Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
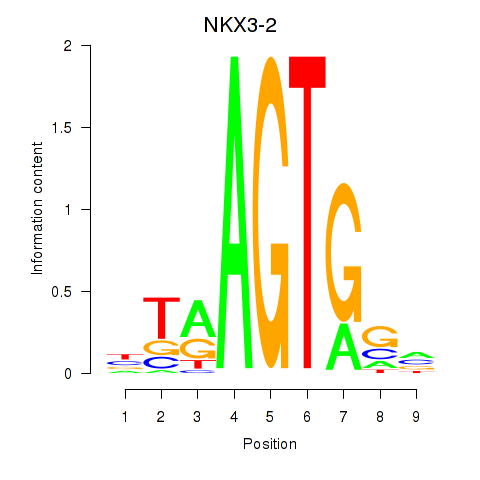
Results for NKX3-2
Z-value: 0.78
Transcription factors associated with NKX3-2
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX3-2
|
ENSG00000109705.7 | NKX3-2 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX3-2 | hg19_v2_chr4_-_13546632_13546674 | -0.04 | 9.3e-01 | Click! |
Activity profile of NKX3-2 motif
Sorted Z-values of NKX3-2 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX3-2
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_98031310 | 1.16 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr10_-_98031265 | 1.08 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr2_+_17721920 | 1.03 |
ENST00000295156.4 |
VSNL1 |
visinin-like 1 |
| chr19_-_6720686 | 0.97 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chr1_+_86889769 | 0.95 |
ENST00000370565.4 |
CLCA2 |
chloride channel accessory 2 |
| chr12_+_4382917 | 0.92 |
ENST00000261254.3 |
CCND2 |
cyclin D2 |
| chr11_-_119993979 | 0.90 |
ENST00000524816.3 ENST00000525327.1 |
TRIM29 |
tripartite motif containing 29 |
| chr8_+_86376081 | 0.88 |
ENST00000285379.5 |
CA2 |
carbonic anhydrase II |
| chr1_+_35247859 | 0.82 |
ENST00000373362.3 |
GJB3 |
gap junction protein, beta 3, 31kDa |
| chr6_-_136788001 | 0.78 |
ENST00000544465.1 |
MAP7 |
microtubule-associated protein 7 |
| chr1_-_21059029 | 0.77 |
ENST00000444387.2 ENST00000375031.1 ENST00000518294.1 |
SH2D5 |
SH2 domain containing 5 |
| chr2_-_70781087 | 0.77 |
ENST00000394241.3 ENST00000295400.6 |
TGFA |
transforming growth factor, alpha |
| chr19_-_6767516 | 0.74 |
ENST00000245908.6 |
SH2D3A |
SH2 domain containing 3A |
| chr14_+_61789382 | 0.71 |
ENST00000555082.1 |
PRKCH |
protein kinase C, eta |
| chr1_+_205473720 | 0.63 |
ENST00000429964.2 ENST00000506784.1 ENST00000360066.2 |
CDK18 |
cyclin-dependent kinase 18 |
| chr1_+_60280458 | 0.63 |
ENST00000455990.1 ENST00000371208.3 |
HOOK1 |
hook microtubule-tethering protein 1 |
| chr3_-_112218205 | 0.58 |
ENST00000383680.4 |
BTLA |
B and T lymphocyte associated |
| chr12_-_53298841 | 0.57 |
ENST00000293308.6 |
KRT8 |
keratin 8 |
| chr11_-_119993734 | 0.57 |
ENST00000533302.1 |
TRIM29 |
tripartite motif containing 29 |
| chr10_+_47746929 | 0.57 |
ENST00000340243.6 ENST00000374277.5 ENST00000449464.2 ENST00000538825.1 ENST00000335083.5 |
ANXA8L2 AL603965.1 |
annexin A8-like 2 Protein LOC100996760 |
| chr11_+_128563652 | 0.57 |
ENST00000527786.2 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chrX_+_105937068 | 0.55 |
ENST00000324342.3 |
RNF128 |
ring finger protein 128, E3 ubiquitin protein ligase |
| chr17_-_46507537 | 0.54 |
ENST00000336915.6 |
SKAP1 |
src kinase associated phosphoprotein 1 |
| chr9_-_23821842 | 0.54 |
ENST00000544538.1 |
ELAVL2 |
ELAV like neuron-specific RNA binding protein 2 |
| chr1_+_109792641 | 0.54 |
ENST00000271332.3 |
CELSR2 |
cadherin, EGF LAG seven-pass G-type receptor 2 |
| chr2_-_70780770 | 0.53 |
ENST00000444975.1 ENST00000445399.1 ENST00000418333.2 |
TGFA |
transforming growth factor, alpha |
| chr16_+_57406368 | 0.49 |
ENST00000006053.6 ENST00000563383.1 |
CX3CL1 |
chemokine (C-X3-C motif) ligand 1 |
| chr7_+_18535786 | 0.48 |
ENST00000406072.1 |
HDAC9 |
histone deacetylase 9 |
| chr2_-_70780572 | 0.45 |
ENST00000450929.1 |
TGFA |
transforming growth factor, alpha |
| chr18_+_55816546 | 0.43 |
ENST00000435432.2 ENST00000357895.5 ENST00000586263.1 |
NEDD4L |
neural precursor cell expressed, developmentally down-regulated 4-like, E3 ubiquitin protein ligase |
| chr6_-_32784687 | 0.43 |
ENST00000447394.1 ENST00000438763.2 |
HLA-DOB |
major histocompatibility complex, class II, DO beta |
| chr10_-_47173994 | 0.40 |
ENST00000414655.2 ENST00000545298.1 ENST00000359178.4 ENST00000358140.4 ENST00000503031.1 |
ANXA8L1 LINC00842 |
annexin A8-like 1 long intergenic non-protein coding RNA 842 |
| chr5_+_68710906 | 0.39 |
ENST00000325631.5 ENST00000454295.2 |
MARVELD2 |
MARVEL domain containing 2 |
| chr5_+_68711209 | 0.39 |
ENST00000512803.1 |
MARVELD2 |
MARVEL domain containing 2 |
| chr8_-_95274536 | 0.38 |
ENST00000297596.2 ENST00000396194.2 |
GEM |
GTP binding protein overexpressed in skeletal muscle |
| chr1_-_153013588 | 0.36 |
ENST00000360379.3 |
SPRR2D |
small proline-rich protein 2D |
| chrX_-_31285042 | 0.35 |
ENST00000378680.2 ENST00000378723.3 |
DMD |
dystrophin |
| chr12_-_50616122 | 0.35 |
ENST00000552823.1 ENST00000552909.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr1_-_24469602 | 0.35 |
ENST00000270800.1 |
IL22RA1 |
interleukin 22 receptor, alpha 1 |
| chr2_-_220436248 | 0.34 |
ENST00000265318.4 |
OBSL1 |
obscurin-like 1 |
| chr17_-_39928106 | 0.33 |
ENST00000540235.1 |
JUP |
junction plakoglobin |
| chr19_-_38747172 | 0.33 |
ENST00000347262.4 ENST00000591585.1 ENST00000301242.4 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr2_+_29336855 | 0.33 |
ENST00000404424.1 |
CLIP4 |
CAP-GLY domain containing linker protein family, member 4 |
| chr3_+_125694347 | 0.32 |
ENST00000505382.1 ENST00000511082.1 |
ROPN1B |
rhophilin associated tail protein 1B |
| chr5_-_35048047 | 0.31 |
ENST00000231420.6 |
AGXT2 |
alanine--glyoxylate aminotransferase 2 |
| chr9_-_124976185 | 0.31 |
ENST00000464484.2 |
LHX6 |
LIM homeobox 6 |
| chr3_+_50192537 | 0.31 |
ENST00000002829.3 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr14_-_21567009 | 0.31 |
ENST00000556174.1 ENST00000554478.1 ENST00000553980.1 ENST00000421093.2 |
ZNF219 |
zinc finger protein 219 |
| chr9_-_124976154 | 0.29 |
ENST00000482062.1 |
LHX6 |
LIM homeobox 6 |
| chr3_+_50192499 | 0.29 |
ENST00000413852.1 |
SEMA3F |
sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3F |
| chr6_-_116381918 | 0.28 |
ENST00000606080.1 |
FRK |
fyn-related kinase |
| chr12_+_57984965 | 0.28 |
ENST00000540759.2 ENST00000551772.1 ENST00000550465.1 ENST00000354947.5 |
PIP4K2C |
phosphatidylinositol-5-phosphate 4-kinase, type II, gamma |
| chrX_-_15683147 | 0.28 |
ENST00000380342.3 |
TMEM27 |
transmembrane protein 27 |
| chr12_+_56473939 | 0.28 |
ENST00000450146.2 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr5_+_147258266 | 0.28 |
ENST00000296694.4 |
SCGB3A2 |
secretoglobin, family 3A, member 2 |
| chrX_-_31285018 | 0.27 |
ENST00000361471.4 |
DMD |
dystrophin |
| chr6_+_18155560 | 0.27 |
ENST00000546309.2 ENST00000388870.2 ENST00000397244.1 |
KDM1B |
lysine (K)-specific demethylase 1B |
| chrX_-_31284974 | 0.27 |
ENST00000378702.4 |
DMD |
dystrophin |
| chr6_-_47010061 | 0.26 |
ENST00000371253.2 |
GPR110 |
G protein-coupled receptor 110 |
| chr3_-_116163830 | 0.26 |
ENST00000333617.4 |
LSAMP |
limbic system-associated membrane protein |
| chr14_-_67859422 | 0.25 |
ENST00000556532.1 |
PLEK2 |
pleckstrin 2 |
| chr1_-_216978709 | 0.24 |
ENST00000360012.3 |
ESRRG |
estrogen-related receptor gamma |
| chr17_+_53828381 | 0.24 |
ENST00000576183.1 |
PCTP |
phosphatidylcholine transfer protein |
| chr1_-_217262969 | 0.24 |
ENST00000361525.3 |
ESRRG |
estrogen-related receptor gamma |
| chr1_+_158223923 | 0.24 |
ENST00000289429.5 |
CD1A |
CD1a molecule |
| chr10_-_131762105 | 0.24 |
ENST00000368648.3 ENST00000355311.5 |
EBF3 |
early B-cell factor 3 |
| chr22_+_38453207 | 0.24 |
ENST00000404072.3 ENST00000424694.1 |
PICK1 |
protein interacting with PRKCA 1 |
| chr1_+_93544791 | 0.24 |
ENST00000545708.1 ENST00000540243.1 ENST00000370298.4 |
MTF2 |
metal response element binding transcription factor 2 |
| chr17_+_55173933 | 0.23 |
ENST00000539273.1 |
AKAP1 |
A kinase (PRKA) anchor protein 1 |
| chr16_-_86532148 | 0.23 |
ENST00000594398.1 |
FENDRR |
FOXF1 adjacent non-coding developmental regulatory RNA |
| chr11_-_19223523 | 0.22 |
ENST00000265968.3 |
CSRP3 |
cysteine and glycine-rich protein 3 (cardiac LIM protein) |
| chr1_+_173793641 | 0.22 |
ENST00000361951.4 |
DARS2 |
aspartyl-tRNA synthetase 2, mitochondrial |
| chr4_+_156587979 | 0.22 |
ENST00000511507.1 |
GUCY1A3 |
guanylate cyclase 1, soluble, alpha 3 |
| chr19_+_1026566 | 0.22 |
ENST00000348419.3 ENST00000565096.2 ENST00000562958.2 ENST00000562075.2 ENST00000607102.1 |
CNN2 |
calponin 2 |
| chr12_-_50616382 | 0.22 |
ENST00000552783.1 |
LIMA1 |
LIM domain and actin binding 1 |
| chr2_-_220435963 | 0.22 |
ENST00000373876.1 ENST00000404537.1 ENST00000603926.1 ENST00000373873.4 ENST00000289656.3 |
OBSL1 |
obscurin-like 1 |
| chr1_-_217262933 | 0.21 |
ENST00000359162.2 |
ESRRG |
estrogen-related receptor gamma |
| chr1_+_173793777 | 0.21 |
ENST00000239457.5 |
DARS2 |
aspartyl-tRNA synthetase 2, mitochondrial |
| chr2_-_208489275 | 0.20 |
ENST00000272839.3 ENST00000426075.1 |
METTL21A |
methyltransferase like 21A |
| chr15_+_52155001 | 0.20 |
ENST00000544199.1 |
TMOD3 |
tropomodulin 3 (ubiquitous) |
| chr1_+_149230680 | 0.20 |
ENST00000443018.1 |
RP11-403I13.5 |
RP11-403I13.5 |
| chr3_-_112693865 | 0.20 |
ENST00000471858.1 ENST00000295863.4 ENST00000308611.3 |
CD200R1 |
CD200 receptor 1 |
| chr9_-_131940526 | 0.20 |
ENST00000372491.2 |
IER5L |
immediate early response 5-like |
| chr1_+_151739131 | 0.20 |
ENST00000400999.1 |
OAZ3 |
ornithine decarboxylase antizyme 3 |
| chr3_+_193853927 | 0.20 |
ENST00000232424.3 |
HES1 |
hes family bHLH transcription factor 1 |
| chr5_+_56111361 | 0.19 |
ENST00000399503.3 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chrX_+_13707235 | 0.19 |
ENST00000464506.1 |
RAB9A |
RAB9A, member RAS oncogene family |
| chr2_+_96931834 | 0.19 |
ENST00000488633.1 |
CIAO1 |
cytosolic iron-sulfur protein assembly 1 |
| chr3_-_112218378 | 0.19 |
ENST00000334529.5 |
BTLA |
B and T lymphocyte associated |
| chr6_+_32821924 | 0.19 |
ENST00000374859.2 ENST00000453265.2 |
PSMB9 |
proteasome (prosome, macropain) subunit, beta type, 9 |
| chr6_+_24357131 | 0.19 |
ENST00000274766.1 |
KAAG1 |
kidney associated antigen 1 |
| chr9_+_124329336 | 0.19 |
ENST00000394340.3 ENST00000436835.1 ENST00000259371.2 |
DAB2IP |
DAB2 interacting protein |
| chr17_-_19771216 | 0.19 |
ENST00000395544.4 |
ULK2 |
unc-51 like autophagy activating kinase 2 |
| chr8_-_27115931 | 0.18 |
ENST00000523048.1 |
STMN4 |
stathmin-like 4 |
| chr17_-_43487741 | 0.18 |
ENST00000455881.1 |
ARHGAP27 |
Rho GTPase activating protein 27 |
| chrX_-_119445306 | 0.18 |
ENST00000371369.4 ENST00000440464.1 ENST00000519908.1 |
TMEM255A |
transmembrane protein 255A |
| chr4_+_144257915 | 0.18 |
ENST00000262995.4 |
GAB1 |
GRB2-associated binding protein 1 |
| chrX_-_119445263 | 0.18 |
ENST00000309720.5 |
TMEM255A |
transmembrane protein 255A |
| chr17_-_43487780 | 0.18 |
ENST00000532038.1 ENST00000528677.1 |
ARHGAP27 |
Rho GTPase activating protein 27 |
| chr12_+_6309517 | 0.18 |
ENST00000382519.4 ENST00000009180.4 |
CD9 |
CD9 molecule |
| chr17_+_53828333 | 0.18 |
ENST00000268896.5 |
PCTP |
phosphatidylcholine transfer protein |
| chr2_+_98703643 | 0.17 |
ENST00000477737.1 |
VWA3B |
von Willebrand factor A domain containing 3B |
| chr7_+_129906660 | 0.17 |
ENST00000222481.4 |
CPA2 |
carboxypeptidase A2 (pancreatic) |
| chr3_-_48481518 | 0.17 |
ENST00000412398.2 ENST00000395696.1 |
CCDC51 |
coiled-coil domain containing 51 |
| chr2_+_145780725 | 0.17 |
ENST00000451478.1 |
TEX41 |
testis expressed 41 (non-protein coding) |
| chr1_-_149814478 | 0.17 |
ENST00000369161.3 |
HIST2H2AA3 |
histone cluster 2, H2aa3 |
| chr4_+_123300591 | 0.17 |
ENST00000439307.1 ENST00000388724.2 |
ADAD1 |
adenosine deaminase domain containing 1 (testis-specific) |
| chr15_-_51535208 | 0.16 |
ENST00000405913.3 ENST00000559878.1 |
CYP19A1 |
cytochrome P450, family 19, subfamily A, polypeptide 1 |
| chr1_+_149822620 | 0.16 |
ENST00000369159.2 |
HIST2H2AA4 |
histone cluster 2, H2aa4 |
| chr1_+_116519112 | 0.16 |
ENST00000369503.4 |
SLC22A15 |
solute carrier family 22, member 15 |
| chrX_+_38420623 | 0.16 |
ENST00000378482.2 |
TSPAN7 |
tetraspanin 7 |
| chr12_+_6494285 | 0.16 |
ENST00000541102.1 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr9_-_34458531 | 0.16 |
ENST00000379089.1 ENST00000379087.1 ENST00000379084.1 ENST00000379081.1 ENST00000379080.1 ENST00000422409.1 ENST00000379078.1 ENST00000445726.1 ENST00000297620.4 |
FAM219A |
family with sequence similarity 219, member A |
| chr6_-_39290316 | 0.16 |
ENST00000425054.2 ENST00000373227.4 ENST00000373229.5 ENST00000437525.2 |
KCNK16 |
potassium channel, subfamily K, member 16 |
| chr19_-_18548921 | 0.16 |
ENST00000545187.1 ENST00000578352.1 |
ISYNA1 |
inositol-3-phosphate synthase 1 |
| chr6_-_47009996 | 0.15 |
ENST00000371243.2 |
GPR110 |
G protein-coupled receptor 110 |
| chr5_-_115872142 | 0.15 |
ENST00000510263.1 |
SEMA6A |
sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6A |
| chr1_-_235098935 | 0.15 |
ENST00000423175.1 |
RP11-443B7.1 |
RP11-443B7.1 |
| chr4_+_144303093 | 0.15 |
ENST00000505913.1 |
GAB1 |
GRB2-associated binding protein 1 |
| chr3_-_52443799 | 0.15 |
ENST00000470173.1 ENST00000296288.5 |
BAP1 |
BRCA1 associated protein-1 (ubiquitin carboxy-terminal hydrolase) |
| chr9_-_34397800 | 0.15 |
ENST00000297623.2 |
C9orf24 |
chromosome 9 open reading frame 24 |
| chr5_+_34915444 | 0.15 |
ENST00000336767.5 |
BRIX1 |
BRX1, biogenesis of ribosomes, homolog (S. cerevisiae) |
| chr11_+_34642656 | 0.15 |
ENST00000257831.3 ENST00000450654.2 |
EHF |
ets homologous factor |
| chr1_+_93544821 | 0.14 |
ENST00000370303.4 |
MTF2 |
metal response element binding transcription factor 2 |
| chr22_+_38453378 | 0.14 |
ENST00000437453.1 ENST00000356976.3 |
PICK1 |
protein interacting with PRKCA 1 |
| chr3_+_23847432 | 0.14 |
ENST00000346855.3 |
UBE2E1 |
ubiquitin-conjugating enzyme E2E 1 |
| chr20_+_42984330 | 0.14 |
ENST00000316673.4 ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A |
hepatocyte nuclear factor 4, alpha |
| chr9_+_116263778 | 0.14 |
ENST00000394646.3 |
RGS3 |
regulator of G-protein signaling 3 |
| chrX_+_44732757 | 0.13 |
ENST00000377967.4 ENST00000536777.1 ENST00000382899.4 ENST00000543216.1 |
KDM6A |
lysine (K)-specific demethylase 6A |
| chr6_+_125304502 | 0.13 |
ENST00000519799.1 ENST00000368414.2 ENST00000359704.2 |
RNF217 |
ring finger protein 217 |
| chr5_-_59481406 | 0.13 |
ENST00000546160.1 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr1_-_156399184 | 0.13 |
ENST00000368243.1 ENST00000357975.4 ENST00000310027.5 ENST00000400991.2 |
C1orf61 |
chromosome 1 open reading frame 61 |
| chr8_+_144295067 | 0.13 |
ENST00000330824.2 |
GPIHBP1 |
glycosylphosphatidylinositol anchored high density lipoprotein binding protein 1 |
| chr2_+_201754050 | 0.13 |
ENST00000426253.1 ENST00000416651.1 ENST00000454952.1 ENST00000409020.1 ENST00000359683.4 |
NIF3L1 |
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr8_+_104033296 | 0.13 |
ENST00000521514.1 ENST00000518738.1 |
ATP6V1C1 |
ATPase, H+ transporting, lysosomal 42kDa, V1 subunit C1 |
| chr14_+_104604916 | 0.13 |
ENST00000423312.2 |
KIF26A |
kinesin family member 26A |
| chr6_+_31082603 | 0.13 |
ENST00000259881.9 |
PSORS1C1 |
psoriasis susceptibility 1 candidate 1 |
| chr19_+_48867652 | 0.13 |
ENST00000344846.2 |
SYNGR4 |
synaptogyrin 4 |
| chr3_+_111697843 | 0.13 |
ENST00000534857.1 ENST00000273359.3 ENST00000494817.1 |
ABHD10 |
abhydrolase domain containing 10 |
| chr19_-_10628117 | 0.13 |
ENST00000333430.4 |
S1PR5 |
sphingosine-1-phosphate receptor 5 |
| chr13_+_43355732 | 0.13 |
ENST00000313851.1 |
FAM216B |
family with sequence similarity 216, member B |
| chr4_+_144258021 | 0.13 |
ENST00000262994.4 |
GAB1 |
GRB2-associated binding protein 1 |
| chr2_+_48541776 | 0.12 |
ENST00000413569.1 ENST00000340553.3 |
FOXN2 |
forkhead box N2 |
| chr2_+_201754135 | 0.12 |
ENST00000409357.1 ENST00000409129.2 |
NIF3L1 |
NIF3 NGG1 interacting factor 3-like 1 (S. cerevisiae) |
| chr19_-_10444188 | 0.12 |
ENST00000293677.6 |
RAVER1 |
ribonucleoprotein, PTB-binding 1 |
| chr17_+_40714092 | 0.12 |
ENST00000420359.1 ENST00000449624.1 ENST00000585811.1 ENST00000585909.1 ENST00000586771.1 ENST00000421097.2 ENST00000591779.1 ENST00000587858.1 ENST00000587214.1 ENST00000587157.1 ENST00000590958.1 ENST00000393818.2 |
COASY |
CoA synthase |
| chrX_+_76709648 | 0.12 |
ENST00000439435.1 |
FGF16 |
fibroblast growth factor 16 |
| chr12_+_6309963 | 0.12 |
ENST00000382515.2 |
CD9 |
CD9 molecule |
| chr16_-_2318055 | 0.12 |
ENST00000561518.1 ENST00000561718.1 ENST00000567147.1 ENST00000562690.1 ENST00000569598.2 |
RNPS1 |
RNA binding protein S1, serine-rich domain |
| chr7_+_39125365 | 0.11 |
ENST00000559001.1 ENST00000464276.2 |
POU6F2 |
POU class 6 homeobox 2 |
| chr1_+_95583479 | 0.11 |
ENST00000455656.1 ENST00000604534.1 |
TMEM56 RP11-57H12.6 |
transmembrane protein 56 TMEM56-RWDD3 readthrough |
| chr17_-_10372875 | 0.11 |
ENST00000255381.2 |
MYH4 |
myosin, heavy chain 4, skeletal muscle |
| chr13_+_43355683 | 0.11 |
ENST00000537894.1 |
FAM216B |
family with sequence similarity 216, member B |
| chr5_+_79331164 | 0.11 |
ENST00000350881.2 |
THBS4 |
thrombospondin 4 |
| chr20_-_634000 | 0.11 |
ENST00000381962.3 |
SRXN1 |
sulfiredoxin 1 |
| chr6_-_32557610 | 0.11 |
ENST00000360004.5 |
HLA-DRB1 |
major histocompatibility complex, class II, DR beta 1 |
| chr11_+_64126614 | 0.11 |
ENST00000528057.1 ENST00000334205.4 ENST00000294261.4 |
RPS6KA4 |
ribosomal protein S6 kinase, 90kDa, polypeptide 4 |
| chr17_-_79623597 | 0.11 |
ENST00000574024.1 ENST00000331056.5 |
PDE6G |
phosphodiesterase 6G, cGMP-specific, rod, gamma |
| chr13_-_47012325 | 0.11 |
ENST00000409879.2 |
KIAA0226L |
KIAA0226-like |
| chr7_+_115862858 | 0.11 |
ENST00000393481.2 |
TES |
testis derived transcript (3 LIM domains) |
| chr17_-_17184605 | 0.11 |
ENST00000268717.5 |
COPS3 |
COP9 signalosome subunit 3 |
| chr19_+_1026298 | 0.11 |
ENST00000263097.4 |
CNN2 |
calponin 2 |
| chr15_-_40401062 | 0.11 |
ENST00000354670.4 ENST00000559701.1 ENST00000557870.1 ENST00000558774.1 |
BMF |
Bcl2 modifying factor |
| chrX_-_44402231 | 0.10 |
ENST00000378045.4 |
FUNDC1 |
FUN14 domain containing 1 |
| chr2_+_196521458 | 0.10 |
ENST00000409086.3 |
SLC39A10 |
solute carrier family 39 (zinc transporter), member 10 |
| chr10_-_121296045 | 0.10 |
ENST00000392865.1 |
RGS10 |
regulator of G-protein signaling 10 |
| chr17_+_68071389 | 0.10 |
ENST00000283936.1 ENST00000392671.1 |
KCNJ16 |
potassium inwardly-rectifying channel, subfamily J, member 16 |
| chr1_-_217250231 | 0.10 |
ENST00000493748.1 ENST00000463665.1 |
ESRRG |
estrogen-related receptor gamma |
| chr12_-_113658892 | 0.10 |
ENST00000299732.2 ENST00000416617.2 |
IQCD |
IQ motif containing D |
| chr7_+_129007964 | 0.10 |
ENST00000460109.1 ENST00000474594.1 ENST00000446212.1 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
| chr20_+_2276639 | 0.10 |
ENST00000381458.5 |
TGM3 |
transglutaminase 3 |
| chr12_+_70219052 | 0.10 |
ENST00000552032.2 ENST00000547771.2 |
MYRFL |
myelin regulatory factor-like |
| chr10_-_99531709 | 0.10 |
ENST00000266066.3 |
SFRP5 |
secreted frizzled-related protein 5 |
| chr9_-_114521783 | 0.10 |
ENST00000394779.3 ENST00000394777.4 |
C9orf84 |
chromosome 9 open reading frame 84 |
| chr7_-_121944491 | 0.10 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr1_+_152974218 | 0.10 |
ENST00000331860.3 ENST00000443178.1 ENST00000295367.4 |
SPRR3 |
small proline-rich protein 3 |
| chr16_-_46723066 | 0.10 |
ENST00000299138.7 |
VPS35 |
vacuolar protein sorting 35 homolog (S. cerevisiae) |
| chr1_-_31769656 | 0.10 |
ENST00000446633.2 |
SNRNP40 |
small nuclear ribonucleoprotein 40kDa (U5) |
| chr1_-_44482979 | 0.10 |
ENST00000360584.2 ENST00000357730.2 ENST00000528803.1 |
SLC6A9 |
solute carrier family 6 (neurotransmitter transporter, glycine), member 9 |
| chr20_+_30063067 | 0.10 |
ENST00000201979.2 |
REM1 |
RAS (RAD and GEM)-like GTP-binding 1 |
| chr6_+_50681541 | 0.09 |
ENST00000008391.3 |
TFAP2D |
transcription factor AP-2 delta (activating enhancer binding protein 2 delta) |
| chr1_-_31769595 | 0.09 |
ENST00000263694.4 |
SNRNP40 |
small nuclear ribonucleoprotein 40kDa (U5) |
| chr1_+_119957554 | 0.09 |
ENST00000543831.1 ENST00000433745.1 ENST00000369416.3 |
HSD3B2 |
hydroxy-delta-5-steroid dehydrogenase, 3 beta- and steroid delta-isomerase 2 |
| chrX_-_131262048 | 0.09 |
ENST00000298542.4 |
FRMD7 |
FERM domain containing 7 |
| chr9_+_116263639 | 0.09 |
ENST00000343817.5 |
RGS3 |
regulator of G-protein signaling 3 |
| chr15_+_88120158 | 0.09 |
ENST00000560153.1 |
LINC00052 |
long intergenic non-protein coding RNA 52 |
| chr9_+_125281420 | 0.09 |
ENST00000340750.1 |
OR1J4 |
olfactory receptor, family 1, subfamily J, member 4 |
| chr17_-_7297519 | 0.09 |
ENST00000576362.1 ENST00000571078.1 |
TMEM256-PLSCR3 |
TMEM256-PLSCR3 readthrough (NMD candidate) |
| chr18_+_28898052 | 0.09 |
ENST00000257192.4 |
DSG1 |
desmoglein 1 |
| chr18_+_59000815 | 0.09 |
ENST00000262717.4 |
CDH20 |
cadherin 20, type 2 |
| chr7_-_135412925 | 0.09 |
ENST00000354042.4 |
SLC13A4 |
solute carrier family 13 (sodium/sulfate symporter), member 4 |
| chr14_+_39583427 | 0.09 |
ENST00000308317.6 ENST00000396249.2 ENST00000250379.8 ENST00000534684.2 ENST00000527381.1 |
GEMIN2 |
gem (nuclear organelle) associated protein 2 |
| chr6_+_5261730 | 0.09 |
ENST00000274680.4 |
FARS2 |
phenylalanyl-tRNA synthetase 2, mitochondrial |
| chr9_-_112970436 | 0.09 |
ENST00000400613.4 |
C9orf152 |
chromosome 9 open reading frame 152 |
| chr8_-_49834299 | 0.09 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr4_+_88529681 | 0.09 |
ENST00000399271.1 |
DSPP |
dentin sialophosphoprotein |
| chr7_+_29237354 | 0.09 |
ENST00000546235.1 |
CHN2 |
chimerin 2 |
| chr2_-_25016251 | 0.09 |
ENST00000328379.5 |
PTRHD1 |
peptidyl-tRNA hydrolase domain containing 1 |
| chr19_+_17337473 | 0.09 |
ENST00000598068.1 |
OCEL1 |
occludin/ELL domain containing 1 |
| chr20_+_52824367 | 0.08 |
ENST00000371419.2 |
PFDN4 |
prefoldin subunit 4 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 1.0 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.3 | 0.9 | GO:0042938 | dipeptide transport(GO:0042938) |
| 0.2 | 0.5 | GO:0050902 | leukocyte adhesive activation(GO:0050902) |
| 0.1 | 0.4 | GO:0006421 | asparaginyl-tRNA aminoacylation(GO:0006421) |
| 0.1 | 0.4 | GO:0002581 | negative regulation of antigen processing and presentation of peptide or polysaccharide antigen via MHC class II(GO:0002581) |
| 0.1 | 0.5 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.1 | 0.3 | GO:0009436 | glyoxylate catabolic process(GO:0009436) |
| 0.1 | 1.7 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.1 | 0.3 | GO:0042668 | auditory receptor cell fate determination(GO:0042668) negative regulation of pro-B cell differentiation(GO:2000974) |
| 0.1 | 0.4 | GO:0071603 | endothelial cell-cell adhesion(GO:0071603) |
| 0.1 | 0.4 | GO:2001288 | positive regulation of caveolin-mediated endocytosis(GO:2001288) |
| 0.1 | 0.9 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.1 | 0.6 | GO:0035290 | trunk segmentation(GO:0035290) trunk neural crest cell migration(GO:0036484) ventral trunk neural crest cell migration(GO:0036486) |
| 0.1 | 0.7 | GO:0034351 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 0.1 | GO:0071878 | negative regulation of adrenergic receptor signaling pathway(GO:0071878) regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901204) negative regulation of adrenergic receptor signaling pathway involved in heart process(GO:1901205) |
| 0.1 | 0.4 | GO:0061086 | negative regulation of histone H3-K27 methylation(GO:0061086) |
| 0.1 | 0.2 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.6 | GO:0021800 | cerebral cortex tangential migration(GO:0021800) |
| 0.1 | 0.2 | GO:2000863 | positive regulation of estrogen secretion(GO:2000863) positive regulation of estradiol secretion(GO:2000866) |
| 0.1 | 0.5 | GO:0021999 | neural plate anterior/posterior regionalization(GO:0021999) |
| 0.1 | 0.4 | GO:0034316 | negative regulation of Arp2/3 complex-mediated actin nucleation(GO:0034316) |
| 0.0 | 0.5 | GO:1904352 | positive regulation of protein catabolic process in the vacuole(GO:1904352) |
| 0.0 | 0.1 | GO:0071557 | histone H3-K27 demethylation(GO:0071557) |
| 0.0 | 0.2 | GO:0035995 | detection of muscle stretch(GO:0035995) |
| 0.0 | 0.3 | GO:0052565 | response to defense-related nitric oxide production by other organism involved in symbiotic interaction(GO:0052551) response to defense-related host nitric oxide production(GO:0052565) |
| 0.0 | 0.1 | GO:0010986 | positive regulation of lipoprotein particle clearance(GO:0010986) |
| 0.0 | 0.1 | GO:0001560 | regulation of cell growth by extracellular stimulus(GO:0001560) |
| 0.0 | 0.6 | GO:0051599 | response to hydrostatic pressure(GO:0051599) |
| 0.0 | 0.2 | GO:1902268 | negative regulation of polyamine transmembrane transport(GO:1902268) |
| 0.0 | 0.5 | GO:0090050 | positive regulation of cell migration involved in sprouting angiogenesis(GO:0090050) |
| 0.0 | 0.2 | GO:0036324 | vascular endothelial growth factor receptor-2 signaling pathway(GO:0036324) |
| 0.0 | 0.1 | GO:0035522 | monoubiquitinated histone deubiquitination(GO:0035521) monoubiquitinated histone H2A deubiquitination(GO:0035522) |
| 0.0 | 0.9 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.1 | GO:0043163 | cell envelope organization(GO:0043163) external encapsulating structure organization(GO:0045229) |
| 0.0 | 0.1 | GO:1903181 | regulation of dopamine biosynthetic process(GO:1903179) positive regulation of dopamine biosynthetic process(GO:1903181) negative regulation of protein catabolic process in the vacuole(GO:1904351) negative regulation of lysosomal protein catabolic process(GO:1905166) |
| 0.0 | 0.1 | GO:0061536 | glycine secretion(GO:0061536) glycine secretion, neurotransmission(GO:0061537) |
| 0.0 | 0.3 | GO:0034720 | histone H3-K4 demethylation(GO:0034720) |
| 0.0 | 0.4 | GO:0071688 | striated muscle myosin thick filament assembly(GO:0071688) |
| 0.0 | 0.6 | GO:0007342 | fusion of sperm to egg plasma membrane(GO:0007342) |
| 0.0 | 0.1 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.0 | 0.3 | GO:0031340 | positive regulation of vesicle fusion(GO:0031340) |
| 0.0 | 0.2 | GO:0052405 | negative regulation by host of symbiont molecular function(GO:0052405) |
| 0.0 | 0.1 | GO:1901876 | regulation of calcium ion binding(GO:1901876) negative regulation of calcium ion binding(GO:1901877) |
| 0.0 | 0.2 | GO:0075044 | autophagy of host cells involved in interaction with symbiont(GO:0075044) autophagy involved in symbiotic interaction(GO:0075071) |
| 0.0 | 0.2 | GO:0006021 | inositol biosynthetic process(GO:0006021) |
| 0.0 | 0.3 | GO:0048003 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 1.0 | GO:0035774 | positive regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0035774) |
| 0.0 | 0.1 | GO:0002399 | MHC class II protein complex assembly(GO:0002399) |
| 0.0 | 0.1 | GO:0043988 | histone H3-S28 phosphorylation(GO:0043988) |
| 0.0 | 0.1 | GO:0000117 | regulation of transcription involved in G2/M transition of mitotic cell cycle(GO:0000117) |
| 0.0 | 0.2 | GO:0051694 | pointed-end actin filament capping(GO:0051694) |
| 0.0 | 0.1 | GO:0021797 | forebrain anterior/posterior pattern specification(GO:0021797) |
| 0.0 | 0.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.2 | GO:0007039 | protein catabolic process in the vacuole(GO:0007039) |
| 0.0 | 0.3 | GO:2000786 | positive regulation of autophagosome assembly(GO:2000786) |
| 0.0 | 0.1 | GO:0016139 | glycoside catabolic process(GO:0016139) |
| 0.0 | 0.1 | GO:0035928 | rRNA import into mitochondrion(GO:0035928) |
| 0.0 | 0.1 | GO:0070346 | positive regulation of fat cell proliferation(GO:0070346) positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.1 | GO:1901594 | detection of temperature stimulus involved in thermoception(GO:0050960) response to capsazepine(GO:1901594) |
| 0.0 | 0.1 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0046967 | cytosol to ER transport(GO:0046967) |
| 0.0 | 0.2 | GO:1901898 | negative regulation of relaxation of cardiac muscle(GO:1901898) |
| 0.0 | 0.0 | GO:0052314 | phytoalexin metabolic process(GO:0052314) |
| 0.0 | 0.1 | GO:0034344 | type III interferon production(GO:0034343) regulation of type III interferon production(GO:0034344) |
| 0.0 | 0.1 | GO:0008272 | sulfate transport(GO:0008272) |
| 0.0 | 0.2 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.3 | GO:0035641 | locomotory exploration behavior(GO:0035641) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 2.2 | GO:0030183 | B cell differentiation(GO:0030183) |
| 0.0 | 0.1 | GO:0032020 | ISG15-protein conjugation(GO:0032020) |
| 0.0 | 0.1 | GO:0042986 | positive regulation of amyloid precursor protein biosynthetic process(GO:0042986) |
| 0.0 | 0.5 | GO:0035728 | response to hepatocyte growth factor(GO:0035728) |
| 0.0 | 0.1 | GO:0060024 | rhythmic synaptic transmission(GO:0060024) |
| 0.0 | 0.0 | GO:0048690 | modulation by virus of host transcription(GO:0019056) regulation of sprouting of injured axon(GO:0048686) positive regulation of sprouting of injured axon(GO:0048687) regulation of axon extension involved in regeneration(GO:0048690) positive regulation of axon extension involved in regeneration(GO:0048691) modulation by symbiont of host transcription(GO:0052026) |
| 0.0 | 0.1 | GO:0007023 | post-chaperonin tubulin folding pathway(GO:0007023) |
| 0.0 | 0.1 | GO:0071163 | DNA replication preinitiation complex assembly(GO:0071163) |
| 0.0 | 0.8 | GO:0061028 | establishment of endothelial barrier(GO:0061028) |
| 0.0 | 0.1 | GO:2000623 | regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000622) negative regulation of nuclear-transcribed mRNA catabolic process, nonsense-mediated decay(GO:2000623) |
| 0.0 | 0.1 | GO:0006432 | phenylalanyl-tRNA aminoacylation(GO:0006432) |
| 0.0 | 0.6 | GO:0045022 | early endosome to late endosome transport(GO:0045022) |
| 0.0 | 0.1 | GO:0038172 | interleukin-33-mediated signaling pathway(GO:0038172) |
| 0.0 | 0.1 | GO:0090625 | mRNA cleavage involved in gene silencing by siRNA(GO:0090625) |
| 0.0 | 0.1 | GO:0001934 | positive regulation of protein phosphorylation(GO:0001934) |
| 0.0 | 0.0 | GO:0046684 | response to pyrethroid(GO:0046684) |
| 0.0 | 0.1 | GO:2000253 | positive regulation of feeding behavior(GO:2000253) |
| 0.0 | 0.1 | GO:0036444 | calcium ion transmembrane import into mitochondrion(GO:0036444) |
| 0.0 | 1.2 | GO:1900181 | negative regulation of protein localization to nucleus(GO:1900181) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.9 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 0.6 | GO:0070695 | FHF complex(GO:0070695) |
| 0.1 | 0.9 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.1 | 0.8 | GO:0061689 | tricellular tight junction(GO:0061689) |
| 0.1 | 0.6 | GO:1990393 | 3M complex(GO:1990393) |
| 0.1 | 0.2 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 0.3 | GO:0071664 | catenin-TCF7L2 complex(GO:0071664) |
| 0.0 | 0.2 | GO:0000221 | vacuolar proton-transporting V-type ATPase, V1 domain(GO:0000221) |
| 0.0 | 0.2 | GO:0071817 | MMXD complex(GO:0071817) |
| 0.0 | 0.6 | GO:0042613 | MHC class II protein complex(GO:0042613) |
| 0.0 | 0.5 | GO:0005922 | connexon complex(GO:0005922) |
| 0.0 | 0.6 | GO:0016010 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 1.6 | GO:0012507 | ER to Golgi transport vesicle membrane(GO:0012507) |
| 0.0 | 0.2 | GO:1990111 | spermatoproteasome complex(GO:1990111) |
| 0.0 | 0.1 | GO:0030906 | retromer, cargo-selective complex(GO:0030906) |
| 0.0 | 1.0 | GO:0009925 | basal plasma membrane(GO:0009925) |
| 0.0 | 0.5 | GO:0042101 | T cell receptor complex(GO:0042101) |
| 0.0 | 0.1 | GO:1990425 | ryanodine receptor complex(GO:1990425) |
| 0.0 | 0.1 | GO:0061574 | ASAP complex(GO:0061574) |
| 0.0 | 0.1 | GO:0030868 | smooth endoplasmic reticulum membrane(GO:0030868) smooth endoplasmic reticulum part(GO:0097425) |
| 0.0 | 0.0 | GO:0097629 | extrinsic component of omegasome membrane(GO:0097629) |
| 0.0 | 0.0 | GO:0042721 | mitochondrial inner membrane protein insertion complex(GO:0042721) |
| 0.0 | 0.4 | GO:0035098 | ESC/E(Z) complex(GO:0035098) |
| 0.0 | 0.6 | GO:0001533 | cornified envelope(GO:0001533) |
| 0.0 | 0.1 | GO:0016272 | prefoldin complex(GO:0016272) |
| 0.0 | 1.3 | GO:0035578 | azurophil granule lumen(GO:0035578) |
| 0.0 | 1.4 | GO:0005913 | cell-cell adherens junction(GO:0005913) |
| 0.0 | 0.1 | GO:0044666 | MLL3/4 complex(GO:0044666) |
| 0.0 | 0.1 | GO:0097504 | Gemini of coiled bodies(GO:0097504) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 0.8 | GO:0050682 | AF-2 domain binding(GO:0050682) |
| 0.1 | 0.9 | GO:0004064 | arylesterase activity(GO:0004064) |
| 0.1 | 2.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.1 | 0.3 | GO:0016309 | 1-phosphatidylinositol-5-phosphate 4-kinase activity(GO:0016309) |
| 0.1 | 0.4 | GO:0004815 | aspartate-tRNA ligase activity(GO:0004815) |
| 0.1 | 0.3 | GO:0008453 | alanine-glyoxylate transaminase activity(GO:0008453) |
| 0.1 | 0.3 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 1.7 | GO:0098641 | cadherin binding involved in cell-cell adhesion(GO:0098641) |
| 0.0 | 0.3 | GO:0034648 | histone demethylase activity (H3-dimethyl-K4 specific)(GO:0034648) |
| 0.0 | 0.1 | GO:0071558 | histone demethylase activity (H3-K27 specific)(GO:0071558) |
| 0.0 | 0.4 | GO:0019870 | potassium channel inhibitor activity(GO:0019870) |
| 0.0 | 0.4 | GO:0008525 | phosphatidylcholine transporter activity(GO:0008525) |
| 0.0 | 0.3 | GO:0008241 | peptidyl-dipeptidase activity(GO:0008241) |
| 0.0 | 0.2 | GO:0008073 | ornithine decarboxylase inhibitor activity(GO:0008073) |
| 0.0 | 0.8 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.6 | GO:0045499 | chemorepellent activity(GO:0045499) |
| 0.0 | 0.3 | GO:0086083 | cell adhesive protein binding involved in bundle of His cell-Purkinje myocyte communication(GO:0086083) |
| 0.0 | 0.5 | GO:0032395 | MHC class II receptor activity(GO:0032395) |
| 0.0 | 0.1 | GO:0004711 | ribosomal protein S6 kinase activity(GO:0004711) |
| 0.0 | 0.4 | GO:0071933 | Arp2/3 complex binding(GO:0071933) |
| 0.0 | 0.1 | GO:0004769 | steroid delta-isomerase activity(GO:0004769) |
| 0.0 | 0.9 | GO:0005229 | intracellular calcium activated chloride channel activity(GO:0005229) |
| 0.0 | 0.1 | GO:0004140 | dephospho-CoA kinase activity(GO:0004140) |
| 0.0 | 0.3 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 1.6 | GO:0005154 | epidermal growth factor receptor binding(GO:0005154) |
| 0.0 | 0.3 | GO:0004969 | histamine receptor activity(GO:0004969) |
| 0.0 | 0.5 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.2 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.0 | 0.1 | GO:0015375 | glycine:sodium symporter activity(GO:0015375) |
| 0.0 | 0.2 | GO:0043325 | phosphatidylinositol-3,4-bisphosphate binding(GO:0043325) |
| 0.0 | 0.1 | GO:0004464 | leukotriene-C4 synthase activity(GO:0004464) |
| 0.0 | 0.1 | GO:0102008 | cytosolic dipeptidase activity(GO:0102008) |
| 0.0 | 0.6 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.2 | GO:0008553 | hydrogen-exporting ATPase activity, phosphorylative mechanism(GO:0008553) |
| 0.0 | 0.4 | GO:0032041 | histone deacetylase activity (H3-K14 specific)(GO:0031078) NAD-dependent histone deacetylase activity (H3-K14 specific)(GO:0032041) |
| 0.0 | 0.1 | GO:0005330 | dopamine:sodium symporter activity(GO:0005330) |
| 0.0 | 0.1 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0042296 | ISG15 transferase activity(GO:0042296) |
| 0.0 | 0.1 | GO:0004013 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.1 | GO:0004517 | nitric-oxide synthase activity(GO:0004517) |
| 0.0 | 0.2 | GO:0031433 | telethonin binding(GO:0031433) |
| 0.0 | 0.6 | GO:0051959 | dynein light intermediate chain binding(GO:0051959) |
| 0.0 | 0.2 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.1 | GO:0004045 | aminoacyl-tRNA hydrolase activity(GO:0004045) |
| 0.0 | 0.0 | GO:0001002 | polymerase III regulatory region sequence-specific DNA binding(GO:0000992) RNA polymerase III type 1 promoter sequence-specific DNA binding(GO:0001002) RNA polymerase III type 2 promoter sequence-specific DNA binding(GO:0001003) |
| 0.0 | 0.1 | GO:0017018 | myosin phosphatase activity(GO:0017018) |
| 0.0 | 0.1 | GO:0004826 | phenylalanine-tRNA ligase activity(GO:0004826) |
| 0.0 | 0.5 | GO:0048020 | CCR chemokine receptor binding(GO:0048020) |
| 0.0 | 0.1 | GO:0002114 | interleukin-33 receptor activity(GO:0002114) |
| 0.0 | 0.6 | GO:0003785 | actin monomer binding(GO:0003785) |
| 0.0 | 0.1 | GO:0015272 | ATP-activated inward rectifier potassium channel activity(GO:0015272) |
| 0.0 | 0.1 | GO:0038036 | sphingosine-1-phosphate receptor activity(GO:0038036) |
| 0.0 | 0.1 | GO:0097322 | 7SK snRNA binding(GO:0097322) |
| 0.0 | 1.1 | GO:0005070 | SH3/SH2 adaptor activity(GO:0005070) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.4 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.1 | 1.9 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.0 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.9 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.1 | PID S1P S1P4 PATHWAY | S1P4 pathway |
| 0.0 | 0.3 | PID ECADHERIN KERATINOCYTE PATHWAY | E-cadherin signaling in keratinocytes |
| 0.0 | 0.5 | PID EPO PATHWAY | EPO signaling pathway |
| 0.0 | 1.0 | PID TAP63 PATHWAY | Validated transcriptional targets of TAp63 isoforms |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 2.2 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.1 | 0.9 | REACTOME REVERSIBLE HYDRATION OF CARBON DIOXIDE | Genes involved in Reversible Hydration of Carbon Dioxide |
| 0.0 | 1.0 | REACTOME INITIAL TRIGGERING OF COMPLEMENT | Genes involved in Initial triggering of complement |
| 0.0 | 0.5 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.3 | REACTOME PYRIMIDINE CATABOLISM | Genes involved in Pyrimidine catabolism |
| 0.0 | 0.5 | REACTOME MITOCHONDRIAL TRNA AMINOACYLATION | Genes involved in Mitochondrial tRNA aminoacylation |
| 0.0 | 0.5 | REACTOME SIGNALING BY CONSTITUTIVELY ACTIVE EGFR | Genes involved in Signaling by constitutively active EGFR |
| 0.0 | 0.6 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.8 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.4 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.8 | REACTOME STRIATED MUSCLE CONTRACTION | Genes involved in Striated Muscle Contraction |
| 0.0 | 0.2 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.9 | REACTOME NUCLEAR RECEPTOR TRANSCRIPTION PATHWAY | Genes involved in Nuclear Receptor transcription pathway |