Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
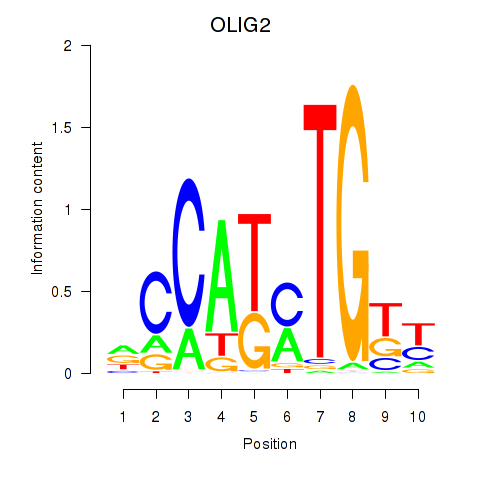
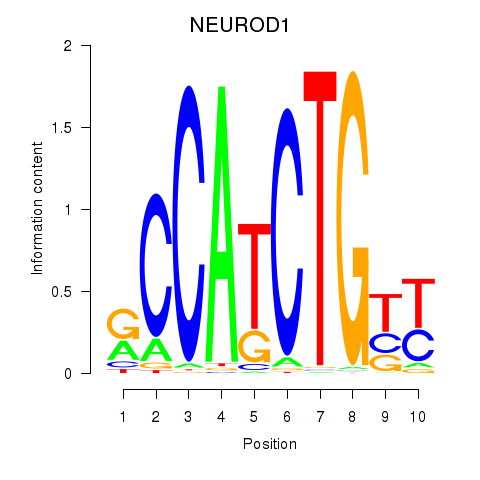
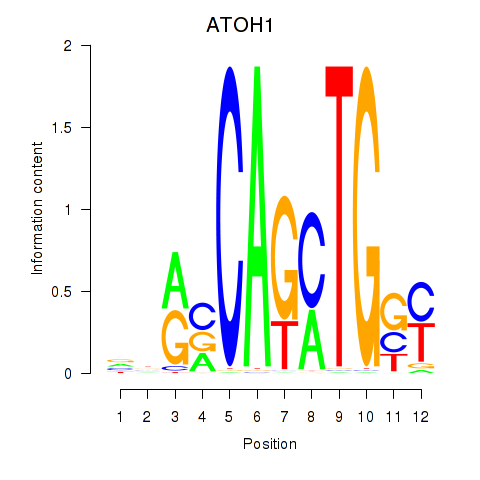
Results for OLIG2_NEUROD1_ATOH1
Z-value: 0.98
Transcription factors associated with OLIG2_NEUROD1_ATOH1
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OLIG2
|
ENSG00000205927.4 | OLIG2 |
|
NEUROD1
|
ENSG00000162992.3 | NEUROD1 |
|
ATOH1
|
ENSG00000172238.3 | ATOH1 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| NEUROD1 | hg19_v2_chr2_-_182545603_182545603 | -0.90 | 2.4e-03 | Click! |
| OLIG2 | hg19_v2_chr21_+_34398153_34398250 | 0.55 | 1.6e-01 | Click! |
| ATOH1 | hg19_v2_chr4_+_94750014_94750042 | 0.43 | 2.9e-01 | Click! |
Activity profile of OLIG2_NEUROD1_ATOH1 motif
Sorted Z-values of OLIG2_NEUROD1_ATOH1 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of OLIG2_NEUROD1_ATOH1
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr19_-_38743878 | 1.32 |
ENST00000587515.1 |
PPP1R14A |
protein phosphatase 1, regulatory (inhibitor) subunit 14A |
| chr12_+_4382917 | 1.09 |
ENST00000261254.3 |
CCND2 |
cyclin D2 |
| chr17_-_56492989 | 1.01 |
ENST00000583753.1 |
RNF43 |
ring finger protein 43 |
| chr4_+_1795012 | 0.99 |
ENST00000481110.2 ENST00000340107.4 ENST00000440486.2 ENST00000412135.2 |
FGFR3 |
fibroblast growth factor receptor 3 |
| chr22_+_45072925 | 0.96 |
ENST00000006251.7 |
PRR5 |
proline rich 5 (renal) |
| chr22_+_45072958 | 0.95 |
ENST00000403581.1 |
PRR5 |
proline rich 5 (renal) |
| chr20_+_58179582 | 0.92 |
ENST00000371015.1 ENST00000395639.4 |
PHACTR3 |
phosphatase and actin regulator 3 |
| chr2_+_69001913 | 0.79 |
ENST00000409030.3 ENST00000409220.1 |
ARHGAP25 |
Rho GTPase activating protein 25 |
| chr6_+_30851840 | 0.77 |
ENST00000511510.1 ENST00000376569.3 ENST00000376575.3 ENST00000376570.4 ENST00000446312.1 ENST00000504927.1 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr12_+_41086297 | 0.73 |
ENST00000551295.2 |
CNTN1 |
contactin 1 |
| chr16_+_23847267 | 0.72 |
ENST00000321728.7 |
PRKCB |
protein kinase C, beta |
| chr12_+_107712173 | 0.69 |
ENST00000280758.5 ENST00000420571.2 |
BTBD11 |
BTB (POZ) domain containing 11 |
| chr8_-_66701319 | 0.66 |
ENST00000379419.4 |
PDE7A |
phosphodiesterase 7A |
| chr7_-_41742697 | 0.65 |
ENST00000242208.4 |
INHBA |
inhibin, beta A |
| chr5_+_167718604 | 0.63 |
ENST00000265293.4 |
WWC1 |
WW and C2 domain containing 1 |
| chr12_-_52845910 | 0.63 |
ENST00000252252.3 |
KRT6B |
keratin 6B |
| chr6_+_150464155 | 0.63 |
ENST00000361131.4 |
PPP1R14C |
protein phosphatase 1, regulatory (inhibitor) subunit 14C |
| chr16_-_84273304 | 0.58 |
ENST00000308251.4 ENST00000568181.1 |
KCNG4 |
potassium voltage-gated channel, subfamily G, member 4 |
| chr1_+_172422026 | 0.56 |
ENST00000367725.4 |
C1orf105 |
chromosome 1 open reading frame 105 |
| chr11_+_128634589 | 0.55 |
ENST00000281428.8 |
FLI1 |
Fli-1 proto-oncogene, ETS transcription factor |
| chr10_-_116444371 | 0.52 |
ENST00000533213.2 ENST00000369252.4 |
ABLIM1 |
actin binding LIM protein 1 |
| chr17_-_77925806 | 0.51 |
ENST00000574241.2 |
TBC1D16 |
TBC1 domain family, member 16 |
| chr8_-_135522425 | 0.51 |
ENST00000521673.1 |
ZFAT |
zinc finger and AT hook domain containing |
| chr3_-_52486841 | 0.50 |
ENST00000496590.1 |
TNNC1 |
troponin C type 1 (slow) |
| chr19_-_11450249 | 0.50 |
ENST00000222120.3 |
RAB3D |
RAB3D, member RAS oncogene family |
| chr19_-_6720686 | 0.49 |
ENST00000245907.6 |
C3 |
complement component 3 |
| chr15_+_40532058 | 0.49 |
ENST00000260404.4 |
PAK6 |
p21 protein (Cdc42/Rac)-activated kinase 6 |
| chr17_-_3595181 | 0.47 |
ENST00000552050.1 |
P2RX5 |
purinergic receptor P2X, ligand-gated ion channel, 5 |
| chr19_-_4338783 | 0.47 |
ENST00000601482.1 ENST00000600324.1 |
STAP2 |
signal transducing adaptor family member 2 |
| chr19_+_45281118 | 0.46 |
ENST00000270279.3 ENST00000341505.4 |
CBLC |
Cbl proto-oncogene C, E3 ubiquitin protein ligase |
| chr19_+_41222998 | 0.43 |
ENST00000263370.2 |
ITPKC |
inositol-trisphosphate 3-kinase C |
| chr6_-_66417107 | 0.42 |
ENST00000370621.3 ENST00000370618.3 ENST00000503581.1 ENST00000393380.2 |
EYS |
eyes shut homolog (Drosophila) |
| chr11_-_108408895 | 0.42 |
ENST00000443411.1 ENST00000533052.1 |
EXPH5 |
exophilin 5 |
| chrX_-_102565932 | 0.41 |
ENST00000372674.1 ENST00000372677.3 |
BEX2 |
brain expressed X-linked 2 |
| chr1_+_28261621 | 0.41 |
ENST00000549094.1 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr10_-_62149433 | 0.40 |
ENST00000280772.2 |
ANK3 |
ankyrin 3, node of Ranvier (ankyrin G) |
| chr3_+_49840685 | 0.40 |
ENST00000333323.4 |
FAM212A |
family with sequence similarity 212, member A |
| chr21_+_30502806 | 0.40 |
ENST00000399928.1 ENST00000399926.1 |
MAP3K7CL |
MAP3K7 C-terminal like |
| chr11_+_124933191 | 0.40 |
ENST00000532000.1 ENST00000308074.4 |
SLC37A2 |
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr1_-_153363452 | 0.40 |
ENST00000368732.1 ENST00000368733.3 |
S100A8 |
S100 calcium binding protein A8 |
| chr1_-_24469602 | 0.39 |
ENST00000270800.1 |
IL22RA1 |
interleukin 22 receptor, alpha 1 |
| chr11_+_124932986 | 0.39 |
ENST00000407458.1 ENST00000298280.5 |
SLC37A2 |
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr9_-_33447584 | 0.39 |
ENST00000297991.4 |
AQP3 |
aquaporin 3 (Gill blood group) |
| chr1_+_155051305 | 0.39 |
ENST00000368408.3 |
EFNA3 |
ephrin-A3 |
| chr11_+_124932955 | 0.39 |
ENST00000403796.2 |
SLC37A2 |
solute carrier family 37 (glucose-6-phosphate transporter), member 2 |
| chr14_+_61788429 | 0.39 |
ENST00000332981.5 |
PRKCH |
protein kinase C, eta |
| chr1_+_28261492 | 0.38 |
ENST00000373894.3 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr17_-_39274606 | 0.38 |
ENST00000391413.2 |
KRTAP4-11 |
keratin associated protein 4-11 |
| chr13_-_20767037 | 0.38 |
ENST00000382848.4 |
GJB2 |
gap junction protein, beta 2, 26kDa |
| chr1_+_92632542 | 0.37 |
ENST00000409154.4 ENST00000370378.4 |
KIAA1107 |
KIAA1107 |
| chr5_+_66124590 | 0.37 |
ENST00000490016.2 ENST00000403666.1 ENST00000450827.1 |
MAST4 |
microtubule associated serine/threonine kinase family member 4 |
| chr17_+_39394250 | 0.36 |
ENST00000254072.6 |
KRTAP9-8 |
keratin associated protein 9-8 |
| chr19_+_15904761 | 0.36 |
ENST00000308940.8 |
OR10H5 |
olfactory receptor, family 10, subfamily H, member 5 |
| chr19_-_1650666 | 0.35 |
ENST00000588136.1 |
TCF3 |
transcription factor 3 |
| chr6_+_159290917 | 0.34 |
ENST00000367072.1 |
C6orf99 |
chromosome 6 open reading frame 99 |
| chr11_+_7597639 | 0.33 |
ENST00000533792.1 |
PPFIBP2 |
PTPRF interacting protein, binding protein 2 (liprin beta 2) |
| chr17_+_39382900 | 0.33 |
ENST00000377721.3 ENST00000455970.2 |
KRTAP9-2 |
keratin associated protein 9-2 |
| chr6_+_30852130 | 0.33 |
ENST00000428153.2 ENST00000376568.3 ENST00000452441.1 ENST00000515219.1 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr21_-_34185944 | 0.33 |
ENST00000479548.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr1_-_207206092 | 0.32 |
ENST00000359470.5 ENST00000461135.2 |
C1orf116 |
chromosome 1 open reading frame 116 |
| chr1_-_205391178 | 0.32 |
ENST00000367153.4 ENST00000367151.2 ENST00000391936.2 ENST00000367149.3 |
LEMD1 |
LEM domain containing 1 |
| chr11_+_76494253 | 0.32 |
ENST00000333090.4 |
TSKU |
tsukushi, small leucine rich proteoglycan |
| chr20_+_42984330 | 0.32 |
ENST00000316673.4 ENST00000609795.1 ENST00000457232.1 ENST00000609262.1 |
HNF4A |
hepatocyte nuclear factor 4, alpha |
| chr16_+_618837 | 0.31 |
ENST00000409439.2 |
PIGQ |
phosphatidylinositol glycan anchor biosynthesis, class Q |
| chr3_+_189349162 | 0.31 |
ENST00000264731.3 ENST00000382063.4 ENST00000418709.2 ENST00000320472.5 ENST00000392460.3 ENST00000440651.2 |
TP63 |
tumor protein p63 |
| chr12_+_56473628 | 0.31 |
ENST00000549282.1 ENST00000549061.1 ENST00000267101.3 |
ERBB3 |
v-erb-b2 avian erythroblastic leukemia viral oncogene homolog 3 |
| chr14_+_61789382 | 0.30 |
ENST00000555082.1 |
PRKCH |
protein kinase C, eta |
| chr11_+_57365150 | 0.30 |
ENST00000457869.1 ENST00000340687.6 ENST00000378323.4 ENST00000378324.2 ENST00000403558.1 |
SERPING1 |
serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 |
| chr16_+_83932684 | 0.29 |
ENST00000262430.4 |
MLYCD |
malonyl-CoA decarboxylase |
| chr1_-_39339777 | 0.29 |
ENST00000397572.2 |
MYCBP |
MYC binding protein |
| chr22_-_37882395 | 0.29 |
ENST00000416983.3 ENST00000424765.2 ENST00000356998.3 |
MFNG |
MFNG O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase |
| chr4_+_77356248 | 0.28 |
ENST00000296043.6 |
SHROOM3 |
shroom family member 3 |
| chr15_+_89182178 | 0.27 |
ENST00000559876.1 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr21_-_34185989 | 0.27 |
ENST00000487113.1 ENST00000382373.4 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr11_-_63933504 | 0.26 |
ENST00000255681.6 |
MACROD1 |
MACRO domain containing 1 |
| chr21_-_34186006 | 0.25 |
ENST00000490358.1 |
C21orf62 |
chromosome 21 open reading frame 62 |
| chr1_-_38273840 | 0.25 |
ENST00000373044.2 |
YRDC |
yrdC N(6)-threonylcarbamoyltransferase domain containing |
| chr7_+_30174426 | 0.25 |
ENST00000324453.8 |
C7orf41 |
maturin, neural progenitor differentiation regulator homolog (Xenopus) |
| chr19_-_49567124 | 0.25 |
ENST00000301411.3 |
NTF4 |
neurotrophin 4 |
| chr4_+_103422471 | 0.24 |
ENST00000226574.4 ENST00000394820.4 |
NFKB1 |
nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| chr6_+_30848557 | 0.24 |
ENST00000460944.2 ENST00000324771.8 |
DDR1 |
discoidin domain receptor tyrosine kinase 1 |
| chr12_-_67072714 | 0.24 |
ENST00000545666.1 ENST00000398016.3 ENST00000359742.4 ENST00000286445.7 ENST00000538211.1 |
GRIP1 |
glutamate receptor interacting protein 1 |
| chr17_-_47841485 | 0.24 |
ENST00000506156.1 ENST00000240364.2 |
FAM117A |
family with sequence similarity 117, member A |
| chr12_+_6309963 | 0.24 |
ENST00000382515.2 |
CD9 |
CD9 molecule |
| chr8_-_20040601 | 0.24 |
ENST00000265808.7 ENST00000522513.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chrX_-_32173579 | 0.24 |
ENST00000359836.1 ENST00000343523.2 ENST00000378707.3 ENST00000541735.1 ENST00000474231.1 |
DMD |
dystrophin |
| chr8_-_20040638 | 0.24 |
ENST00000519026.1 ENST00000276373.5 ENST00000440926.1 ENST00000437980.1 |
SLC18A1 |
solute carrier family 18 (vesicular monoamine transporter), member 1 |
| chr11_-_72070206 | 0.23 |
ENST00000544382.1 |
CLPB |
ClpB caseinolytic peptidase B homolog (E. coli) |
| chr22_-_37823468 | 0.23 |
ENST00000402918.2 |
ELFN2 |
extracellular leucine-rich repeat and fibronectin type III domain containing 2 |
| chr12_+_32655110 | 0.23 |
ENST00000546442.1 ENST00000583694.1 |
FGD4 |
FYVE, RhoGEF and PH domain containing 4 |
| chr8_-_80993010 | 0.23 |
ENST00000537855.1 ENST00000520527.1 ENST00000517427.1 ENST00000448733.2 ENST00000379097.3 |
TPD52 |
tumor protein D52 |
| chrX_-_30993201 | 0.23 |
ENST00000288422.2 ENST00000378932.2 |
TAB3 |
TGF-beta activated kinase 1/MAP3K7 binding protein 3 |
| chr3_-_71774516 | 0.23 |
ENST00000425534.3 |
EIF4E3 |
eukaryotic translation initiation factor 4E family member 3 |
| chr1_-_171621815 | 0.23 |
ENST00000037502.6 |
MYOC |
myocilin, trabecular meshwork inducible glucocorticoid response |
| chr3_+_111717511 | 0.23 |
ENST00000478951.1 ENST00000393917.2 |
TAGLN3 |
transgelin 3 |
| chrX_+_135279179 | 0.22 |
ENST00000370676.3 |
FHL1 |
four and a half LIM domains 1 |
| chr10_-_98031265 | 0.22 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr15_+_89181974 | 0.22 |
ENST00000306072.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chrX_+_135278908 | 0.22 |
ENST00000539015.1 ENST00000370683.1 |
FHL1 |
four and a half LIM domains 1 |
| chr6_+_25279651 | 0.22 |
ENST00000329474.6 |
LRRC16A |
leucine rich repeat containing 16A |
| chr12_+_56415100 | 0.22 |
ENST00000547791.1 |
IKZF4 |
IKAROS family zinc finger 4 (Eos) |
| chr8_-_49834299 | 0.22 |
ENST00000396822.1 |
SNAI2 |
snail family zinc finger 2 |
| chr15_+_89182156 | 0.22 |
ENST00000379224.5 |
ISG20 |
interferon stimulated exonuclease gene 20kDa |
| chr20_+_30640004 | 0.21 |
ENST00000520553.1 ENST00000518730.1 ENST00000375852.2 |
HCK |
hemopoietic cell kinase |
| chr17_+_38333263 | 0.21 |
ENST00000456989.2 ENST00000543876.1 ENST00000544503.1 ENST00000264644.6 ENST00000538884.1 |
RAPGEFL1 |
Rap guanine nucleotide exchange factor (GEF)-like 1 |
| chr4_-_24914508 | 0.21 |
ENST00000504487.1 |
CCDC149 |
coiled-coil domain containing 149 |
| chr11_-_125648690 | 0.21 |
ENST00000436890.2 ENST00000358524.3 |
PATE2 |
prostate and testis expressed 2 |
| chr8_-_49833978 | 0.21 |
ENST00000020945.1 |
SNAI2 |
snail family zinc finger 2 |
| chr1_+_17559776 | 0.20 |
ENST00000537499.1 ENST00000413717.2 ENST00000536552.1 |
PADI1 |
peptidyl arginine deiminase, type I |
| chr20_-_31124186 | 0.20 |
ENST00000375678.3 |
C20orf112 |
chromosome 20 open reading frame 112 |
| chr16_+_29674540 | 0.20 |
ENST00000436527.1 ENST00000360121.3 ENST00000449759.1 |
SPN QPRT |
sialophorin quinolinate phosphoribosyltransferase |
| chr5_-_16936340 | 0.20 |
ENST00000507288.1 ENST00000513610.1 |
MYO10 |
myosin X |
| chr1_+_15480197 | 0.20 |
ENST00000400796.3 ENST00000434578.2 ENST00000376008.2 |
TMEM51 |
transmembrane protein 51 |
| chr19_+_17416457 | 0.20 |
ENST00000252602.1 |
MRPL34 |
mitochondrial ribosomal protein L34 |
| chr1_+_28261533 | 0.20 |
ENST00000411604.1 ENST00000373888.4 |
SMPDL3B |
sphingomyelin phosphodiesterase, acid-like 3B |
| chr10_-_98031310 | 0.20 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr19_+_35485682 | 0.20 |
ENST00000599564.1 |
GRAMD1A |
GRAM domain containing 1A |
| chr12_+_69201923 | 0.20 |
ENST00000462284.1 ENST00000258149.5 ENST00000356290.4 ENST00000540827.1 ENST00000428863.2 ENST00000393412.3 |
MDM2 |
MDM2 oncogene, E3 ubiquitin protein ligase |
| chr1_+_55013889 | 0.19 |
ENST00000343744.2 ENST00000371316.3 |
ACOT11 |
acyl-CoA thioesterase 11 |
| chrX_-_77914825 | 0.19 |
ENST00000321110.1 |
ZCCHC5 |
zinc finger, CCHC domain containing 5 |
| chr11_-_119252359 | 0.19 |
ENST00000455332.2 |
USP2 |
ubiquitin specific peptidase 2 |
| chr6_+_135502501 | 0.19 |
ENST00000527615.1 ENST00000420123.2 ENST00000525369.1 ENST00000528774.1 ENST00000534121.1 ENST00000534044.1 ENST00000533624.1 |
MYB |
v-myb avian myeloblastosis viral oncogene homolog |
| chr4_-_24914576 | 0.19 |
ENST00000502801.1 ENST00000428116.2 |
CCDC149 |
coiled-coil domain containing 149 |
| chr1_+_81771806 | 0.18 |
ENST00000370721.1 ENST00000370727.1 ENST00000370725.1 ENST00000370723.1 ENST00000370728.1 ENST00000370730.1 |
LPHN2 |
latrophilin 2 |
| chr17_-_39296739 | 0.18 |
ENST00000345847.4 |
KRTAP4-6 |
keratin associated protein 4-6 |
| chr13_-_101327028 | 0.18 |
ENST00000328767.5 ENST00000342624.5 ENST00000376234.3 ENST00000423847.1 |
TMTC4 |
transmembrane and tetratricopeptide repeat containing 4 |
| chr6_-_30043539 | 0.18 |
ENST00000376751.3 ENST00000244360.6 |
RNF39 |
ring finger protein 39 |
| chr9_-_140351928 | 0.18 |
ENST00000339554.3 |
NSMF |
NMDA receptor synaptonuclear signaling and neuronal migration factor |
| chr17_-_41738931 | 0.18 |
ENST00000329168.3 ENST00000549132.1 |
MEOX1 |
mesenchyme homeobox 1 |
| chr12_+_6309517 | 0.18 |
ENST00000382519.4 ENST00000009180.4 |
CD9 |
CD9 molecule |
| chr17_+_7792101 | 0.18 |
ENST00000358181.4 ENST00000330494.7 |
CHD3 |
chromodomain helicase DNA binding protein 3 |
| chr19_+_54371114 | 0.18 |
ENST00000448420.1 ENST00000439000.1 ENST00000391770.4 ENST00000391771.1 |
MYADM |
myeloid-associated differentiation marker |
| chr1_-_154842741 | 0.18 |
ENST00000271915.4 |
KCNN3 |
potassium intermediate/small conductance calcium-activated channel, subfamily N, member 3 |
| chr17_-_41739283 | 0.17 |
ENST00000393661.2 ENST00000318579.4 |
MEOX1 |
mesenchyme homeobox 1 |
| chr13_-_95364389 | 0.17 |
ENST00000376945.2 |
SOX21 |
SRY (sex determining region Y)-box 21 |
| chr8_-_8243968 | 0.17 |
ENST00000520004.1 |
SGK223 |
Tyrosine-protein kinase SgK223 |
| chr17_-_37009882 | 0.17 |
ENST00000378096.3 ENST00000394332.1 ENST00000394333.1 ENST00000577407.1 ENST00000479035.2 |
RPL23 |
ribosomal protein L23 |
| chr7_+_75931861 | 0.17 |
ENST00000248553.6 |
HSPB1 |
heat shock 27kDa protein 1 |
| chr9_+_116298778 | 0.17 |
ENST00000462143.1 |
RGS3 |
regulator of G-protein signaling 3 |
| chr19_+_17416609 | 0.17 |
ENST00000602206.1 |
MRPL34 |
mitochondrial ribosomal protein L34 |
| chr1_+_33231268 | 0.17 |
ENST00000373480.1 |
KIAA1522 |
KIAA1522 |
| chr12_-_57328187 | 0.17 |
ENST00000293502.1 |
SDR9C7 |
short chain dehydrogenase/reductase family 9C, member 7 |
| chr1_-_74663825 | 0.17 |
ENST00000370911.3 ENST00000370909.2 ENST00000354431.4 |
LRRIQ3 |
leucine-rich repeats and IQ motif containing 3 |
| chr17_-_47286729 | 0.17 |
ENST00000300406.2 ENST00000511277.1 ENST00000511673.1 |
GNGT2 |
guanine nucleotide binding protein (G protein), gamma transducing activity polypeptide 2 |
| chr18_+_47088401 | 0.17 |
ENST00000261292.4 ENST00000427224.2 ENST00000580036.1 |
LIPG |
lipase, endothelial |
| chr17_-_39165366 | 0.16 |
ENST00000391588.1 |
KRTAP3-1 |
keratin associated protein 3-1 |
| chr2_+_220325441 | 0.16 |
ENST00000396688.1 |
SPEG |
SPEG complex locus |
| chr12_+_113416191 | 0.16 |
ENST00000342315.4 ENST00000392583.2 |
OAS2 |
2'-5'-oligoadenylate synthetase 2, 69/71kDa |
| chr19_-_42759300 | 0.16 |
ENST00000222329.4 |
ERF |
Ets2 repressor factor |
| chr17_-_41132088 | 0.16 |
ENST00000591916.1 ENST00000451885.2 ENST00000454303.1 |
PTGES3L PTGES3L-AARSD1 |
prostaglandin E synthase 3 (cytosolic)-like PTGES3L-AARSD1 readthrough |
| chr1_+_180897269 | 0.16 |
ENST00000367587.1 |
KIAA1614 |
KIAA1614 |
| chr1_+_15272271 | 0.16 |
ENST00000400797.3 |
KAZN |
kazrin, periplakin interacting protein |
| chr15_+_75940218 | 0.16 |
ENST00000308527.5 |
SNX33 |
sorting nexin 33 |
| chr10_-_97321112 | 0.15 |
ENST00000607232.1 ENST00000371227.4 ENST00000371249.2 ENST00000371247.2 ENST00000371246.2 ENST00000393949.1 ENST00000353505.5 ENST00000347291.4 |
SORBS1 |
sorbin and SH3 domain containing 1 |
| chr18_-_52989525 | 0.15 |
ENST00000457482.3 |
TCF4 |
transcription factor 4 |
| chr7_-_11871815 | 0.15 |
ENST00000423059.4 |
THSD7A |
thrombospondin, type I, domain containing 7A |
| chr11_-_124180733 | 0.15 |
ENST00000357821.2 |
OR8D1 |
olfactory receptor, family 8, subfamily D, member 1 |
| chr17_-_46657473 | 0.15 |
ENST00000332503.5 |
HOXB4 |
homeobox B4 |
| chrX_-_49089771 | 0.15 |
ENST00000376251.1 ENST00000323022.5 ENST00000376265.2 |
CACNA1F |
calcium channel, voltage-dependent, L type, alpha 1F subunit |
| chr18_-_52989217 | 0.15 |
ENST00000570287.2 |
TCF4 |
transcription factor 4 |
| chr6_+_29364416 | 0.15 |
ENST00000383555.2 |
OR12D2 |
olfactory receptor, family 12, subfamily D, member 2 (gene/pseudogene) |
| chr7_-_31380502 | 0.15 |
ENST00000297142.3 |
NEUROD6 |
neuronal differentiation 6 |
| chr7_-_44105158 | 0.15 |
ENST00000297283.3 |
PGAM2 |
phosphoglycerate mutase 2 (muscle) |
| chr8_-_41522779 | 0.15 |
ENST00000522231.1 ENST00000314214.8 ENST00000348036.4 ENST00000457297.1 ENST00000522543.1 |
ANK1 |
ankyrin 1, erythrocytic |
| chr3_-_27763803 | 0.15 |
ENST00000449599.1 |
EOMES |
eomesodermin |
| chr9_-_131644202 | 0.14 |
ENST00000320665.6 ENST00000436267.2 |
CCBL1 |
cysteine conjugate-beta lyase, cytoplasmic |
| chr10_-_97321165 | 0.14 |
ENST00000306402.6 |
SORBS1 |
sorbin and SH3 domain containing 1 |
| chr16_+_57662138 | 0.14 |
ENST00000562414.1 ENST00000561969.1 ENST00000562631.1 ENST00000563445.1 ENST00000565338.1 ENST00000567702.1 |
GPR56 |
G protein-coupled receptor 56 |
| chr4_-_153303658 | 0.14 |
ENST00000296555.5 |
FBXW7 |
F-box and WD repeat domain containing 7, E3 ubiquitin protein ligase |
| chr1_+_110881945 | 0.14 |
ENST00000602849.1 ENST00000487146.2 |
RBM15 |
RNA binding motif protein 15 |
| chr17_-_39254391 | 0.14 |
ENST00000333822.4 |
KRTAP4-8 |
keratin associated protein 4-8 |
| chr12_+_41086215 | 0.14 |
ENST00000547702.1 ENST00000551424.1 |
CNTN1 |
contactin 1 |
| chr16_+_71660079 | 0.14 |
ENST00000565261.1 ENST00000268485.3 ENST00000299952.4 |
MARVELD3 |
MARVEL domain containing 3 |
| chr17_-_7197881 | 0.14 |
ENST00000007699.5 |
YBX2 |
Y box binding protein 2 |
| chr8_+_98788003 | 0.14 |
ENST00000521545.2 |
LAPTM4B |
lysosomal protein transmembrane 4 beta |
| chr7_-_76255444 | 0.14 |
ENST00000454397.1 |
POMZP3 |
POM121 and ZP3 fusion |
| chr19_+_15160130 | 0.14 |
ENST00000427043.3 |
CASP14 |
caspase 14, apoptosis-related cysteine peptidase |
| chr11_-_14913765 | 0.14 |
ENST00000334636.5 |
CYP2R1 |
cytochrome P450, family 2, subfamily R, polypeptide 1 |
| chr19_+_50084561 | 0.13 |
ENST00000246794.5 |
PRRG2 |
proline rich Gla (G-carboxyglutamic acid) 2 |
| chr19_+_16059818 | 0.13 |
ENST00000322107.1 |
OR10H4 |
olfactory receptor, family 10, subfamily H, member 4 |
| chr2_+_191002486 | 0.13 |
ENST00000396974.2 |
C2orf88 |
chromosome 2 open reading frame 88 |
| chr12_-_109221160 | 0.13 |
ENST00000326470.5 |
SSH1 |
slingshot protein phosphatase 1 |
| chr10_+_102759045 | 0.13 |
ENST00000370220.1 |
LZTS2 |
leucine zipper, putative tumor suppressor 2 |
| chr20_+_55205825 | 0.13 |
ENST00000544508.1 |
TFAP2C |
transcription factor AP-2 gamma (activating enhancer binding protein 2 gamma) |
| chr8_+_98788057 | 0.13 |
ENST00000517924.1 |
LAPTM4B |
lysosomal protein transmembrane 4 beta |
| chr16_+_57662419 | 0.13 |
ENST00000388812.4 ENST00000538815.1 ENST00000456916.1 ENST00000567154.1 ENST00000388813.5 ENST00000562558.1 ENST00000566271.2 |
GPR56 |
G protein-coupled receptor 56 |
| chr9_+_131644388 | 0.13 |
ENST00000372600.4 |
LRRC8A |
leucine rich repeat containing 8 family, member A |
| chr10_-_72141330 | 0.13 |
ENST00000395011.1 ENST00000395010.1 |
LRRC20 |
leucine rich repeat containing 20 |
| chr20_-_14318248 | 0.13 |
ENST00000378053.3 ENST00000341420.4 |
FLRT3 |
fibronectin leucine rich transmembrane protein 3 |
| chr8_+_107460147 | 0.13 |
ENST00000442977.2 |
OXR1 |
oxidation resistance 1 |
| chr18_-_53068911 | 0.13 |
ENST00000537856.3 |
TCF4 |
transcription factor 4 |
| chr1_-_116383738 | 0.12 |
ENST00000320238.3 |
NHLH2 |
nescient helix loop helix 2 |
| chr12_-_64616019 | 0.12 |
ENST00000311915.8 ENST00000398055.3 ENST00000544871.1 |
C12orf66 |
chromosome 12 open reading frame 66 |
| chr6_+_26124373 | 0.12 |
ENST00000377791.2 ENST00000602637.1 |
HIST1H2AC |
histone cluster 1, H2ac |
| chr3_-_27764190 | 0.12 |
ENST00000537516.1 |
EOMES |
eomesodermin |
| chr4_-_186877806 | 0.12 |
ENST00000355634.5 |
SORBS2 |
sorbin and SH3 domain containing 2 |
| chr5_+_56111361 | 0.12 |
ENST00000399503.3 |
MAP3K1 |
mitogen-activated protein kinase kinase kinase 1, E3 ubiquitin protein ligase |
| chr19_-_46272106 | 0.12 |
ENST00000560168.1 |
SIX5 |
SIX homeobox 5 |
| chr9_+_131644781 | 0.12 |
ENST00000259324.5 |
LRRC8A |
leucine rich repeat containing 8 family, member A |
| chr9_-_131644306 | 0.12 |
ENST00000302586.3 |
CCBL1 |
cysteine conjugate-beta lyase, cytoplasmic |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0035408 | histone H3-T6 phosphorylation(GO:0035408) |
| 0.2 | 0.6 | GO:0060279 | regulation of ovulation(GO:0060278) positive regulation of ovulation(GO:0060279) |
| 0.2 | 1.0 | GO:0038018 | Wnt receptor catabolic process(GO:0038018) |
| 0.2 | 0.5 | GO:0001798 | positive regulation of type IIa hypersensitivity(GO:0001798) positive regulation of type II hypersensitivity(GO:0002894) |
| 0.1 | 2.0 | GO:0038203 | TORC2 signaling(GO:0038203) |
| 0.1 | 0.7 | GO:0000738 | DNA catabolic process, exonucleolytic(GO:0000738) |
| 0.1 | 0.4 | GO:0070563 | negative regulation of vitamin D receptor signaling pathway(GO:0070563) |
| 0.1 | 0.5 | GO:0032972 | regulation of muscle filament sliding speed(GO:0032972) |
| 0.1 | 1.2 | GO:0015760 | hexose phosphate transport(GO:0015712) glucose-6-phosphate transport(GO:0015760) |
| 0.1 | 0.3 | GO:0002522 | leukocyte migration involved in immune response(GO:0002522) |
| 0.1 | 0.3 | GO:0061073 | ciliary body morphogenesis(GO:0061073) |
| 0.1 | 0.4 | GO:0072658 | maintenance of protein location in membrane(GO:0072658) maintenance of protein location in plasma membrane(GO:0072660) positive regulation of membrane depolarization during cardiac muscle cell action potential(GO:1900827) |
| 0.1 | 0.5 | GO:0018125 | peptidyl-cysteine methylation(GO:0018125) |
| 0.1 | 1.3 | GO:0038063 | collagen-activated tyrosine kinase receptor signaling pathway(GO:0038063) |
| 0.1 | 0.4 | GO:0044752 | response to human chorionic gonadotropin(GO:0044752) |
| 0.1 | 0.3 | GO:0002302 | CD8-positive, alpha-beta T cell differentiation involved in immune response(GO:0002302) |
| 0.1 | 0.3 | GO:0002949 | tRNA threonylcarbamoyladenosine modification(GO:0002949) |
| 0.1 | 1.0 | GO:0006685 | sphingomyelin catabolic process(GO:0006685) |
| 0.1 | 0.5 | GO:0015842 | aminergic neurotransmitter loading into synaptic vesicle(GO:0015842) |
| 0.1 | 0.1 | GO:0010609 | mRNA localization resulting in posttranscriptional regulation of gene expression(GO:0010609) |
| 0.1 | 0.3 | GO:0007499 | ectoderm and mesoderm interaction(GO:0007499) |
| 0.1 | 0.4 | GO:0070295 | renal water absorption(GO:0070295) |
| 0.1 | 0.7 | GO:0034350 | regulation of glial cell apoptotic process(GO:0034350) negative regulation of glial cell apoptotic process(GO:0034351) |
| 0.1 | 0.4 | GO:0072717 | cellular response to actinomycin D(GO:0072717) |
| 0.1 | 0.4 | GO:0001757 | somite specification(GO:0001757) |
| 0.1 | 0.2 | GO:1904899 | regulation of hepatic stellate cell proliferation(GO:1904897) positive regulation of hepatic stellate cell proliferation(GO:1904899) hepatic stellate cell proliferation(GO:1990922) |
| 0.1 | 0.2 | GO:0007402 | ganglion mother cell fate determination(GO:0007402) |
| 0.1 | 0.3 | GO:0036115 | fatty-acyl-CoA catabolic process(GO:0036115) malonyl-CoA metabolic process(GO:2001293) |
| 0.1 | 0.6 | GO:0035331 | negative regulation of hippo signaling(GO:0035331) |
| 0.1 | 0.2 | GO:0014734 | skeletal muscle hypertrophy(GO:0014734) |
| 0.1 | 0.4 | GO:0002329 | pre-B cell differentiation(GO:0002329) |
| 0.1 | 0.2 | GO:0072709 | cellular response to sorbitol(GO:0072709) |
| 0.1 | 1.1 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.1 | 0.2 | GO:0002184 | cytoplasmic translational termination(GO:0002184) |
| 0.1 | 0.4 | GO:0002326 | B cell lineage commitment(GO:0002326) |
| 0.1 | 0.2 | GO:0046521 | sphingoid catabolic process(GO:0046521) |
| 0.1 | 0.2 | GO:0001808 | negative regulation of type IV hypersensitivity(GO:0001808) |
| 0.0 | 1.2 | GO:0035988 | chondrocyte proliferation(GO:0035988) |
| 0.0 | 0.2 | GO:0036378 | calcitriol biosynthetic process from calciol(GO:0036378) |
| 0.0 | 0.2 | GO:0033123 | positive regulation of cyclic nucleotide catabolic process(GO:0030807) positive regulation of cAMP catabolic process(GO:0030822) positive regulation of purine nucleotide catabolic process(GO:0033123) |
| 0.0 | 0.2 | GO:0090038 | negative regulation of protein kinase C signaling(GO:0090038) |
| 0.0 | 0.1 | GO:0098976 | excitatory chemical synaptic transmission(GO:0098976) regulation of AMPA glutamate receptor clustering(GO:1904717) positive regulation of AMPA glutamate receptor clustering(GO:1904719) |
| 0.0 | 0.2 | GO:0099640 | axo-dendritic protein transport(GO:0099640) |
| 0.0 | 0.1 | GO:0042137 | sequestering of neurotransmitter(GO:0042137) |
| 0.0 | 0.2 | GO:0010982 | regulation of high-density lipoprotein particle clearance(GO:0010982) |
| 0.0 | 0.2 | GO:0010956 | negative regulation of calcidiol 1-monooxygenase activity(GO:0010956) |
| 0.0 | 0.2 | GO:0018101 | protein citrullination(GO:0018101) histone citrullination(GO:0036414) |
| 0.0 | 0.3 | GO:2000189 | positive regulation of cholesterol homeostasis(GO:2000189) |
| 0.0 | 0.1 | GO:0018057 | peptidyl-lysine oxidation(GO:0018057) |
| 0.0 | 0.4 | GO:0009414 | response to water deprivation(GO:0009414) |
| 0.0 | 0.3 | GO:0051725 | protein de-ADP-ribosylation(GO:0051725) |
| 0.0 | 0.1 | GO:0043456 | regulation of pentose-phosphate shunt(GO:0043456) |
| 0.0 | 0.1 | GO:0009386 | translational attenuation(GO:0009386) |
| 0.0 | 0.4 | GO:1905247 | positive regulation of aspartic-type endopeptidase activity involved in amyloid precursor protein catabolic process(GO:1902961) positive regulation of aspartic-type peptidase activity(GO:1905247) |
| 0.0 | 0.4 | GO:0032119 | sequestering of zinc ion(GO:0032119) |
| 0.0 | 0.3 | GO:0018377 | protein myristoylation(GO:0018377) |
| 0.0 | 0.1 | GO:0007206 | phospholipase C-activating G-protein coupled glutamate receptor signaling pathway(GO:0007206) |
| 0.0 | 0.1 | GO:1902723 | negative regulation of skeletal muscle cell proliferation(GO:0014859) negative regulation of skeletal muscle satellite cell proliferation(GO:1902723) |
| 0.0 | 0.1 | GO:0098968 | neurotransmitter receptor transport postsynaptic membrane to endosome(GO:0098968) |
| 0.0 | 0.6 | GO:0007175 | negative regulation of epidermal growth factor-activated receptor activity(GO:0007175) |
| 0.0 | 0.1 | GO:0038043 | interleukin-5-mediated signaling pathway(GO:0038043) |
| 0.0 | 0.1 | GO:2000639 | regulation of SREBP signaling pathway(GO:2000638) negative regulation of SREBP signaling pathway(GO:2000639) |
| 0.0 | 0.1 | GO:0035803 | egg coat formation(GO:0035803) |
| 0.0 | 0.2 | GO:1904637 | response to ionomycin(GO:1904636) cellular response to ionomycin(GO:1904637) |
| 0.0 | 0.1 | GO:0032510 | endosome to lysosome transport via multivesicular body sorting pathway(GO:0032510) |
| 0.0 | 0.1 | GO:0015917 | aminophospholipid transport(GO:0015917) |
| 0.0 | 0.6 | GO:0050962 | detection of light stimulus involved in visual perception(GO:0050908) detection of light stimulus involved in sensory perception(GO:0050962) |
| 0.0 | 0.0 | GO:2000297 | negative regulation of synapse maturation(GO:2000297) |
| 0.0 | 0.2 | GO:1902414 | protein localization to cell junction(GO:1902414) |
| 0.0 | 0.1 | GO:0097212 | cadmium ion homeostasis(GO:0055073) lysosomal membrane organization(GO:0097212) negative regulation of hydrogen peroxide catabolic process(GO:2000296) regulation of oxygen metabolic process(GO:2000374) |
| 0.0 | 0.3 | GO:0001867 | complement activation, lectin pathway(GO:0001867) |
| 0.0 | 0.0 | GO:0048807 | female genitalia morphogenesis(GO:0048807) |
| 0.0 | 0.1 | GO:1904397 | negative regulation of neuromuscular junction development(GO:1904397) |
| 0.0 | 0.1 | GO:0060373 | regulation of ventricular cardiac muscle cell membrane depolarization(GO:0060373) |
| 0.0 | 0.2 | GO:0048539 | bone marrow development(GO:0048539) |
| 0.0 | 0.1 | GO:0002337 | B-1a B cell differentiation(GO:0002337) |
| 0.0 | 0.2 | GO:0014809 | regulation of skeletal muscle contraction by regulation of release of sequestered calcium ion(GO:0014809) |
| 0.0 | 0.5 | GO:0003334 | keratinocyte development(GO:0003334) |
| 0.0 | 0.6 | GO:0035855 | megakaryocyte development(GO:0035855) |
| 0.0 | 0.3 | GO:0045176 | apical protein localization(GO:0045176) |
| 0.0 | 0.3 | GO:0045475 | locomotor rhythm(GO:0045475) |
| 0.0 | 0.1 | GO:0034970 | histone H3-R2 methylation(GO:0034970) |
| 0.0 | 0.3 | GO:2001224 | positive regulation of neuron migration(GO:2001224) |
| 0.0 | 0.4 | GO:0060712 | spongiotrophoblast layer development(GO:0060712) |
| 0.0 | 0.1 | GO:0000379 | tRNA-type intron splice site recognition and cleavage(GO:0000379) |
| 0.0 | 0.7 | GO:0010765 | positive regulation of sodium ion transport(GO:0010765) |
| 0.0 | 0.1 | GO:2000980 | regulation of auditory receptor cell differentiation(GO:0045607) regulation of mechanoreceptor differentiation(GO:0045631) regulation of inner ear receptor cell differentiation(GO:2000980) |
| 0.0 | 0.6 | GO:0007398 | ectoderm development(GO:0007398) |
| 0.0 | 0.1 | GO:0007221 | positive regulation of transcription of Notch receptor target(GO:0007221) |
| 0.0 | 0.0 | GO:0030950 | establishment or maintenance of actin cytoskeleton polarity(GO:0030950) |
| 0.0 | 0.1 | GO:0044333 | Wnt signaling pathway involved in digestive tract morphogenesis(GO:0044333) |
| 0.0 | 0.1 | GO:0007538 | primary sex determination(GO:0007538) |
| 0.0 | 0.4 | GO:0006198 | cAMP catabolic process(GO:0006198) |
| 0.0 | 0.2 | GO:0036089 | cleavage furrow formation(GO:0036089) |
| 0.0 | 0.0 | GO:0019836 | hemolysis by symbiont of host erythrocytes(GO:0019836) hemolysis in other organism(GO:0044179) hemolysis in other organism involved in symbiotic interaction(GO:0052331) |
| 0.0 | 0.8 | GO:0050850 | positive regulation of calcium-mediated signaling(GO:0050850) |
| 0.0 | 0.1 | GO:1904262 | negative regulation of TORC1 signaling(GO:1904262) |
| 0.0 | 0.1 | GO:0061051 | positive regulation of cell growth involved in cardiac muscle cell development(GO:0061051) |
| 0.0 | 0.1 | GO:0086024 | adrenergic receptor signaling pathway involved in positive regulation of heart rate(GO:0086024) |
| 0.0 | 0.1 | GO:1901842 | negative regulation of high voltage-gated calcium channel activity(GO:1901842) |
| 0.0 | 0.0 | GO:0043449 | olfactory learning(GO:0008355) cellular alkene metabolic process(GO:0043449) |
| 0.0 | 0.3 | GO:0016254 | preassembly of GPI anchor in ER membrane(GO:0016254) |
| 0.0 | 0.1 | GO:1904627 | response to phorbol 13-acetate 12-myristate(GO:1904627) cellular response to phorbol 13-acetate 12-myristate(GO:1904628) |
| 0.0 | 0.1 | GO:0003344 | pericardium morphogenesis(GO:0003344) |
| 0.0 | 0.1 | GO:0030046 | parallel actin filament bundle assembly(GO:0030046) |
| 0.0 | 0.1 | GO:0061739 | protein lipidation involved in autophagosome assembly(GO:0061739) |
| 0.0 | 0.2 | GO:0003301 | physiological muscle hypertrophy(GO:0003298) physiological cardiac muscle hypertrophy(GO:0003301) cell growth involved in cardiac muscle cell development(GO:0061049) |
| 0.0 | 1.2 | GO:0035690 | cellular response to drug(GO:0035690) |
| 0.0 | 0.0 | GO:0030311 | poly-N-acetyllactosamine biosynthetic process(GO:0030311) |
| 0.0 | 0.0 | GO:0042560 | folic acid-containing compound catabolic process(GO:0009397) pteridine-containing compound catabolic process(GO:0042560) |
| 0.0 | 0.0 | GO:0048170 | positive regulation of long-term neuronal synaptic plasticity(GO:0048170) |
| 0.0 | 0.1 | GO:0032224 | regulation of synaptic transmission, cholinergic(GO:0032222) positive regulation of synaptic transmission, cholinergic(GO:0032224) |
| 0.0 | 0.0 | GO:0016185 | synaptic vesicle budding from presynaptic endocytic zone membrane(GO:0016185) synaptic vesicle budding(GO:0070142) |
| 0.0 | 0.0 | GO:0097032 | respiratory chain complex II assembly(GO:0034552) mitochondrial respiratory chain complex II assembly(GO:0034553) mitochondrial respiratory chain complex II biogenesis(GO:0097032) |
| 0.0 | 0.1 | GO:0046886 | positive regulation of hormone biosynthetic process(GO:0046886) |
| 0.0 | 0.0 | GO:0002215 | defense response to nematode(GO:0002215) |
| 0.0 | 0.0 | GO:0019072 | viral genome packaging(GO:0019072) viral RNA genome packaging(GO:0019074) |
| 0.0 | 0.1 | GO:0032049 | cardiolipin biosynthetic process(GO:0032049) |
| 0.0 | 0.1 | GO:0033353 | S-adenosylmethionine cycle(GO:0033353) |
| 0.0 | 0.0 | GO:0072061 | inner medullary collecting duct development(GO:0072061) |
| 0.0 | 0.1 | GO:0019230 | proprioception(GO:0019230) |
| 0.0 | 0.1 | GO:0044240 | multicellular organism lipid catabolic process(GO:0044240) |
| 0.0 | 0.0 | GO:0021633 | optic nerve structural organization(GO:0021633) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.2 | 0.6 | GO:0043511 | inhibin complex(GO:0043511) inhibin A complex(GO:0043512) |
| 0.2 | 1.3 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.1 | 2.0 | GO:0031932 | TORC2 complex(GO:0031932) |
| 0.1 | 0.5 | GO:1990584 | cardiac Troponin complex(GO:1990584) |
| 0.1 | 0.1 | GO:0060200 | clathrin-sculpted acetylcholine transport vesicle(GO:0060200) clathrin-sculpted acetylcholine transport vesicle membrane(GO:0060201) |
| 0.1 | 0.3 | GO:0005899 | insulin receptor complex(GO:0005899) |
| 0.1 | 0.5 | GO:0070083 | clathrin-sculpted monoamine transport vesicle(GO:0070081) clathrin-sculpted monoamine transport vesicle membrane(GO:0070083) |
| 0.1 | 0.2 | GO:0018444 | translation release factor complex(GO:0018444) |
| 0.0 | 0.1 | GO:0060203 | clathrin-sculpted glutamate transport vesicle(GO:0060199) clathrin-sculpted glutamate transport vesicle membrane(GO:0060203) |
| 0.0 | 0.1 | GO:0000814 | ESCRT II complex(GO:0000814) |
| 0.0 | 0.4 | GO:0097449 | astrocyte projection(GO:0097449) |
| 0.0 | 0.6 | GO:0042588 | zymogen granule(GO:0042588) |
| 0.0 | 0.6 | GO:0014731 | spectrin-associated cytoskeleton(GO:0014731) |
| 0.0 | 0.1 | GO:1990032 | parallel fiber(GO:1990032) |
| 0.0 | 2.1 | GO:0045095 | keratin filament(GO:0045095) |
| 0.0 | 0.3 | GO:0000506 | glycosylphosphatidylinositol-N-acetylglucosaminyltransferase (GPI-GnT) complex(GO:0000506) |
| 0.0 | 0.2 | GO:0016013 | syntrophin complex(GO:0016013) |
| 0.0 | 0.2 | GO:0033256 | I-kappaB/NF-kappaB complex(GO:0033256) |
| 0.0 | 0.1 | GO:1990452 | Parkin-FBXW7-Cul1 ubiquitin ligase complex(GO:1990452) |
| 0.0 | 0.5 | GO:0005639 | integral component of nuclear inner membrane(GO:0005639) intrinsic component of nuclear inner membrane(GO:0031229) |
| 0.0 | 0.1 | GO:0034750 | Scrib-APC-beta-catenin complex(GO:0034750) |
| 0.0 | 0.3 | GO:0090665 | dystrophin-associated glycoprotein complex(GO:0016010) glycoprotein complex(GO:0090665) |
| 0.0 | 0.1 | GO:0000214 | tRNA-intron endonuclease complex(GO:0000214) |
| 0.0 | 0.4 | GO:0031092 | platelet alpha granule membrane(GO:0031092) |
| 0.0 | 0.2 | GO:0032433 | filopodium tip(GO:0032433) |
| 0.0 | 0.2 | GO:0034518 | mRNA cap binding complex(GO:0005845) RNA cap binding complex(GO:0034518) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.3 | 0.8 | GO:0035403 | histone kinase activity (H3-T6 specific)(GO:0035403) |
| 0.3 | 1.3 | GO:0038062 | protein tyrosine kinase collagen receptor activity(GO:0038062) |
| 0.2 | 0.7 | GO:0008859 | exoribonuclease II activity(GO:0008859) |
| 0.2 | 0.6 | GO:0070699 | type II activin receptor binding(GO:0070699) |
| 0.2 | 1.2 | GO:0015526 | hexose phosphate transmembrane transporter activity(GO:0015119) organophosphate:inorganic phosphate antiporter activity(GO:0015315) hexose-phosphate:inorganic phosphate antiporter activity(GO:0015526) glucose 6-phosphate:inorganic phosphate antiporter activity(GO:0061513) |
| 0.2 | 0.5 | GO:0015222 | serotonin transmembrane transporter activity(GO:0015222) |
| 0.1 | 0.7 | GO:0004699 | calcium-independent protein kinase C activity(GO:0004699) |
| 0.1 | 1.0 | GO:0005007 | fibroblast growth factor-activated receptor activity(GO:0005007) |
| 0.1 | 0.4 | GO:0015254 | glycerol channel activity(GO:0015254) |
| 0.1 | 0.3 | GO:0033829 | O-fucosylpeptide 3-beta-N-acetylglucosaminyltransferase activity(GO:0033829) |
| 0.1 | 1.0 | GO:0004767 | sphingomyelin phosphodiesterase activity(GO:0004767) |
| 0.1 | 0.4 | GO:0070644 | vitamin D response element binding(GO:0070644) |
| 0.1 | 0.5 | GO:0035662 | Toll-like receptor 4 binding(GO:0035662) |
| 0.1 | 0.4 | GO:0042015 | interleukin-20 binding(GO:0042015) |
| 0.1 | 0.2 | GO:0061663 | NEDD8 ligase activity(GO:0061663) |
| 0.1 | 0.2 | GO:0004514 | nicotinate-nucleotide diphosphorylase (carboxylating) activity(GO:0004514) |
| 0.0 | 0.4 | GO:0005005 | transmembrane-ephrin receptor activity(GO:0005005) |
| 0.0 | 0.2 | GO:0070643 | vitamin D3 25-hydroxylase activity(GO:0030343) vitamin D 25-hydroxylase activity(GO:0070643) |
| 0.0 | 0.4 | GO:0008440 | inositol-1,4,5-trisphosphate 3-kinase activity(GO:0008440) |
| 0.0 | 3.1 | GO:0004864 | protein phosphatase inhibitor activity(GO:0004864) |
| 0.0 | 0.2 | GO:0070180 | large ribosomal subunit rRNA binding(GO:0070180) |
| 0.0 | 0.5 | GO:0004931 | extracellular ATP-gated cation channel activity(GO:0004931) ATP-gated ion channel activity(GO:0035381) |
| 0.0 | 0.2 | GO:0008426 | protein kinase C inhibitor activity(GO:0008426) |
| 0.0 | 0.5 | GO:0001011 | transcription factor activity, sequence-specific DNA binding, RNA polymerase recruiting(GO:0001011) transcription factor activity, TFIIB-class binding(GO:0001087) |
| 0.0 | 0.5 | GO:0031013 | troponin I binding(GO:0031013) |
| 0.0 | 0.2 | GO:0004668 | protein-arginine deiminase activity(GO:0004668) |
| 0.0 | 0.1 | GO:0046538 | bisphosphoglycerate mutase activity(GO:0004082) phosphoglycerate mutase activity(GO:0004619) 2,3-bisphosphoglycerate-dependent phosphoglycerate mutase activity(GO:0046538) |
| 0.0 | 0.2 | GO:0005166 | neurotrophin p75 receptor binding(GO:0005166) |
| 0.0 | 0.2 | GO:0016286 | small conductance calcium-activated potassium channel activity(GO:0016286) |
| 0.0 | 0.1 | GO:0004874 | aryl hydrocarbon receptor activity(GO:0004874) |
| 0.0 | 0.1 | GO:0004914 | interleukin-5 receptor activity(GO:0004914) |
| 0.0 | 1.2 | GO:0005109 | frizzled binding(GO:0005109) |
| 0.0 | 0.3 | GO:0017176 | phosphatidylinositol N-acetylglucosaminyltransferase activity(GO:0017176) |
| 0.0 | 0.2 | GO:0008481 | sphinganine kinase activity(GO:0008481) D-erythro-sphingosine kinase activity(GO:0017050) |
| 0.0 | 0.5 | GO:0004993 | G-protein coupled serotonin receptor activity(GO:0004993) |
| 0.0 | 0.3 | GO:0097371 | MDM2/MDM4 family protein binding(GO:0097371) |
| 0.0 | 0.3 | GO:0038132 | neuregulin binding(GO:0038132) |
| 0.0 | 0.3 | GO:0001730 | 2'-5'-oligoadenylate synthetase activity(GO:0001730) |
| 0.0 | 0.1 | GO:0004991 | parathyroid hormone receptor activity(GO:0004991) |
| 0.0 | 0.1 | GO:0004720 | protein-lysine 6-oxidase activity(GO:0004720) |
| 0.0 | 0.2 | GO:0060002 | plus-end directed microfilament motor activity(GO:0060002) |
| 0.0 | 0.1 | GO:0015319 | sodium:inorganic phosphate symporter activity(GO:0015319) |
| 0.0 | 0.2 | GO:0004465 | lipoprotein lipase activity(GO:0004465) |
| 0.0 | 0.4 | GO:0005225 | volume-sensitive anion channel activity(GO:0005225) |
| 0.0 | 0.4 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.2 | GO:0008079 | translation release factor activity(GO:0003747) translation termination factor activity(GO:0008079) |
| 0.0 | 0.6 | GO:0005251 | delayed rectifier potassium channel activity(GO:0005251) |
| 0.0 | 0.1 | GO:0050816 | phosphothreonine binding(GO:0050816) |
| 0.0 | 0.1 | GO:0098770 | FBXO family protein binding(GO:0098770) |
| 0.0 | 0.0 | GO:0030272 | 5-formyltetrahydrofolate cyclo-ligase activity(GO:0030272) |
| 0.0 | 0.2 | GO:0001135 | transcription factor activity, RNA polymerase II transcription factor recruiting(GO:0001135) |
| 0.0 | 0.3 | GO:0005243 | gap junction channel activity(GO:0005243) |
| 0.0 | 0.3 | GO:0050998 | nitric-oxide synthase binding(GO:0050998) |
| 0.0 | 0.0 | GO:0005111 | type 2 fibroblast growth factor receptor binding(GO:0005111) |
| 0.0 | 0.2 | GO:0017034 | Rap guanyl-nucleotide exchange factor activity(GO:0017034) |
| 0.0 | 0.4 | GO:0008093 | cytoskeletal adaptor activity(GO:0008093) |
| 0.0 | 0.1 | GO:1901375 | acetylcholine transmembrane transporter activity(GO:0005277) acetate ester transmembrane transporter activity(GO:1901375) |
| 0.0 | 0.8 | GO:0001784 | phosphotyrosine binding(GO:0001784) |
| 0.0 | 0.1 | GO:0008607 | phosphorylase kinase regulator activity(GO:0008607) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.0 | GO:0031859 | platelet activating factor receptor binding(GO:0031859) |
| 0.0 | 0.1 | GO:0032190 | acrosin binding(GO:0032190) |
| 0.0 | 0.4 | GO:0031489 | myosin V binding(GO:0031489) |
| 0.0 | 0.0 | GO:0033878 | hormone-sensitive lipase activity(GO:0033878) |
| 0.0 | 0.1 | GO:0035241 | protein-arginine omega-N monomethyltransferase activity(GO:0035241) |
| 0.0 | 0.0 | GO:0016262 | protein N-acetylglucosaminyltransferase activity(GO:0016262) |
| 0.0 | 0.0 | GO:0005105 | type 1 fibroblast growth factor receptor binding(GO:0005105) |
| 0.0 | 0.1 | GO:0099583 | neurotransmitter receptor activity involved in regulation of postsynaptic cytosolic calcium ion concentration(GO:0099583) |
| 0.0 | 0.5 | GO:0004115 | 3',5'-cyclic-AMP phosphodiesterase activity(GO:0004115) |
| 0.0 | 0.5 | GO:0030332 | cyclin binding(GO:0030332) |
| 0.0 | 0.1 | GO:0000213 | tRNA-intron endonuclease activity(GO:0000213) |
| 0.0 | 0.1 | GO:0001227 | transcriptional repressor activity, RNA polymerase II transcription regulatory region sequence-specific binding(GO:0001227) |
| 0.0 | 0.2 | GO:0047617 | acyl-CoA hydrolase activity(GO:0047617) |
| 0.0 | 0.2 | GO:0005549 | odorant binding(GO:0005549) |
| 0.0 | 0.1 | GO:0005025 | transforming growth factor beta receptor activity, type I(GO:0005025) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID TCR RAS PATHWAY | Ras signaling in the CD4+ TCR pathway |
| 0.0 | 0.5 | SA B CELL RECEPTOR COMPLEXES | Antigen binding to B cell receptors activates protein tyrosine kinases, such as the Src family, which ultimate activate MAP kinases. |
| 0.0 | 1.2 | PID IL2 STAT5 PATHWAY | IL2 signaling events mediated by STAT5 |
| 0.0 | 0.4 | PID IL2 PI3K PATHWAY | IL2 signaling events mediated by PI3K |
| 0.0 | 0.4 | PID ERB GENOMIC PATHWAY | Validated nuclear estrogen receptor beta network |
| 0.0 | 0.5 | PID P38 MK2 PATHWAY | p38 signaling mediated by MAPKAP kinases |
| 0.0 | 1.8 | PID MTOR 4PATHWAY | mTOR signaling pathway |
| 0.0 | 1.6 | PID ILK PATHWAY | Integrin-linked kinase signaling |
| 0.0 | 0.7 | PID ALK1 PATHWAY | ALK1 signaling events |
| 0.0 | 0.3 | SIG INSULIN RECEPTOR PATHWAY IN CARDIAC MYOCYTES | Genes related to the insulin receptor pathway |
| 0.0 | 1.1 | PID FGF PATHWAY | FGF signaling pathway |
| 0.0 | 0.5 | PID EPHA FWDPATHWAY | EPHA forward signaling |
| 0.0 | 0.2 | PID RANBP2 PATHWAY | Sumoylation by RanBP2 regulates transcriptional repression |
| 0.0 | 0.7 | PID TXA2PATHWAY | Thromboxane A2 receptor signaling |
| 0.0 | 1.2 | PID NOTCH PATHWAY | Notch signaling pathway |
| 0.0 | 0.3 | PID ERBB NETWORK PATHWAY | ErbB receptor signaling network |
| 0.0 | 1.3 | PID P73PATHWAY | p73 transcription factor network |
| 0.0 | 0.2 | ST G ALPHA S PATHWAY | G alpha s Pathway |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 1.0 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.1 | 0.7 | REACTOME GLYCOPROTEIN HORMONES | Genes involved in Glycoprotein hormones |
| 0.0 | 0.2 | REACTOME IRAK2 MEDIATED ACTIVATION OF TAK1 COMPLEX UPON TLR7 8 OR 9 STIMULATION | Genes involved in IRAK2 mediated activation of TAK1 complex upon TLR7/8 or 9 stimulation |
| 0.0 | 0.7 | REACTOME TRAFFICKING OF GLUR2 CONTAINING AMPA RECEPTORS | Genes involved in Trafficking of GluR2-containing AMPA receptors |
| 0.0 | 0.8 | REACTOME REGULATION OF SIGNALING BY CBL | Genes involved in Regulation of signaling by CBL |
| 0.0 | 1.3 | REACTOME G1 PHASE | Genes involved in G1 Phase |
| 0.0 | 0.9 | REACTOME ACTIVATED NOTCH1 TRANSMITS SIGNAL TO THE NUCLEUS | Genes involved in Activated NOTCH1 Transmits Signal to the Nucleus |
| 0.0 | 0.5 | REACTOME ACTIVATION OF RAC | Genes involved in Activation of Rac |
| 0.0 | 0.5 | REACTOME NA CL DEPENDENT NEUROTRANSMITTER TRANSPORTERS | Genes involved in Na+/Cl- dependent neurotransmitter transporters |
| 0.0 | 0.3 | REACTOME PASSIVE TRANSPORT BY AQUAPORINS | Genes involved in Passive Transport by Aquaporins |
| 0.0 | 0.6 | REACTOME SIGNALING BY HIPPO | Genes involved in Signaling by Hippo |
| 0.0 | 0.9 | REACTOME MYOGENESIS | Genes involved in Myogenesis |
| 0.0 | 0.7 | REACTOME EFFECTS OF PIP2 HYDROLYSIS | Genes involved in Effects of PIP2 hydrolysis |
| 0.0 | 0.3 | REACTOME INTRINSIC PATHWAY | Genes involved in Intrinsic Pathway |
| 0.0 | 0.3 | REACTOME DOWNREGULATION OF ERBB2 ERBB3 SIGNALING | Genes involved in Downregulation of ERBB2:ERBB3 signaling |
| 0.0 | 0.3 | REACTOME GAP JUNCTION ASSEMBLY | Genes involved in Gap junction assembly |
| 0.0 | 0.4 | REACTOME REGULATION OF COMPLEMENT CASCADE | Genes involved in Regulation of Complement cascade |
| 0.0 | 0.3 | REACTOME PRE NOTCH PROCESSING IN GOLGI | Genes involved in Pre-NOTCH Processing in Golgi |
| 0.0 | 0.5 | REACTOME INTERACTION BETWEEN L1 AND ANKYRINS | Genes involved in Interaction between L1 and Ankyrins |
| 0.0 | 0.2 | REACTOME AKT PHOSPHORYLATES TARGETS IN THE CYTOSOL | Genes involved in AKT phosphorylates targets in the cytosol |
| 0.0 | 1.0 | REACTOME INTERFERON ALPHA BETA SIGNALING | Genes involved in Interferon alpha/beta signaling |
| 0.0 | 0.3 | REACTOME SYNTHESIS OF GLYCOSYLPHOSPHATIDYLINOSITOL GPI | Genes involved in Synthesis of glycosylphosphatidylinositol (GPI) |