Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
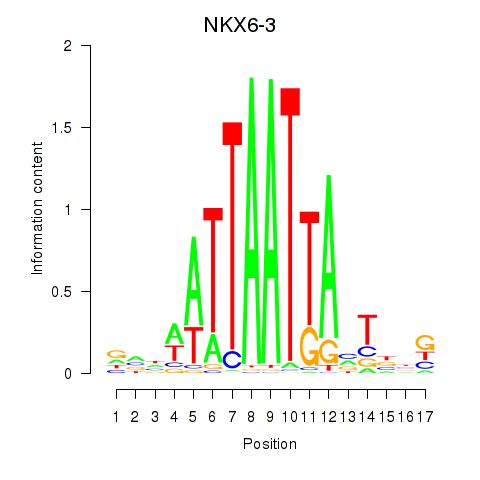
Results for NKX6-3
Z-value: 0.14
Transcription factors associated with NKX6-3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NKX6-3
|
ENSG00000165066.11 | NKX6-3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NKX6-3 | hg19_v2_chr8_-_41504878_41504878 | 0.07 | 8.7e-01 | Click! |
Activity profile of NKX6-3 motif
Sorted Z-values of NKX6-3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NKX6-3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_27529486 | 0.09 |
ENST00000375888.1 |
ACBD5 |
acyl-CoA binding domain containing 5 |
| chr1_-_232651312 | 0.07 |
ENST00000262861.4 |
SIPA1L2 |
signal-induced proliferation-associated 1 like 2 |
| chr19_+_50016610 | 0.07 |
ENST00000596975.1 |
FCGRT |
Fc fragment of IgG, receptor, transporter, alpha |
| chr6_+_114178512 | 0.06 |
ENST00000368635.4 |
MARCKS |
myristoylated alanine-rich protein kinase C substrate |
| chr20_-_33735070 | 0.06 |
ENST00000374491.3 ENST00000542871.1 ENST00000374492.3 |
EDEM2 |
ER degradation enhancer, mannosidase alpha-like 2 |
| chr14_-_51027838 | 0.05 |
ENST00000555216.1 |
MAP4K5 |
mitogen-activated protein kinase kinase kinase kinase 5 |
| chr9_+_125133315 | 0.04 |
ENST00000223423.4 ENST00000362012.2 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr8_+_77593448 | 0.04 |
ENST00000521891.2 |
ZFHX4 |
zinc finger homeobox 4 |
| chr8_+_77593474 | 0.03 |
ENST00000455469.2 ENST00000050961.6 |
ZFHX4 |
zinc finger homeobox 4 |
| chr15_+_93443419 | 0.03 |
ENST00000557381.1 ENST00000420239.2 |
CHD2 |
chromodomain helicase DNA binding protein 2 |
| chr1_-_204183071 | 0.03 |
ENST00000308302.3 |
GOLT1A |
golgi transport 1A |
| chr14_+_88852059 | 0.02 |
ENST00000045347.7 |
SPATA7 |
spermatogenesis associated 7 |
| chr14_+_88851874 | 0.02 |
ENST00000393545.4 ENST00000356583.5 ENST00000555401.1 ENST00000553885.1 |
SPATA7 |
spermatogenesis associated 7 |
| chr13_+_53602894 | 0.02 |
ENST00000219022.2 |
OLFM4 |
olfactomedin 4 |
| chr3_+_138340067 | 0.02 |
ENST00000479848.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr9_+_125132803 | 0.02 |
ENST00000540753.1 |
PTGS1 |
prostaglandin-endoperoxide synthase 1 (prostaglandin G/H synthase and cyclooxygenase) |
| chr1_-_151431647 | 0.02 |
ENST00000368863.2 ENST00000409503.1 ENST00000491586.1 ENST00000533351.1 ENST00000540984.1 |
POGZ |
pogo transposable element with ZNF domain |
| chr1_-_208417620 | 0.02 |
ENST00000367033.3 |
PLXNA2 |
plexin A2 |
| chr20_+_11008408 | 0.02 |
ENST00000378252.1 |
C20orf187 |
chromosome 20 open reading frame 187 |
| chr7_+_138145076 | 0.02 |
ENST00000343526.4 |
TRIM24 |
tripartite motif containing 24 |
| chr18_+_32558208 | 0.02 |
ENST00000436190.2 |
MAPRE2 |
microtubule-associated protein, RP/EB family, member 2 |
| chr15_+_92006567 | 0.02 |
ENST00000554333.1 |
RP11-661P17.1 |
RP11-661P17.1 |
| chr5_-_58295712 | 0.02 |
ENST00000317118.8 |
PDE4D |
phosphodiesterase 4D, cAMP-specific |
| chr5_-_142780280 | 0.01 |
ENST00000424646.2 |
NR3C1 |
nuclear receptor subfamily 3, group C, member 1 (glucocorticoid receptor) |
| chr1_-_151431909 | 0.01 |
ENST00000361398.3 ENST00000271715.2 |
POGZ |
pogo transposable element with ZNF domain |
| chr10_+_18549645 | 0.01 |
ENST00000396576.2 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr17_+_4692230 | 0.01 |
ENST00000331264.7 |
GLTPD2 |
glycolipid transfer protein domain containing 2 |
| chr9_-_215744 | 0.01 |
ENST00000382387.2 |
C9orf66 |
chromosome 9 open reading frame 66 |
| chr3_+_138340049 | 0.01 |
ENST00000464668.1 |
FAIM |
Fas apoptotic inhibitory molecule |
| chr20_-_29978286 | 0.01 |
ENST00000376315.2 |
DEFB119 |
defensin, beta 119 |
| chr4_+_80584903 | 0.00 |
ENST00000506460.1 |
RP11-452C8.1 |
RP11-452C8.1 |
| chr3_+_136649311 | 0.00 |
ENST00000469404.1 ENST00000467911.1 |
NCK1 |
NCK adaptor protein 1 |
| chr8_-_90769422 | 0.00 |
ENST00000524190.1 ENST00000523859.1 |
RP11-37B2.1 |
RP11-37B2.1 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0036512 | trimming of terminal mannose on B branch(GO:0036509) trimming of first mannose on A branch(GO:0036511) trimming of second mannose on A branch(GO:0036512) |
| 0.0 | 0.1 | GO:0030242 | pexophagy(GO:0030242) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0042585 | germinal vesicle(GO:0042585) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0019770 | IgG receptor activity(GO:0019770) |
| 0.0 | 0.1 | GO:0004666 | prostaglandin-endoperoxide synthase activity(GO:0004666) |