Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
Results for NR2F1
Z-value: 0.39
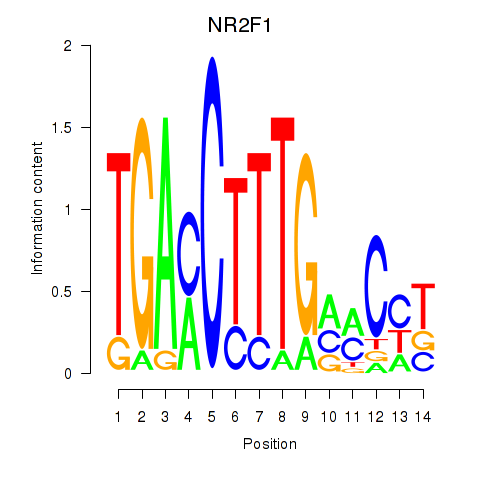
Transcription factors associated with NR2F1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
NR2F1
|
ENSG00000175745.7 | NR2F1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NR2F1 | hg19_v2_chr5_+_92919043_92919082 | -0.23 | 5.9e-01 | Click! |
Activity profile of NR2F1 motif
Sorted Z-values of NR2F1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of NR2F1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr10_-_129691195 | 0.28 |
ENST00000368671.3 |
CLRN3 |
clarin 3 |
| chr7_+_121513143 | 0.19 |
ENST00000393386.2 |
PTPRZ1 |
protein tyrosine phosphatase, receptor-type, Z polypeptide 1 |
| chr15_+_43885252 | 0.14 |
ENST00000453782.1 ENST00000300283.6 ENST00000437924.1 ENST00000450086.2 |
CKMT1B |
creatine kinase, mitochondrial 1B |
| chr1_+_158149737 | 0.14 |
ENST00000368171.3 |
CD1D |
CD1d molecule |
| chr8_+_103563792 | 0.14 |
ENST00000285402.3 |
ODF1 |
outer dense fiber of sperm tails 1 |
| chr13_-_103046837 | 0.13 |
ENST00000607251.1 |
FGF14-IT1 |
FGF14 intronic transcript 1 (non-protein coding) |
| chr15_+_43985084 | 0.13 |
ENST00000434505.1 ENST00000411750.1 |
CKMT1A |
creatine kinase, mitochondrial 1A |
| chr2_+_220491973 | 0.13 |
ENST00000358055.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr12_+_132413765 | 0.12 |
ENST00000376649.3 ENST00000322060.5 |
PUS1 |
pseudouridylate synthase 1 |
| chr9_-_139137648 | 0.11 |
ENST00000358701.5 |
QSOX2 |
quiescin Q6 sulfhydryl oxidase 2 |
| chr2_-_70780572 | 0.11 |
ENST00000450929.1 |
TGFA |
transforming growth factor, alpha |
| chr12_+_132413739 | 0.11 |
ENST00000443358.2 |
PUS1 |
pseudouridylate synthase 1 |
| chr3_+_124355944 | 0.11 |
ENST00000459915.1 |
KALRN |
kalirin, RhoGEF kinase |
| chr19_-_10687907 | 0.10 |
ENST00000589348.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr11_+_72929319 | 0.10 |
ENST00000393597.2 ENST00000311131.2 |
P2RY2 |
purinergic receptor P2Y, G-protein coupled, 2 |
| chr11_+_72929402 | 0.10 |
ENST00000393596.2 |
P2RY2 |
purinergic receptor P2Y, G-protein coupled, 2 |
| chr12_+_132413798 | 0.10 |
ENST00000440818.2 ENST00000542167.2 ENST00000538037.1 ENST00000456665.2 |
PUS1 |
pseudouridylate synthase 1 |
| chr15_+_90728145 | 0.10 |
ENST00000561085.1 ENST00000379122.3 ENST00000332496.6 |
SEMA4B |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4B |
| chr19_-_10687948 | 0.10 |
ENST00000592285.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr2_+_220492116 | 0.10 |
ENST00000373760.2 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr1_+_110163682 | 0.10 |
ENST00000358729.4 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr2_-_165698521 | 0.10 |
ENST00000409184.3 ENST00000392717.2 ENST00000456693.1 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr1_+_62439037 | 0.09 |
ENST00000545929.1 |
INADL |
InaD-like (Drosophila) |
| chr19_-_10687983 | 0.09 |
ENST00000587069.1 |
AP1M2 |
adaptor-related protein complex 1, mu 2 subunit |
| chr17_-_2614927 | 0.09 |
ENST00000435359.1 |
CLUH |
clustered mitochondria (cluA/CLU1) homolog |
| chr19_-_633576 | 0.09 |
ENST00000588649.2 |
POLRMT |
polymerase (RNA) mitochondrial (DNA directed) |
| chr12_-_53074182 | 0.09 |
ENST00000252244.3 |
KRT1 |
keratin 1 |
| chr11_-_116708302 | 0.09 |
ENST00000375320.1 ENST00000359492.2 ENST00000375329.2 ENST00000375323.1 |
APOA1 |
apolipoprotein A-I |
| chr19_+_45394477 | 0.09 |
ENST00000252487.5 ENST00000405636.2 ENST00000592434.1 ENST00000426677.2 ENST00000589649.1 |
TOMM40 |
translocase of outer mitochondrial membrane 40 homolog (yeast) |
| chrX_+_135388147 | 0.09 |
ENST00000394141.1 |
GPR112 |
G protein-coupled receptor 112 |
| chr2_-_165698662 | 0.09 |
ENST00000194871.6 ENST00000445474.2 |
COBLL1 |
cordon-bleu WH2 repeat protein-like 1 |
| chr17_+_7531281 | 0.08 |
ENST00000575729.1 ENST00000340624.5 |
SHBG |
sex hormone-binding globulin |
| chr2_-_70780770 | 0.08 |
ENST00000444975.1 ENST00000445399.1 ENST00000418333.2 |
TGFA |
transforming growth factor, alpha |
| chr12_-_21757774 | 0.08 |
ENST00000261195.2 |
GYS2 |
glycogen synthase 2 (liver) |
| chr14_-_92333873 | 0.08 |
ENST00000435962.2 |
TC2N |
tandem C2 domains, nuclear |
| chr1_-_53793725 | 0.08 |
ENST00000371454.2 |
LRP8 |
low density lipoprotein receptor-related protein 8, apolipoprotein e receptor |
| chr4_-_11431389 | 0.08 |
ENST00000002596.5 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr1_-_36235529 | 0.08 |
ENST00000318121.3 ENST00000373220.3 ENST00000520551.1 |
CLSPN |
claspin |
| chr2_+_220492373 | 0.08 |
ENST00000317151.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chr2_+_219433588 | 0.08 |
ENST00000295701.5 |
RQCD1 |
RCD1 required for cell differentiation1 homolog (S. pombe) |
| chr8_-_56685859 | 0.08 |
ENST00000523423.1 ENST00000523073.1 ENST00000519784.1 ENST00000434581.2 ENST00000519780.1 ENST00000521229.1 ENST00000522576.1 ENST00000523180.1 ENST00000522090.1 |
TMEM68 |
transmembrane protein 68 |
| chr19_-_6767431 | 0.08 |
ENST00000437152.3 ENST00000597687.1 |
SH2D3A |
SH2 domain containing 3A |
| chr19_-_51472031 | 0.07 |
ENST00000391808.1 |
KLK6 |
kallikrein-related peptidase 6 |
| chr2_+_220492287 | 0.07 |
ENST00000273063.6 ENST00000373762.3 |
SLC4A3 |
solute carrier family 4 (anion exchanger), member 3 |
| chrX_-_114253536 | 0.07 |
ENST00000371936.1 |
IL13RA2 |
interleukin 13 receptor, alpha 2 |
| chr19_-_51472222 | 0.07 |
ENST00000376851.3 |
KLK6 |
kallikrein-related peptidase 6 |
| chr12_-_7125770 | 0.07 |
ENST00000261407.4 |
LPCAT3 |
lysophosphatidylcholine acyltransferase 3 |
| chr19_+_751122 | 0.07 |
ENST00000215582.6 |
MISP |
mitotic spindle positioning |
| chr16_+_3014217 | 0.07 |
ENST00000572045.1 |
KREMEN2 |
kringle containing transmembrane protein 2 |
| chr3_-_48229846 | 0.07 |
ENST00000302506.3 ENST00000351231.3 ENST00000437972.1 |
CDC25A |
cell division cycle 25A |
| chr8_-_56685966 | 0.07 |
ENST00000334667.2 |
TMEM68 |
transmembrane protein 68 |
| chr3_+_37284824 | 0.07 |
ENST00000431105.1 |
GOLGA4 |
golgin A4 |
| chr1_+_205473720 | 0.07 |
ENST00000429964.2 ENST00000506784.1 ENST00000360066.2 |
CDK18 |
cyclin-dependent kinase 18 |
| chr19_-_46418033 | 0.06 |
ENST00000341294.2 |
NANOS2 |
nanos homolog 2 (Drosophila) |
| chr12_-_51419924 | 0.06 |
ENST00000541174.2 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr1_+_110163202 | 0.06 |
ENST00000531203.1 ENST00000256578.3 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr12_-_51420108 | 0.06 |
ENST00000547198.1 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr19_-_16045619 | 0.06 |
ENST00000402119.4 |
CYP4F11 |
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr19_-_16045665 | 0.06 |
ENST00000248041.8 |
CYP4F11 |
cytochrome P450, family 4, subfamily F, polypeptide 11 |
| chr4_-_681114 | 0.06 |
ENST00000503156.1 |
MFSD7 |
major facilitator superfamily domain containing 7 |
| chr11_-_45939565 | 0.06 |
ENST00000525192.1 ENST00000378750.5 |
PEX16 |
peroxisomal biogenesis factor 16 |
| chr9_-_27005686 | 0.06 |
ENST00000380055.5 |
LRRC19 |
leucine rich repeat containing 19 |
| chr21_+_43919710 | 0.06 |
ENST00000398341.3 |
SLC37A1 |
solute carrier family 37 (glucose-6-phosphate transporter), member 1 |
| chr1_+_110163709 | 0.06 |
ENST00000369840.2 ENST00000527846.1 |
AMPD2 |
adenosine monophosphate deaminase 2 |
| chr15_-_41120896 | 0.06 |
ENST00000299174.5 ENST00000427255.2 |
PPP1R14D |
protein phosphatase 1, regulatory (inhibitor) subunit 14D |
| chr17_+_1646130 | 0.06 |
ENST00000453066.1 ENST00000324015.3 ENST00000450523.2 ENST00000453723.1 ENST00000382061.4 |
SERPINF2 |
serpin peptidase inhibitor, clade F (alpha-2 antiplasmin, pigment epithelium derived factor), member 2 |
| chr1_+_156123359 | 0.06 |
ENST00000368284.1 ENST00000368286.2 ENST00000438830.1 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr19_-_1174226 | 0.06 |
ENST00000587024.1 ENST00000361757.3 |
SBNO2 |
strawberry notch homolog 2 (Drosophila) |
| chr14_-_21492251 | 0.06 |
ENST00000554398.1 |
NDRG2 |
NDRG family member 2 |
| chr7_+_100663353 | 0.05 |
ENST00000306151.4 |
MUC17 |
mucin 17, cell surface associated |
| chr1_+_156123318 | 0.05 |
ENST00000368285.3 |
SEMA4A |
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4A |
| chr10_+_81107271 | 0.05 |
ENST00000448165.1 |
PPIF |
peptidylprolyl isomerase F |
| chr3_+_62936098 | 0.05 |
ENST00000475886.1 ENST00000465684.1 ENST00000465262.1 ENST00000468072.1 |
LINC00698 |
long intergenic non-protein coding RNA 698 |
| chr2_+_197577841 | 0.05 |
ENST00000409270.1 |
CCDC150 |
coiled-coil domain containing 150 |
| chr14_-_21492113 | 0.05 |
ENST00000554094.1 |
NDRG2 |
NDRG family member 2 |
| chr19_+_39390587 | 0.05 |
ENST00000572515.1 ENST00000392079.3 ENST00000575359.1 ENST00000313582.5 |
NFKBIB |
nuclear factor of kappa light polypeptide gene enhancer in B-cells inhibitor, beta |
| chrX_-_117119243 | 0.05 |
ENST00000539496.1 ENST00000469946.1 |
KLHL13 |
kelch-like family member 13 |
| chr15_+_40453204 | 0.05 |
ENST00000287598.6 ENST00000412359.3 |
BUB1B |
BUB1 mitotic checkpoint serine/threonine kinase B |
| chr1_+_196857144 | 0.05 |
ENST00000367416.2 ENST00000251424.4 ENST00000367418.2 |
CFHR4 |
complement factor H-related 4 |
| chr14_-_23285011 | 0.05 |
ENST00000397532.3 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr3_-_172428959 | 0.05 |
ENST00000475381.1 ENST00000538775.1 ENST00000273512.3 ENST00000543711.1 |
NCEH1 |
neutral cholesterol ester hydrolase 1 |
| chr12_-_54779511 | 0.05 |
ENST00000551109.1 ENST00000546970.1 |
ZNF385A |
zinc finger protein 385A |
| chr6_+_31926857 | 0.05 |
ENST00000375394.2 ENST00000544581.1 |
SKIV2L |
superkiller viralicidic activity 2-like (S. cerevisiae) |
| chr19_+_6464243 | 0.05 |
ENST00000600229.1 ENST00000356762.3 |
CRB3 |
crumbs homolog 3 (Drosophila) |
| chr14_-_23285069 | 0.05 |
ENST00000554758.1 ENST00000397528.4 |
SLC7A7 |
solute carrier family 7 (amino acid transporter light chain, y+L system), member 7 |
| chr9_-_39239171 | 0.05 |
ENST00000358144.2 |
CNTNAP3 |
contactin associated protein-like 3 |
| chr12_-_109219937 | 0.05 |
ENST00000546697.1 |
SSH1 |
slingshot protein phosphatase 1 |
| chr2_+_113763031 | 0.05 |
ENST00000259211.6 |
IL36A |
interleukin 36, alpha |
| chr3_+_37284668 | 0.05 |
ENST00000361924.2 ENST00000444882.1 ENST00000356847.4 ENST00000450863.2 ENST00000429018.1 |
GOLGA4 |
golgin A4 |
| chr8_-_16859690 | 0.05 |
ENST00000180166.5 |
FGF20 |
fibroblast growth factor 20 |
| chr19_+_6464502 | 0.05 |
ENST00000308243.7 |
CRB3 |
crumbs homolog 3 (Drosophila) |
| chr3_-_149510553 | 0.04 |
ENST00000462519.2 ENST00000446160.1 ENST00000383050.3 |
ANKUB1 |
ankyrin repeat and ubiquitin domain containing 1 |
| chr20_-_62130474 | 0.04 |
ENST00000217182.3 |
EEF1A2 |
eukaryotic translation elongation factor 1 alpha 2 |
| chr12_+_50898881 | 0.04 |
ENST00000301180.5 |
DIP2B |
DIP2 disco-interacting protein 2 homolog B (Drosophila) |
| chr19_-_42133420 | 0.04 |
ENST00000221954.2 ENST00000600925.1 |
CEACAM4 |
carcinoembryonic antigen-related cell adhesion molecule 4 |
| chr3_-_194393206 | 0.04 |
ENST00000265245.5 |
LSG1 |
large 60S subunit nuclear export GTPase 1 |
| chr10_+_101542462 | 0.04 |
ENST00000370449.4 ENST00000370434.1 |
ABCC2 |
ATP-binding cassette, sub-family C (CFTR/MRP), member 2 |
| chr11_-_45939374 | 0.04 |
ENST00000533151.1 ENST00000241041.3 |
PEX16 |
peroxisomal biogenesis factor 16 |
| chr22_-_29784519 | 0.04 |
ENST00000357586.2 ENST00000356015.2 ENST00000432560.2 ENST00000317368.7 |
AP1B1 |
adaptor-related protein complex 1, beta 1 subunit |
| chr10_-_103815874 | 0.04 |
ENST00000370033.4 ENST00000311122.5 |
C10orf76 |
chromosome 10 open reading frame 76 |
| chr9_+_133971863 | 0.04 |
ENST00000372309.3 |
AIF1L |
allograft inflammatory factor 1-like |
| chr18_+_29769978 | 0.04 |
ENST00000269202.6 ENST00000581447.1 |
MEP1B |
meprin A, beta |
| chrX_+_138612889 | 0.04 |
ENST00000218099.2 ENST00000394090.2 |
F9 |
coagulation factor IX |
| chr17_+_7533439 | 0.04 |
ENST00000441599.2 ENST00000380450.4 ENST00000416273.3 ENST00000575903.1 ENST00000576830.1 ENST00000571153.1 ENST00000575618.1 ENST00000576152.1 |
SHBG |
sex hormone-binding globulin |
| chr12_+_6494285 | 0.04 |
ENST00000541102.1 |
LTBR |
lymphotoxin beta receptor (TNFR superfamily, member 3) |
| chr5_+_140557371 | 0.04 |
ENST00000239444.2 |
PCDHB8 |
protocadherin beta 8 |
| chr12_+_53497263 | 0.04 |
ENST00000551896.1 ENST00000301466.3 |
SOAT2 |
sterol O-acyltransferase 2 |
| chr17_+_48423453 | 0.04 |
ENST00000017003.2 ENST00000509778.1 ENST00000507602.1 |
XYLT2 |
xylosyltransferase II |
| chr2_-_159313214 | 0.04 |
ENST00000409889.1 ENST00000283233.5 ENST00000536771.1 |
CCDC148 |
coiled-coil domain containing 148 |
| chr8_-_144815966 | 0.04 |
ENST00000388913.3 |
FAM83H |
family with sequence similarity 83, member H |
| chr2_+_73441350 | 0.04 |
ENST00000389501.4 |
SMYD5 |
SMYD family member 5 |
| chr11_+_63974135 | 0.04 |
ENST00000544997.1 ENST00000345728.5 ENST00000279227.5 |
FERMT3 |
fermitin family member 3 |
| chr16_+_57673430 | 0.04 |
ENST00000540164.2 ENST00000568531.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr17_-_73285293 | 0.04 |
ENST00000582778.1 ENST00000581988.1 ENST00000579207.1 ENST00000583332.1 ENST00000416858.2 ENST00000442286.2 ENST00000580151.1 ENST00000580994.1 ENST00000584438.1 ENST00000320362.3 ENST00000580273.1 |
SLC25A19 |
solute carrier family 25 (mitochondrial thiamine pyrophosphate carrier), member 19 |
| chr3_+_186560476 | 0.04 |
ENST00000320741.2 ENST00000444204.2 |
ADIPOQ |
adiponectin, C1Q and collagen domain containing |
| chr3_+_186560462 | 0.04 |
ENST00000412955.2 |
ADIPOQ |
adiponectin, C1Q and collagen domain containing |
| chr1_+_43855545 | 0.04 |
ENST00000372450.4 ENST00000310739.4 |
SZT2 |
seizure threshold 2 homolog (mouse) |
| chrX_-_49041242 | 0.04 |
ENST00000453382.1 ENST00000540849.1 ENST00000536904.1 ENST00000432913.1 |
PRICKLE3 |
prickle homolog 3 (Drosophila) |
| chr5_+_121297650 | 0.04 |
ENST00000339397.4 |
SRFBP1 |
serum response factor binding protein 1 |
| chr6_+_43737939 | 0.04 |
ENST00000372067.3 |
VEGFA |
vascular endothelial growth factor A |
| chr17_-_4463856 | 0.04 |
ENST00000574584.1 ENST00000381550.3 ENST00000301395.3 |
GGT6 |
gamma-glutamyltransferase 6 |
| chr19_+_10222189 | 0.04 |
ENST00000321826.4 |
P2RY11 |
purinergic receptor P2Y, G-protein coupled, 11 |
| chr21_-_45682099 | 0.04 |
ENST00000270172.3 ENST00000418993.1 |
DNMT3L |
DNA (cytosine-5-)-methyltransferase 3-like |
| chr10_+_18689637 | 0.04 |
ENST00000377315.4 |
CACNB2 |
calcium channel, voltage-dependent, beta 2 subunit |
| chr2_+_159313452 | 0.04 |
ENST00000389757.3 ENST00000389759.3 |
PKP4 |
plakophilin 4 |
| chr16_-_1275257 | 0.03 |
ENST00000234798.4 |
TPSG1 |
tryptase gamma 1 |
| chr12_+_49717081 | 0.03 |
ENST00000547807.1 ENST00000551567.1 |
TROAP |
trophinin associated protein |
| chr17_+_77893135 | 0.03 |
ENST00000574526.1 ENST00000572353.1 |
RP11-353N14.4 |
RP11-353N14.4 |
| chr6_+_24775153 | 0.03 |
ENST00000356509.3 ENST00000230056.3 |
GMNN |
geminin, DNA replication inhibitor |
| chr16_-_69448 | 0.03 |
ENST00000326592.9 |
WASH4P |
WAS protein family homolog 4 pseudogene |
| chr11_-_123756334 | 0.03 |
ENST00000528595.1 ENST00000375026.2 |
TMEM225 |
transmembrane protein 225 |
| chr12_+_67663056 | 0.03 |
ENST00000545606.1 |
CAND1 |
cullin-associated and neddylation-dissociated 1 |
| chr13_+_113777105 | 0.03 |
ENST00000409306.1 ENST00000375551.3 ENST00000375559.3 |
F10 |
coagulation factor X |
| chr2_-_152382500 | 0.03 |
ENST00000434685.1 |
NEB |
nebulin |
| chr5_-_1801408 | 0.03 |
ENST00000505818.1 |
MRPL36 |
mitochondrial ribosomal protein L36 |
| chr22_+_24105394 | 0.03 |
ENST00000305199.5 ENST00000382821.3 |
C22orf15 |
chromosome 22 open reading frame 15 |
| chr1_-_36235559 | 0.03 |
ENST00000251195.5 |
CLSPN |
claspin |
| chr20_-_3644046 | 0.03 |
ENST00000290417.2 ENST00000319242.3 |
GFRA4 |
GDNF family receptor alpha 4 |
| chr22_+_31518938 | 0.03 |
ENST00000412985.1 ENST00000331075.5 ENST00000412277.2 ENST00000420017.1 ENST00000400294.2 ENST00000405300.1 ENST00000404390.3 |
INPP5J |
inositol polyphosphate-5-phosphatase J |
| chr3_+_128444994 | 0.03 |
ENST00000482525.1 |
RAB7A |
RAB7A, member RAS oncogene family |
| chr19_+_39616410 | 0.03 |
ENST00000602004.1 ENST00000599470.1 ENST00000321944.4 ENST00000593480.1 ENST00000358301.3 ENST00000593690.1 ENST00000599386.1 |
PAK4 |
p21 protein (Cdc42/Rac)-activated kinase 4 |
| chr19_-_36304201 | 0.03 |
ENST00000301175.3 |
PRODH2 |
proline dehydrogenase (oxidase) 2 |
| chr10_+_26986582 | 0.03 |
ENST00000376215.5 ENST00000376203.5 |
PDSS1 |
prenyl (decaprenyl) diphosphate synthase, subunit 1 |
| chr6_+_7108210 | 0.03 |
ENST00000467782.1 ENST00000334984.6 ENST00000349384.6 |
RREB1 |
ras responsive element binding protein 1 |
| chrX_-_43741594 | 0.03 |
ENST00000536181.1 ENST00000378069.4 |
MAOB |
monoamine oxidase B |
| chr10_-_17171817 | 0.03 |
ENST00000377833.4 |
CUBN |
cubilin (intrinsic factor-cobalamin receptor) |
| chr12_-_53729525 | 0.03 |
ENST00000303846.3 |
SP7 |
Sp7 transcription factor |
| chr2_+_138721850 | 0.03 |
ENST00000329366.4 ENST00000280097.3 |
HNMT |
histamine N-methyltransferase |
| chr7_+_150782945 | 0.03 |
ENST00000463381.1 |
AGAP3 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 3 |
| chr13_+_73302047 | 0.03 |
ENST00000377814.2 ENST00000377815.3 ENST00000390667.5 |
BORA |
bora, aurora kinase A activator |
| chr11_+_66624527 | 0.03 |
ENST00000393952.3 |
LRFN4 |
leucine rich repeat and fibronectin type III domain containing 4 |
| chr1_+_109234907 | 0.03 |
ENST00000370025.4 ENST00000370022.5 ENST00000370021.1 |
PRPF38B |
pre-mRNA processing factor 38B |
| chr15_-_73925651 | 0.03 |
ENST00000545878.1 ENST00000287226.8 ENST00000345330.4 |
NPTN |
neuroplastin |
| chr10_-_54531406 | 0.03 |
ENST00000373968.3 |
MBL2 |
mannose-binding lectin (protein C) 2, soluble |
| chr6_+_7107999 | 0.03 |
ENST00000491191.1 ENST00000379938.2 ENST00000471433.1 |
RREB1 |
ras responsive element binding protein 1 |
| chr10_-_104211294 | 0.03 |
ENST00000239125.1 |
C10orf95 |
chromosome 10 open reading frame 95 |
| chr18_-_61034743 | 0.03 |
ENST00000406396.3 |
KDSR |
3-ketodihydrosphingosine reductase |
| chr17_+_27071002 | 0.03 |
ENST00000262395.5 ENST00000422344.1 ENST00000444415.3 ENST00000262396.6 |
TRAF4 |
TNF receptor-associated factor 4 |
| chr1_-_229569834 | 0.03 |
ENST00000366684.3 ENST00000366683.2 |
ACTA1 |
actin, alpha 1, skeletal muscle |
| chr2_-_207024134 | 0.03 |
ENST00000457011.1 ENST00000440274.1 ENST00000432169.1 |
NDUFS1 |
NADH dehydrogenase (ubiquinone) Fe-S protein 1, 75kDa (NADH-coenzyme Q reductase) |
| chr19_+_7660716 | 0.03 |
ENST00000160298.4 ENST00000446248.2 |
CAMSAP3 |
calmodulin regulated spectrin-associated protein family, member 3 |
| chr1_-_161193349 | 0.03 |
ENST00000469730.2 ENST00000463273.1 ENST00000464492.1 ENST00000367990.3 ENST00000470459.2 ENST00000468465.1 ENST00000463812.1 |
APOA2 |
apolipoprotein A-II |
| chr2_-_219134822 | 0.03 |
ENST00000444053.1 ENST00000248450.4 |
AAMP |
angio-associated, migratory cell protein |
| chr22_-_47882857 | 0.03 |
ENST00000405369.3 |
LL22NC03-75H12.2 |
Novel protein; Uncharacterized protein |
| chr7_-_150779995 | 0.03 |
ENST00000462940.1 ENST00000492838.1 ENST00000392818.3 ENST00000488752.1 ENST00000476627.1 |
TMUB1 |
transmembrane and ubiquitin-like domain containing 1 |
| chr7_-_75452673 | 0.03 |
ENST00000416943.1 |
CCL24 |
chemokine (C-C motif) ligand 24 |
| chr1_-_43751230 | 0.03 |
ENST00000523677.1 |
C1orf210 |
chromosome 1 open reading frame 210 |
| chr11_+_18417813 | 0.03 |
ENST00000540430.1 ENST00000379412.5 |
LDHA |
lactate dehydrogenase A |
| chr10_-_30638090 | 0.03 |
ENST00000421701.1 ENST00000263063.4 |
MTPAP |
mitochondrial poly(A) polymerase |
| chr7_-_150780609 | 0.03 |
ENST00000297533.4 |
TMUB1 |
transmembrane and ubiquitin-like domain containing 1 |
| chr12_+_121416489 | 0.03 |
ENST00000541395.1 ENST00000544413.1 |
HNF1A |
HNF1 homeobox A |
| chr12_-_118810688 | 0.02 |
ENST00000542532.1 ENST00000392533.3 |
TAOK3 |
TAO kinase 3 |
| chr6_-_30080876 | 0.02 |
ENST00000376734.3 |
TRIM31 |
tripartite motif containing 31 |
| chr8_-_37594944 | 0.02 |
ENST00000330539.1 |
RP11-863K10.7 |
Uncharacterized protein |
| chr19_+_10362882 | 0.02 |
ENST00000393733.2 ENST00000588502.1 |
MRPL4 |
mitochondrial ribosomal protein L4 |
| chr12_-_51420128 | 0.02 |
ENST00000262051.7 ENST00000547732.1 ENST00000262052.5 ENST00000546488.1 ENST00000550714.1 ENST00000548193.1 ENST00000547579.1 ENST00000546743.1 |
SLC11A2 |
solute carrier family 11 (proton-coupled divalent metal ion transporter), member 2 |
| chr3_-_113465065 | 0.02 |
ENST00000497255.1 ENST00000478020.1 ENST00000240922.3 ENST00000493900.1 |
NAA50 |
N(alpha)-acetyltransferase 50, NatE catalytic subunit |
| chr1_-_155270770 | 0.02 |
ENST00000392414.3 |
PKLR |
pyruvate kinase, liver and RBC |
| chr8_-_104153703 | 0.02 |
ENST00000521246.1 |
C8orf56 |
chromosome 8 open reading frame 56 |
| chr20_+_31755934 | 0.02 |
ENST00000354932.5 |
BPIFA2 |
BPI fold containing family A, member 2 |
| chr8_-_70016408 | 0.02 |
ENST00000518540.1 |
RP11-600K15.1 |
RP11-600K15.1 |
| chr19_+_589893 | 0.02 |
ENST00000251287.2 |
HCN2 |
hyperpolarization activated cyclic nucleotide-gated potassium channel 2 |
| chr8_+_22853345 | 0.02 |
ENST00000522948.1 |
RHOBTB2 |
Rho-related BTB domain containing 2 |
| chr7_-_15601595 | 0.02 |
ENST00000342526.3 |
AGMO |
alkylglycerol monooxygenase |
| chr3_-_53878644 | 0.02 |
ENST00000481668.1 ENST00000467802.1 |
CHDH |
choline dehydrogenase |
| chr5_-_42812143 | 0.02 |
ENST00000514985.1 |
SEPP1 |
selenoprotein P, plasma, 1 |
| chr4_-_11431188 | 0.02 |
ENST00000510712.1 |
HS3ST1 |
heparan sulfate (glucosamine) 3-O-sulfotransferase 1 |
| chr1_-_151138323 | 0.02 |
ENST00000368908.5 |
LYSMD1 |
LysM, putative peptidoglycan-binding, domain containing 1 |
| chr16_-_31439735 | 0.02 |
ENST00000287490.4 |
COX6A2 |
cytochrome c oxidase subunit VIa polypeptide 2 |
| chr6_+_97010424 | 0.02 |
ENST00000541107.1 ENST00000450218.1 ENST00000326771.2 |
FHL5 |
four and a half LIM domains 5 |
| chr1_-_169555779 | 0.02 |
ENST00000367797.3 ENST00000367796.3 |
F5 |
coagulation factor V (proaccelerin, labile factor) |
| chr12_-_31158902 | 0.02 |
ENST00000544329.1 ENST00000418254.2 ENST00000222396.5 |
RP11-551L14.4 |
RP11-551L14.4 |
| chr12_-_53594227 | 0.02 |
ENST00000550743.2 |
ITGB7 |
integrin, beta 7 |
| chr15_+_75105170 | 0.02 |
ENST00000379709.3 |
LMAN1L |
lectin, mannose-binding, 1 like |
| chr19_-_1021113 | 0.02 |
ENST00000333175.5 ENST00000356663.3 |
TMEM259 |
transmembrane protein 259 |
| chr6_-_44265411 | 0.02 |
ENST00000371505.4 |
TCTE1 |
t-complex-associated-testis-expressed 1 |
| chr1_+_156785425 | 0.02 |
ENST00000392302.2 |
NTRK1 |
neurotrophic tyrosine kinase, receptor, type 1 |
| chr19_+_41594377 | 0.02 |
ENST00000330436.3 |
CYP2A13 |
cytochrome P450, family 2, subfamily A, polypeptide 13 |
| chr2_-_241725781 | 0.02 |
ENST00000428768.1 |
KIF1A |
kinesin family member 1A |
| chrX_-_108868390 | 0.02 |
ENST00000372101.2 |
KCNE1L |
KCNE1-like |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070902 | mitochondrial tRNA pseudouridine synthesis(GO:0070902) |
| 0.0 | 0.1 | GO:0032581 | ER-dependent peroxisome organization(GO:0032581) |
| 0.0 | 0.1 | GO:0042377 | menaquinone catabolic process(GO:0042361) vitamin K catabolic process(GO:0042377) |
| 0.0 | 0.2 | GO:0015692 | lead ion transport(GO:0015692) |
| 0.0 | 0.1 | GO:0010903 | negative regulation of very-low-density lipoprotein particle remodeling(GO:0010903) |
| 0.0 | 0.1 | GO:2000532 | renal albumin absorption(GO:0097018) regulation of renal albumin absorption(GO:2000532) regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000583) negative regulation of platelet-derived growth factor receptor-alpha signaling pathway(GO:2000584) |
| 0.0 | 0.1 | GO:0006391 | transcription initiation from mitochondrial promoter(GO:0006391) |
| 0.0 | 0.2 | GO:0032264 | IMP salvage(GO:0032264) |
| 0.0 | 0.3 | GO:0070257 | positive regulation of mucus secretion(GO:0070257) |
| 0.0 | 0.2 | GO:0070444 | oligodendrocyte progenitor proliferation(GO:0070444) regulation of oligodendrocyte progenitor proliferation(GO:0070445) |
| 0.0 | 0.1 | GO:1903691 | positive regulation of wound healing, spreading of epidermal cells(GO:1903691) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.1 | GO:0035565 | regulation of pronephros size(GO:0035565) renal glucose absorption(GO:0035623) |
| 0.0 | 0.1 | GO:0000821 | regulation of arginine metabolic process(GO:0000821) |
| 0.0 | 0.1 | GO:0030718 | germ-line stem cell population maintenance(GO:0030718) |
| 0.0 | 0.0 | GO:1904719 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.1 | GO:0060125 | negative regulation of growth hormone secretion(GO:0060125) |
| 0.0 | 0.0 | GO:0050787 | antibiotic metabolic process(GO:0016999) detoxification of mercury ion(GO:0050787) |
| 0.0 | 0.0 | GO:1903542 | negative regulation of exosomal secretion(GO:1903542) |
| 0.0 | 0.1 | GO:0006600 | creatine metabolic process(GO:0006600) |
| 0.0 | 0.1 | GO:0051754 | meiotic sister chromatid cohesion, centromeric(GO:0051754) |
| 0.0 | 0.0 | GO:1903570 | regulation of protein kinase D signaling(GO:1903570) positive regulation of protein kinase D signaling(GO:1903572) |
| 0.0 | 0.2 | GO:0045741 | positive regulation of epidermal growth factor-activated receptor activity(GO:0045741) |
| 0.0 | 0.1 | GO:0048007 | antigen processing and presentation of lipid antigen via MHC class Ib(GO:0048003) antigen processing and presentation, exogenous lipid antigen via MHC class Ib(GO:0048007) |
| 0.0 | 0.2 | GO:0051639 | actin filament network formation(GO:0051639) |
| 0.0 | 0.1 | GO:0071105 | response to interleukin-11(GO:0071105) |
| 0.0 | 0.1 | GO:0038026 | reelin-mediated signaling pathway(GO:0038026) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.2 | GO:0072534 | perineuronal net(GO:0072534) |
| 0.0 | 0.2 | GO:0070826 | paraferritin complex(GO:0070826) |
| 0.0 | 0.1 | GO:0034365 | discoidal high-density lipoprotein particle(GO:0034365) |
| 0.0 | 0.0 | GO:0055087 | Ski complex(GO:0055087) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0004730 | pseudouridylate synthase activity(GO:0004730) |
| 0.0 | 0.2 | GO:0015639 | cadmium ion transmembrane transporter activity(GO:0015086) cobalt ion transmembrane transporter activity(GO:0015087) lead ion transmembrane transporter activity(GO:0015094) ferrous iron uptake transmembrane transporter activity(GO:0015639) |
| 0.0 | 0.1 | GO:0070653 | high-density lipoprotein particle receptor binding(GO:0070653) |
| 0.0 | 0.2 | GO:0003876 | AMP deaminase activity(GO:0003876) adenosine-phosphate deaminase activity(GO:0047623) |
| 0.0 | 0.1 | GO:0016971 | flavin-linked sulfhydryl oxidase activity(GO:0016971) |
| 0.0 | 0.1 | GO:0038025 | reelin receptor activity(GO:0038025) |
| 0.0 | 0.1 | GO:0004111 | creatine kinase activity(GO:0004111) |
| 0.0 | 0.4 | GO:0008510 | sodium:bicarbonate symporter activity(GO:0008510) |
| 0.0 | 0.1 | GO:0047184 | 1-acylglycerophosphocholine O-acyltransferase activity(GO:0047184) |
| 0.0 | 0.1 | GO:0010997 | anaphase-promoting complex binding(GO:0010997) |
| 0.0 | 0.1 | GO:0030882 | lipid antigen binding(GO:0030882) endogenous lipid antigen binding(GO:0030883) exogenous lipid antigen binding(GO:0030884) |
| 0.0 | 0.0 | GO:0030158 | protein xylosyltransferase activity(GO:0030158) |
| 0.0 | 0.1 | GO:0008467 | [heparan sulfate]-glucosamine 3-sulfotransferase 1 activity(GO:0008467) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | PID AR NONGENOMIC PATHWAY | Nongenotropic Androgen signaling |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | REACTOME NEF MEDIATED DOWNREGULATION OF MHC CLASS I COMPLEX CELL SURFACE EXPRESSION | Genes involved in Nef mediated downregulation of MHC class I complex cell surface expression |