENST00000508522.1 ENST00000448069.2 |
HARS2 |
histidyl-tRNA synthetase 2, mitochondrial |
| chr17_-_55911970 | 0.15 |
ENST00000581805.1 ENST00000580960.1 |
RP11-60A24.3 |
RP11-60A24.3 |
| chr6_+_10528560 | 0.15 |
ENST00000379597.3 |
GCNT2 |
glucosaminyl (N-acetyl) transferase 2, I-branching enzyme (I blood group) |
| chr7_+_100466433 | 0.15 |
ENST00000429658.1 |
TRIP6 |
thyroid hormone receptor interactor 6 |
| chr16_+_69796209 | 0.14 |
ENST00000359154.2 ENST00000561780.1 ENST00000563659.1 ENST00000448661.1 |
WWP2 |
WW domain containing E3 ubiquitin protein ligase 2 |
| chr20_+_32254286 | 0.14 |
ENST00000330271.4 |
ACTL10 |
actin-like 10 |
| chr12_-_109531264 | 0.14 |
ENST00000429722.2 ENST00000536242.1 ENST00000343075.3 ENST00000536358.1 |
ALKBH2 |
alkB, alkylation repair homolog 2 (E. coli) |
| chr5_+_140071011 | 0.14 |
ENST00000230771.3 ENST00000509299.1 ENST00000503873.1 ENST00000435019.2 ENST00000437649.2 ENST00000432671.2 |
HARS2 |
histidyl-tRNA synthetase 2, mitochondrial |
| chr3_+_114012819 | 0.14 |
ENST00000383671.3 |
TIGIT |
T cell immunoreceptor with Ig and ITIM domains |
| chr6_-_159466042 | 0.14 |
ENST00000338313.5 |
TAGAP |
T-cell activation RhoGTPase activating protein |
| chr10_+_38717074 | 0.14 |
ENST00000423687.1 |
LINC00999 |
long intergenic non-protein coding RNA 999 |
| chr16_-_23724518 | 0.14 |
ENST00000457008.2 |
ERN2 |
endoplasmic reticulum to nucleus signaling 2 |
| chr20_-_32262165 | 0.13 |
ENST00000606690.1 ENST00000246190.6 ENST00000439478.1 ENST00000375238.4 |
NECAB3 |
N-terminal EF-hand calcium binding protein 3 |
| chr8_+_42396712 | 0.13 |
ENST00000518574.1 ENST00000417410.2 ENST00000414154.2 |
SMIM19 |
small integral membrane protein 19 |
| chr8_-_62602327 | 0.13 |
ENST00000445642.3 ENST00000517847.2 ENST00000389204.4 ENST00000517661.1 ENST00000517903.1 ENST00000522603.1 ENST00000522349.1 ENST00000522835.1 ENST00000541428.1 ENST00000518306.1 |
ASPH |
aspartate beta-hydroxylase |
| chr3_-_48130707 | 0.13 |
ENST00000360240.6 ENST00000383737.4 |
MAP4 |
microtubule-associated protein 4 |
| chr9_-_215744 | 0.13 |
ENST00000382387.2 |
C9orf66 |
chromosome 9 open reading frame 66 |
| chr13_+_24883703 | 0.13 |
ENST00000332018.4 |
C1QTNF9 |
C1q and tumor necrosis factor related protein 9 |
| chr17_-_2966901 | 0.13 |
ENST00000575751.1 |
OR1D5 |
olfactory receptor, family 1, subfamily D, member 5 |
| chr14_+_31494841 | 0.13 |
ENST00000556232.1 ENST00000216366.4 ENST00000334725.4 ENST00000554609.1 ENST00000554345.1 |
AP4S1 |
adaptor-related protein complex 4, sigma 1 subunit |
| chr1_-_21113105 | 0.13 |
ENST00000375000.1 ENST00000419490.1 ENST00000414993.1 ENST00000443615.1 ENST00000312239.5 |
HP1BP3 |
heterochromatin protein 1, binding protein 3 |
| chr9_+_131452239 | 0.13 |
ENST00000372688.4 ENST00000372686.5 |
SET |
SET nuclear oncogene |
| chr3_-_39196049 | 0.12 |
ENST00000514182.1 |
CSRNP1 |
cysteine-serine-rich nuclear protein 1 |
| chr21_-_31852663 | 0.12 |
ENST00000390689.2 |
KRTAP19-1 |
keratin associated protein 19-1 |
| chr19_-_49552363 | 0.12 |
ENST00000448456.3 ENST00000355414.2 |
CGB8 |
chorionic gonadotropin, beta polypeptide 8 |
| chr11_-_104480019 | 0.12 |
ENST00000536529.1 ENST00000545630.1 ENST00000538641.1 |
RP11-886D15.1 |
RP11-886D15.1 |
| chr19_+_10563567 | 0.12 |
ENST00000344979.3 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chr5_-_176889381 | 0.12 |
ENST00000393563.4 ENST00000512501.1 |
DBN1 |
drebrin 1 |
| chr3_+_186358148 | 0.12 |
ENST00000382134.3 ENST00000265029.3 |
FETUB |
fetuin B |
| chr2_-_25565377 | 0.11 |
ENST00000264709.3 ENST00000406659.3 |
DNMT3A |
DNA (cytosine-5-)-methyltransferase 3 alpha |
| chr19_-_39881669 | 0.11 |
ENST00000221266.7 |
PAF1 |
Paf1, RNA polymerase II associated factor, homolog (S. cerevisiae) |
| chr1_-_115259337 | 0.11 |
ENST00000369535.4 |
NRAS |
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr2_+_203776937 | 0.11 |
ENST00000402905.3 ENST00000414490.1 ENST00000431787.1 ENST00000444724.1 ENST00000414857.1 ENST00000430899.1 ENST00000445120.1 ENST00000441569.1 ENST00000432024.1 ENST00000443740.1 ENST00000414439.1 ENST00000428585.1 ENST00000545253.1 ENST00000545262.1 ENST00000447539.1 ENST00000456821.2 ENST00000434998.1 ENST00000320443.8 |
CARF |
calcium responsive transcription factor |
| chr1_-_203144941 | 0.11 |
ENST00000255416.4 |
MYBPH |
myosin binding protein H |
| chr7_-_56101826 | 0.11 |
ENST00000421626.1 |
PSPH |
phosphoserine phosphatase |
| chr5_-_175612149 | 0.10 |
ENST00000515403.1 |
RP11-844P9.2 |
RP11-844P9.2 |
| chr4_-_186317034 | 0.10 |
ENST00000505916.1 |
LRP2BP |
LRP2 binding protein |
| chr20_-_1373682 | 0.10 |
ENST00000381724.3 |
FKBP1A |
FK506 binding protein 1A, 12kDa |
| chr14_+_100531615 | 0.10 |
ENST00000392920.3 |
EVL |
Enah/Vasp-like |
| chr16_-_47493041 | 0.10 |
ENST00000565940.2 |
ITFG1 |
integrin alpha FG-GAP repeat containing 1 |
| chr10_+_18629628 | 0.10 |
ENST00000377329.4 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr19_+_496454 | 0.10 |
ENST00000346144.4 ENST00000215637.3 ENST00000382683.4 |
MADCAM1 |
mucosal vascular addressin cell adhesion molecule 1 |
| chr13_-_44453826 | 0.10 |
ENST00000444614.3 |
CCDC122 |
coiled-coil domain containing 122 |
| chr10_+_115939008 | 0.10 |
ENST00000369282.1 ENST00000251864.2 ENST00000369281.2 ENST00000422662.1 |
TDRD1 |
tudor domain containing 1 |
| chr5_+_52083730 | 0.10 |
ENST00000282588.6 ENST00000274311.2 |
ITGA1 PELO |
integrin, alpha 1 pelota homolog (Drosophila) |
| chr1_+_110162448 | 0.10 |
ENST00000342115.4 ENST00000469039.2 ENST00000474459.1 ENST00000528667.1 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr12_-_89919965 | 0.10 |
ENST00000548729.1 |
POC1B-GALNT4 |
POC1B-GALNT4 readthrough |
| chr17_+_44370099 | 0.10 |
ENST00000496930.1 |
LRRC37A |
leucine rich repeat containing 37A |
| chr9_+_125132803 | 0.09 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr1_-_173793458 | 0.09 |
ENST00000356198.2 |
CENPL |
centromere protein L |
| chr2_-_69870747 | 0.09 |
ENST00000409068.1 |
AAK1 |
AP2 associated kinase 1 |
| chr1_-_173793246 | 0.09 |
ENST00000345664.6 ENST00000367710.3 |
CENPL |
centromere protein L |
| chr12_-_50294033 | 0.09 |
ENST00000552669.1 |
FAIM2 |
Fas apoptotic inhibitory molecule 2 |
| chr22_+_22749343 | 0.09 |
ENST00000390298.2 |
IGLV7-43 |
immunoglobulin lambda variable 7-43 |
| chr7_-_150777949 | 0.09 |
ENST00000482571.1 |
FASTK |
Fas-activated serine/threonine kinase |
| chr14_+_22265444 | 0.09 |
ENST00000390430.2 |
TRAV8-1 |
T cell receptor alpha variable 8-1 |
| chr17_-_57229155 | 0.09 |
ENST00000584089.1 |
SKA2 |
spindle and kinetochore associated complex subunit 2 |
| chr20_-_21494654 | 0.09 |
ENST00000377142.4 |
NKX2-2 |
NK2 homeobox 2 |
| chr7_-_150777874 | 0.09 |
ENST00000540185.1 |
FASTK |
Fas-activated serine/threonine kinase |
| chr3_+_37035289 | 0.08 |
ENST00000455445.2 ENST00000441265.1 ENST00000435176.1 ENST00000429117.1 ENST00000536378.1 |
MLH1 |
mutL homolog 1 |
| chr16_-_69373396 | 0.08 |
ENST00000562595.1 ENST00000562081.1 ENST00000306875.4 |
COG8 |
component of oligomeric golgi complex 8 |
| chr8_+_42396936 | 0.08 |
ENST00000416469.2 |
SMIM19 |
small integral membrane protein 19 |
| chr11_+_118958689 | 0.08 |
ENST00000535253.1 ENST00000392841.1 |
HMBS |
hydroxymethylbilane synthase |
| chr11_-_129817356 | 0.08 |
ENST00000526082.1 |
PRDM10 |
PR domain containing 10 |
| chr4_+_169418255 | 0.08 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr10_+_135204338 | 0.08 |
ENST00000468317.2 |
RP11-108K14.8 |
Mitochondrial GTPase 1 |
| chr2_+_132160448 | 0.08 |
ENST00000437751.1 |
AC073869.19 |
long intergenic non-protein coding RNA 1120 |
| chr15_+_84841242 | 0.08 |
ENST00000558195.1 |
RP11-182J1.16 |
ubiquitin-conjugating enzyme E2Q family member 2-like |
| chr19_-_37663332 | 0.08 |
ENST00000392157.2 |
ZNF585A |
zinc finger protein 585A |
| chr5_-_42825983 | 0.08 |
ENST00000506577.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr19_+_58144529 | 0.08 |
ENST00000347302.3 ENST00000254182.7 ENST00000391703.3 ENST00000541801.1 ENST00000299871.5 ENST00000544273.1 |
ZNF211 |
zinc finger protein 211 |
| chr22_+_23229960 | 0.08 |
ENST00000526893.1 ENST00000532223.2 ENST00000531372.1 |
IGLL5 |
immunoglobulin lambda-like polypeptide 5 |
| chr17_+_25621102 | 0.07 |
ENST00000581440.1 ENST00000262394.2 ENST00000583742.1 ENST00000579733.1 ENST00000583193.1 ENST00000581185.1 ENST00000427287.2 ENST00000348811.2 |
WSB1 |
WD repeat and SOCS box containing 1 |
| chr11_+_27062272 | 0.07 |
ENST00000529202.1 ENST00000533566.1 |
BBOX1 |
butyrobetaine (gamma), 2-oxoglutarate dioxygenase (gamma-butyrobetaine hydroxylase) 1 |
| chr15_+_65843130 | 0.07 |
ENST00000569894.1 |
PTPLAD1 |
protein tyrosine phosphatase-like A domain containing 1 |
| chr7_-_38389573 | 0.07 |
ENST00000390344.2 |
TRGV5 |
T cell receptor gamma variable 5 |
| chr20_-_36661826 | 0.07 |
ENST00000373448.2 ENST00000373447.3 |
TTI1 |
TELO2 interacting protein 1 |
| chr19_-_37663572 | 0.07 |
ENST00000588354.1 ENST00000292841.5 ENST00000355533.2 ENST00000356958.4 |
ZNF585A |
zinc finger protein 585A |
| chr9_+_131447342 | 0.07 |
ENST00000409104.3 |
SET |
SET nuclear oncogene |
| chrX_-_47341928 | 0.07 |
ENST00000313116.7 |
ZNF41 |
zinc finger protein 41 |
| chr21_-_43735628 | 0.07 |
ENST00000291525.10 ENST00000518498.1 |
TFF3 |
trefoil factor 3 (intestinal) |
| chrX_+_47053208 | 0.07 |
ENST00000442035.1 ENST00000457753.1 ENST00000335972.6 |
UBA1 |
ubiquitin-like modifier activating enzyme 1 |
| chr2_+_219246746 | 0.06 |
ENST00000233202.6 |
SLC11A1 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 1 |
| chr7_+_101917407 | 0.06 |
ENST00000487284.1 |
CUX1 |
cut-like homeobox 1 |
| chr9_-_111882195 | 0.06 |
ENST00000374586.3 |
TMEM245 |
transmembrane protein 245 |
| chr14_+_64680854 | 0.06 |
ENST00000458046.2 |
SYNE2 |
spectrin repeat containing, nuclear envelope 2 |
| chr9_-_33402506 | 0.06 |
ENST00000377425.4 ENST00000537089.1 ENST00000297988.1 ENST00000539936.1 ENST00000541274.1 |
AQP7 |
aquaporin 7 |
| chr1_-_169337176 | 0.06 |
ENST00000472647.1 ENST00000367811.3 |
NME7 |
NME/NM23 family member 7 |
| chr1_-_198509804 | 0.06 |
ENST00000489986.1 ENST00000367382.1 |
ATP6V1G3 |
ATPase, H+ transporting, lysosomal 13kDa, V1 subunit G3 |
| chr7_-_150777920 | 0.06 |
ENST00000353841.2 ENST00000297532.6 |
FASTK |
Fas-activated serine/threonine kinase |
| chr2_+_86426478 | 0.06 |
ENST00000254644.8 ENST00000605125.1 ENST00000337109.4 ENST00000409180.1 |
MRPL35 |
mitochondrial ribosomal protein L35 |
| chr12_-_75603482 | 0.06 |
ENST00000341669.3 ENST00000298972.1 ENST00000350228.2 |
KCNC2 |
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr4_+_169418195 | 0.06 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr22_+_42017987 | 0.06 |
ENST00000405506.1 |
XRCC6 |
X-ray repair complementing defective repair in Chinese hamster cells 6 |
| chr15_-_55563072 | 0.06 |
ENST00000567380.1 ENST00000565972.1 ENST00000569493.1 |
RAB27A |
RAB27A, member RAS oncogene family |
| chr7_+_128116783 | 0.06 |
ENST00000262432.8 ENST00000480046.1 |
METTL2B |
methyltransferase like 2B |
| chr5_+_173763250 | 0.06 |
ENST00000515513.1 ENST00000507361.1 ENST00000510234.1 |
RP11-267A15.1 |
RP11-267A15.1 |
| chr3_-_157824292 | 0.06 |
ENST00000483851.2 |
SHOX2 |
short stature homeobox 2 |
| chr17_-_34329084 | 0.05 |
ENST00000354059.4 ENST00000536149.1 |
CCL15 CCL14 |
chemokine (C-C motif) ligand 15 chemokine (C-C motif) ligand 14 |
| chr6_-_110012380 | 0.05 |
ENST00000424296.2 ENST00000341338.6 ENST00000368948.2 ENST00000285397.5 |
AK9 |
adenylate kinase 9 |
| chr11_+_122709200 | 0.05 |
ENST00000227348.4 |
CRTAM |
cytotoxic and regulatory T cell molecule |
| chrX_+_139791917 | 0.05 |
ENST00000607004.1 ENST00000370535.3 |
LINC00632 |
long intergenic non-protein coding RNA 632 |
| chr17_-_34890709 | 0.05 |
ENST00000544606.1 |
MYO19 |
myosin XIX |
| chr12_-_50290839 | 0.05 |
ENST00000552863.1 |
FAIM2 |
Fas apoptotic inhibitory molecule 2 |
| chr3_+_100428188 | 0.05 |
ENST00000418917.2 ENST00000490574.1 |
TFG |
TRK-fused gene |
| chr17_-_47723943 | 0.05 |
ENST00000510476.1 ENST00000503676.1 |
SPOP |
speckle-type POZ protein |
| chr5_-_43412418 | 0.05 |
ENST00000537013.1 ENST00000361115.4 |
CCL28 |
chemokine (C-C motif) ligand 28 |
| chr2_+_109335929 | 0.05 |
ENST00000283195.6 |
RANBP2 |
RAN binding protein 2 |
| chr1_+_113010056 | 0.05 |
ENST00000369686.5 |
WNT2B |
wingless-type MMTV integration site family, member 2B |
| chr11_-_66360548 | 0.05 |
ENST00000333861.3 |
CCDC87 |
coiled-coil domain containing 87 |
| chr22_+_44577237 | 0.04 |
ENST00000415224.1 ENST00000417767.1 |
PARVG |
parvin, gamma |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0097325 | melanocyte proliferation(GO:0097325) |
| 0.2 | 1.5 | GO:0001554 | luteolysis(GO:0001554) |
| 0.2 | 0.5 | GO:0006427 | histidyl-tRNA aminoacylation(GO:0006427) |
| 0.1 | 0.4 | GO:0097089 | methyl-branched fatty acid metabolic process(GO:0097089) |
| 0.1 | 0.3 | GO:0036371 | protein localization to T-tubule(GO:0036371) |
| 0.1 | 0.3 | GO:0006579 | amino-acid betaine catabolic process(GO:0006579) |
| 0.1 | 0.3 | GO:0030070 | insulin processing(GO:0030070) |
| 0.1 | 0.5 | GO:0034421 | post-translational protein acetylation(GO:0034421) |
| 0.1 | 0.2 | GO:0006419 | alanyl-tRNA aminoacylation(GO:0006419) |
| 0.1 | 0.5 | GO:0030242 | pexophagy(GO:0030242) |
| 0.1 | 0.4 | GO:0099590 | neurotransmitter receptor internalization(GO:0099590) |
| 0.1 | 1.0 | GO:0034638 | phosphatidylcholine catabolic process(GO:0034638) |
| 0.1 | 0.2 | GO:0002752 | cell surface pattern recognition receptor signaling pathway(GO:0002752) |
| 0.1 | 0.5 | GO:0044211 | CTP salvage(GO:0044211) |
| 0.1 | 0.2 | GO:0072429 | response to intra-S DNA damage checkpoint signaling(GO:0072429) |
| 0.0 | 0.3 | GO:0003383 | apical constriction(GO:0003383) |
| 0.0 | 0.4 | GO:1901475 | pyruvate transport(GO:0006848) pyruvate transmembrane transport(GO:1901475) |
| 0.0 | 0.3 | GO:0051510 | regulation of unidimensional cell growth(GO:0051510) negative regulation of unidimensional cell growth(GO:0051511) establishment of cell polarity regulating cell shape(GO:0071964) regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000769) positive regulation of establishment or maintenance of cell polarity regulating cell shape(GO:2000771) regulation of establishment of cell polarity regulating cell shape(GO:2000782) positive regulation of establishment of cell polarity regulating cell shape(GO:2000784) positive regulation of barbed-end actin filament capping(GO:2000814) |
| 0.0 | 0.2 | GO:2000124 | regulation of endocannabinoid signaling pathway(GO:2000124) |
| 0.0 | 0.4 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.0 | 0.1 | GO:0042264 | peptidyl-aspartic acid hydroxylation(GO:0042264) |
| 0.0 | 0.2 | GO:0072233 | thick ascending limb development(GO:0072023) metanephric thick ascending limb development(GO:0072233) |
| 0.0 | 0.1 | GO:0051311 | meiotic metaphase I plate congression(GO:0043060) meiotic spindle midzone assembly(GO:0051257) meiotic metaphase plate congression(GO:0051311) |
| 0.0 | 1.4 | GO:0050832 | defense response to fungus(GO:0050832) |
| 0.0 | 0.1 | GO:0008588 | release of cytoplasmic sequestered NF-kappaB(GO:0008588) |
| 0.0 | 0.1 | GO:0021530 | spinal cord oligodendrocyte cell differentiation(GO:0021529) spinal cord oligodendrocyte cell fate specification(GO:0021530) |
| 0.0 | 0.1 | GO:0010643 | cell communication by chemical coupling(GO:0010643) |
| 0.0 | 0.1 | GO:0051012 | microtubule sliding(GO:0051012) |
| 0.0 | 0.1 | GO:0055073 | cadmium ion homeostasis(GO:0055073) divalent metal ion export(GO:0070839) |
| 0.0 | 0.1 | GO:0035511 | oxidative DNA demethylation(GO:0035511) |
| 0.0 | 0.3 | GO:0060155 | platelet dense granule organization(GO:0060155) |
| 0.0 | 0.1 | GO:0090116 | C-5 methylation of cytosine(GO:0090116) |
| 0.0 | 0.2 | GO:0021891 | olfactory bulb interneuron development(GO:0021891) |
| 0.0 | 0.1 | GO:0036438 | maintenance of lens transparency(GO:0036438) |
| 0.0 | 0.1 | GO:0061566 | dADP phosphorylation(GO:0006174) dGDP phosphorylation(GO:0006186) AMP phosphorylation(GO:0006756) CDP phosphorylation(GO:0061508) dAMP phosphorylation(GO:0061565) CMP phosphorylation(GO:0061566) dCMP phosphorylation(GO:0061567) GDP phosphorylation(GO:0061568) UDP phosphorylation(GO:0061569) dCDP phosphorylation(GO:0061570) TDP phosphorylation(GO:0061571) |
| 0.0 | 0.3 | GO:0070986 | left/right axis specification(GO:0070986) |
| 0.0 | 0.3 | GO:0019371 | cyclooxygenase pathway(GO:0019371) |
| 0.0 | 0.1 | GO:1904879 | positive regulation of calcium ion transmembrane transport via high voltage-gated calcium channel(GO:1904879) |
| 0.0 | 0.1 | GO:0070966 | nuclear-transcribed mRNA catabolic process, no-go decay(GO:0070966) |
| 0.0 | 0.2 | GO:0010265 | SCF complex assembly(GO:0010265) |
| 0.0 | 0.1 | GO:0030263 | apoptotic chromosome condensation(GO:0030263) |
| 0.0 | 0.3 | GO:0034162 | toll-like receptor 9 signaling pathway(GO:0034162) |
| 0.0 | 0.0 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.0 | 0.1 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.4 | GO:0010842 | retina layer formation(GO:0010842) |
| 0.0 | 0.2 | GO:0046007 | negative regulation of activated T cell proliferation(GO:0046007) |
| 0.0 | 0.3 | GO:0047497 | establishment of mitochondrion localization, microtubule-mediated(GO:0034643) mitochondrion transport along microtubule(GO:0047497) |
| 0.0 | 0.0 | GO:0090526 | regulation of gluconeogenesis involved in cellular glucose homeostasis(GO:0090526) |
| 0.0 | 0.1 | GO:0045329 | carnitine biosynthetic process(GO:0045329) |
| 0.0 | 0.2 | GO:1902083 | negative regulation of peptidyl-cysteine S-nitrosylation(GO:1902083) |
| 0.0 | 0.1 | GO:0035865 | cellular response to potassium ion(GO:0035865) |
| 0.0 | 0.1 | GO:0031022 | nuclear migration along microfilament(GO:0031022) |
| 0.0 | 0.1 | GO:0042984 | amyloid precursor protein biosynthetic process(GO:0042983) regulation of amyloid precursor protein biosynthetic process(GO:0042984) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0097224 | sperm connecting piece(GO:0097224) |
| 0.1 | 0.2 | GO:1990876 | cytoplasmic side of nuclear pore(GO:1990876) |
| 0.1 | 0.2 | GO:0048476 | Holliday junction resolvase complex(GO:0048476) |
| 0.0 | 0.3 | GO:0071438 | invadopodium membrane(GO:0071438) |
| 0.0 | 0.1 | GO:0034665 | integrin alpha1-beta1 complex(GO:0034665) |
| 0.0 | 0.1 | GO:0005715 | late recombination nodule(GO:0005715) |
| 0.0 | 0.8 | GO:0097440 | apical dendrite(GO:0097440) |
| 0.0 | 0.1 | GO:1990604 | IRE1-TRAF2-ASK1 complex(GO:1990604) |
| 0.0 | 0.5 | GO:0031618 | nuclear pericentric heterochromatin(GO:0031618) |
| 0.0 | 0.1 | GO:0045323 | interleukin-1 receptor complex(GO:0045323) |
| 0.0 | 0.2 | GO:0031232 | extrinsic component of external side of plasma membrane(GO:0031232) |
| 0.0 | 0.4 | GO:0036020 | endolysosome membrane(GO:0036020) |
| 0.0 | 0.1 | GO:0032541 | cortical endoplasmic reticulum(GO:0032541) |
| 0.0 | 0.2 | GO:0031467 | Cul7-RING ubiquitin ligase complex(GO:0031467) |
| 0.0 | 0.5 | GO:0005680 | anaphase-promoting complex(GO:0005680) |
| 0.0 | 0.3 | GO:0045009 | melanosome membrane(GO:0033162) chitosome(GO:0045009) |
| 0.0 | 0.1 | GO:0000137 | Golgi cis cisterna(GO:0000137) |
| 0.0 | 0.5 | GO:0042571 | immunoglobulin complex, circulating(GO:0042571) |
| 0.0 | 2.1 | GO:0030176 | integral component of endoplasmic reticulum membrane(GO:0030176) |
| 0.0 | 0.3 | GO:0000930 | gamma-tubulin complex(GO:0000930) |
| 0.0 | 0.1 | GO:0071546 | pi-body(GO:0071546) |
| 0.0 | 0.2 | GO:0005952 | cAMP-dependent protein kinase complex(GO:0005952) |
| 0.0 | 0.2 | GO:0045263 | proton-transporting ATP synthase complex, coupling factor F(o)(GO:0045263) |
| 0.0 | 0.1 | GO:0016593 | Cdc73/Paf1 complex(GO:0016593) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0047391 | alkylglycerophosphoethanolamine phosphodiesterase activity(GO:0047391) |
| 0.2 | 0.5 | GO:0004821 | histidine-tRNA ligase activity(GO:0004821) |
| 0.1 | 0.3 | GO:0047150 | betaine-homocysteine S-methyltransferase activity(GO:0047150) |
| 0.1 | 0.4 | GO:0005477 | pyruvate secondary active transmembrane transporter activity(GO:0005477) |
| 0.1 | 0.2 | GO:0033867 | Fas-activated serine/threonine kinase activity(GO:0033867) |
| 0.1 | 0.2 | GO:0010698 | acetyltransferase activator activity(GO:0010698) |
| 0.1 | 0.3 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |
| 0.1 | 0.2 | GO:0004813 | alanine-tRNA ligase activity(GO:0004813) |
| 0.1 | 0.3 | GO:0008184 | glycogen phosphorylase activity(GO:0008184) |
| 0.1 | 0.5 | GO:0004468 | lysine N-acetyltransferase activity, acting on acetyl phosphate as donor(GO:0004468) |
| 0.1 | 0.4 | GO:0015315 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.0 | 0.1 | GO:0035514 | DNA demethylase activity(GO:0035514) |
| 0.0 | 0.8 | GO:0005522 | profilin binding(GO:0005522) |
| 0.0 | 0.1 | GO:0004597 | peptide-aspartate beta-dioxygenase activity(GO:0004597) |
| 0.0 | 0.5 | GO:0004849 | uridine kinase activity(GO:0004849) |
| 0.0 | 1.0 | GO:0008239 | dipeptidyl-peptidase activity(GO:0008239) |
| 0.0 | 0.1 | GO:0098640 | integrin binding involved in cell-matrix adhesion(GO:0098640) |
| 0.0 | 0.1 | GO:0008336 | gamma-butyrobetaine dioxygenase activity(GO:0008336) |
| 0.0 | 0.2 | GO:0048256 | flap endonuclease activity(GO:0048256) |
| 0.0 | 0.3 | GO:0004185 | serine-type carboxypeptidase activity(GO:0004185) |
| 0.0 | 0.1 | GO:0051139 | metal ion:proton antiporter activity(GO:0051139) |
| 0.0 | 0.1 | GO:0008109 | N-acetyllactosaminide beta-1,6-N-acetylglucosaminyltransferase activity(GO:0008109) |
| 0.0 | 0.5 | GO:0005149 | interleukin-1 receptor binding(GO:0005149) |
| 0.0 | 0.1 | GO:0003886 | DNA (cytosine-5-)-methyltransferase activity(GO:0003886) |
| 0.0 | 0.1 | GO:0004647 | phosphoserine phosphatase activity(GO:0004647) |
| 0.0 | 0.5 | GO:0000062 | fatty-acyl-CoA binding(GO:0000062) |
| 0.0 | 0.1 | GO:0098634 | protein binding involved in cell-matrix adhesion(GO:0098634) collagen binding involved in cell-matrix adhesion(GO:0098639) |
| 0.0 | 0.4 | GO:0098748 | clathrin adaptor activity(GO:0035615) endocytic adaptor activity(GO:0098748) |
| 0.0 | 0.4 | GO:0031418 | L-ascorbic acid binding(GO:0031418) |
| 0.0 | 0.1 | GO:0047623 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.2 | GO:0047372 | acylglycerol lipase activity(GO:0047372) |
| 0.0 | 0.3 | GO:0005095 | GTPase inhibitor activity(GO:0005095) |
| 0.0 | 0.2 | GO:0033691 | sialic acid binding(GO:0033691) |
| 0.0 | 0.0 | GO:0061752 | telomeric repeat-containing RNA binding(GO:0061752) |
| 0.0 | 0.1 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.0 | 0.1 | GO:0086056 | voltage-gated calcium channel activity involved in AV node cell action potential(GO:0086056) |
| 0.0 | 0.2 | GO:0005049 | nuclear export signal receptor activity(GO:0005049) |
| 0.0 | 0.2 | GO:0032027 | myosin light chain binding(GO:0032027) |
| 0.0 | 0.2 | GO:0008603 | cAMP-dependent protein kinase regulator activity(GO:0008603) |
| 0.0 | 0.1 | GO:0032137 | guanine/thymine mispair binding(GO:0032137) |
| 0.0 | 0.8 | GO:0015485 | cholesterol binding(GO:0015485) |
| 0.0 | 0.1 | GO:0004839 | ubiquitin activating enzyme activity(GO:0004839) |
| 0.0 | 0.1 | GO:0016427 | tRNA (cytosine) methyltransferase activity(GO:0016427) |
| 0.0 | 1.4 | GO:0004222 | metalloendopeptidase activity(GO:0004222) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.4 | REACTOME BILE SALT AND ORGANIC ANION SLC TRANSPORTERS | Genes involved in Bile salt and organic anion SLC transporters |
| 0.0 | 0.4 | REACTOME RETROGRADE NEUROTROPHIN SIGNALLING | Genes involved in Retrograde neurotrophin signalling |
| 0.0 | 0.5 | REACTOME CONVERSION FROM APC C CDC20 TO APC C CDH1 IN LATE ANAPHASE | Genes involved in Conversion from APC/C:Cdc20 to APC/C:Cdh1 in late anaphase |
| 0.0 | 0.5 | REACTOME PYRIMIDINE METABOLISM | Genes involved in Pyrimidine metabolism |
| 0.0 | 0.4 | REACTOME PEROXISOMAL LIPID METABOLISM | Genes involved in Peroxisomal lipid metabolism |
| 0.0 | 0.1 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |