Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
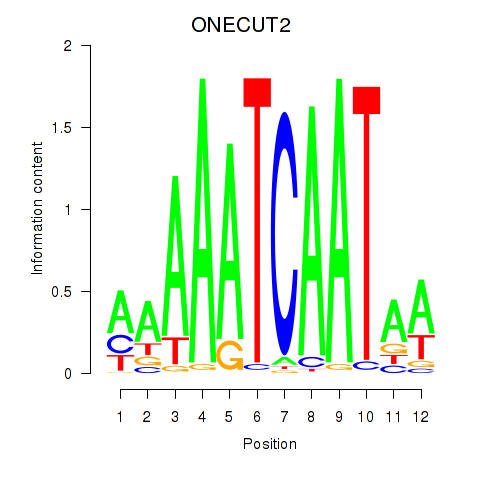
Results for ONECUT2_ONECUT3
Z-value: 0.32
Transcription factors associated with ONECUT2_ONECUT3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
ONECUT2
|
ENSG00000119547.5 | ONECUT2 |
|
ONECUT3
|
ENSG00000205922.4 | ONECUT3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| ONECUT3 | hg19_v2_chr19_+_1752372_1752372 | 0.55 | 1.6e-01 | Click! |
| ONECUT2 | hg19_v2_chr18_+_55102917_55102985 | 0.45 | 2.6e-01 | Click! |
Activity profile of ONECUT2_ONECUT3 motif
Sorted Z-values of ONECUT2_ONECUT3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of ONECUT2_ONECUT3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr12_+_4385230 | 0.41 |
ENST00000536537.1 |
CCND2 |
cyclin D2 |
| chr1_-_116383738 | 0.28 |
ENST00000320238.3 |
NHLH2 |
nescient helix loop helix 2 |
| chr17_+_17685422 | 0.22 |
ENST00000395774.1 |
RAI1 |
retinoic acid induced 1 |
| chr15_-_83837983 | 0.21 |
ENST00000562702.1 |
HDGFRP3 |
Hepatoma-derived growth factor-related protein 3 |
| chr1_-_116383322 | 0.18 |
ENST00000429731.1 |
NHLH2 |
nescient helix loop helix 2 |
| chr5_-_82969405 | 0.18 |
ENST00000510978.1 |
HAPLN1 |
hyaluronan and proteoglycan link protein 1 |
| chr17_+_7461613 | 0.17 |
ENST00000438470.1 ENST00000436057.1 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr11_+_63137251 | 0.16 |
ENST00000310969.4 ENST00000279178.3 |
SLC22A9 |
solute carrier family 22 (organic anion transporter), member 9 |
| chr20_-_1309809 | 0.16 |
ENST00000360779.3 |
SDCBP2 |
syndecan binding protein (syntenin) 2 |
| chr1_+_206730484 | 0.15 |
ENST00000304534.8 |
RASSF5 |
Ras association (RalGDS/AF-6) domain family member 5 |
| chr1_+_159557607 | 0.15 |
ENST00000255040.2 |
APCS |
amyloid P component, serum |
| chr13_-_36429763 | 0.15 |
ENST00000379893.1 |
DCLK1 |
doublecortin-like kinase 1 |
| chr3_+_159557637 | 0.14 |
ENST00000445224.2 |
SCHIP1 |
schwannomin interacting protein 1 |
| chrX_-_117119243 | 0.14 |
ENST00000539496.1 ENST00000469946.1 |
KLHL13 |
kelch-like family member 13 |
| chr3_-_141747439 | 0.14 |
ENST00000467667.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr2_-_192016316 | 0.13 |
ENST00000358470.4 ENST00000432798.1 ENST00000450994.1 |
STAT4 |
signal transducer and activator of transcription 4 |
| chr9_-_27529726 | 0.12 |
ENST00000262244.5 |
MOB3B |
MOB kinase activator 3B |
| chr14_+_37126765 | 0.12 |
ENST00000402703.2 |
PAX9 |
paired box 9 |
| chr10_-_98031265 | 0.12 |
ENST00000224337.5 ENST00000371176.2 |
BLNK |
B-cell linker |
| chr3_-_24207039 | 0.11 |
ENST00000280696.5 |
THRB |
thyroid hormone receptor, beta |
| chr20_-_22566089 | 0.11 |
ENST00000377115.4 |
FOXA2 |
forkhead box A2 |
| chr10_-_98031310 | 0.11 |
ENST00000427367.2 ENST00000413476.2 |
BLNK |
B-cell linker |
| chr12_+_6833237 | 0.11 |
ENST00000229251.3 ENST00000539735.1 ENST00000538410.1 |
COPS7A |
COP9 signalosome subunit 7A |
| chr8_-_145754428 | 0.11 |
ENST00000527462.1 ENST00000313465.5 ENST00000524821.1 |
C8orf82 |
chromosome 8 open reading frame 82 |
| chr14_-_23426322 | 0.11 |
ENST00000555367.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr7_-_104909435 | 0.11 |
ENST00000357311.3 |
SRPK2 |
SRSF protein kinase 2 |
| chr14_-_23426270 | 0.10 |
ENST00000557591.1 ENST00000397409.4 ENST00000490506.1 ENST00000554406.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chr14_-_23426337 | 0.10 |
ENST00000342454.8 ENST00000555986.1 ENST00000541587.1 ENST00000554516.1 ENST00000347758.2 ENST00000206474.7 ENST00000555040.1 |
HAUS4 |
HAUS augmin-like complex, subunit 4 |
| chrX_-_33229636 | 0.10 |
ENST00000357033.4 |
DMD |
dystrophin |
| chr14_+_23012122 | 0.10 |
ENST00000390534.1 |
TRAJ3 |
T cell receptor alpha joining 3 |
| chr4_+_175839506 | 0.09 |
ENST00000505141.1 ENST00000359240.3 ENST00000445694.1 |
ADAM29 |
ADAM metallopeptidase domain 29 |
| chr4_+_175839551 | 0.09 |
ENST00000404450.4 ENST00000514159.1 |
ADAM29 |
ADAM metallopeptidase domain 29 |
| chr3_-_141747459 | 0.09 |
ENST00000477292.1 ENST00000478006.1 ENST00000495310.1 ENST00000486111.1 |
TFDP2 |
transcription factor Dp-2 (E2F dimerization partner 2) |
| chr1_-_17766198 | 0.09 |
ENST00000375436.4 |
RCC2 |
regulator of chromosome condensation 2 |
| chr13_+_97928395 | 0.09 |
ENST00000445661.2 |
MBNL2 |
muscleblind-like splicing regulator 2 |
| chr19_+_13135386 | 0.09 |
ENST00000360105.4 ENST00000588228.1 ENST00000591028.1 |
NFIX |
nuclear factor I/X (CCAAT-binding transcription factor) |
| chr17_-_41623716 | 0.09 |
ENST00000319349.5 |
ETV4 |
ets variant 4 |
| chr8_+_31497271 | 0.08 |
ENST00000520407.1 |
NRG1 |
neuregulin 1 |
| chr17_-_41623691 | 0.08 |
ENST00000545954.1 |
ETV4 |
ets variant 4 |
| chr10_-_127505167 | 0.08 |
ENST00000368786.1 |
UROS |
uroporphyrinogen III synthase |
| chr3_+_63428752 | 0.08 |
ENST00000295894.5 |
SYNPR |
synaptoporin |
| chr10_+_71561704 | 0.08 |
ENST00000520267.1 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr11_+_18154059 | 0.08 |
ENST00000531264.1 |
MRGPRX3 |
MAS-related GPR, member X3 |
| chrX_-_64196307 | 0.08 |
ENST00000545618.1 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chrX_-_64196351 | 0.07 |
ENST00000374839.3 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chrX_-_64196376 | 0.07 |
ENST00000447788.2 |
ZC4H2 |
zinc finger, C4H2 domain containing |
| chr16_-_8962200 | 0.07 |
ENST00000562843.1 ENST00000561530.1 ENST00000396593.2 |
CARHSP1 |
calcium regulated heat stable protein 1, 24kDa |
| chr9_-_138853156 | 0.07 |
ENST00000371756.3 |
UBAC1 |
UBA domain containing 1 |
| chr5_+_139554227 | 0.07 |
ENST00000261811.4 |
CYSTM1 |
cysteine-rich transmembrane module containing 1 |
| chr20_-_1974692 | 0.07 |
ENST00000217305.2 ENST00000539905.1 |
PDYN |
prodynorphin |
| chr4_-_69434245 | 0.07 |
ENST00000317746.2 |
UGT2B17 |
UDP glucuronosyltransferase 2 family, polypeptide B17 |
| chr7_+_129007964 | 0.07 |
ENST00000460109.1 ENST00000474594.1 ENST00000446212.1 |
AHCYL2 |
adenosylhomocysteinase-like 2 |
| chr15_-_99789736 | 0.07 |
ENST00000560235.1 ENST00000394132.2 ENST00000560860.1 ENST00000558078.1 ENST00000394136.1 ENST00000262074.4 ENST00000558613.1 ENST00000394130.1 ENST00000560772.1 |
TTC23 |
tetratricopeptide repeat domain 23 |
| chr10_+_71561649 | 0.07 |
ENST00000398978.3 ENST00000354547.3 ENST00000357811.3 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr12_+_4699244 | 0.06 |
ENST00000540757.2 |
DYRK4 |
dual-specificity tyrosine-(Y)-phosphorylation regulated kinase 4 |
| chr17_-_202579 | 0.06 |
ENST00000577079.1 ENST00000331302.7 ENST00000536489.2 |
RPH3AL |
rabphilin 3A-like (without C2 domains) |
| chr12_+_147052 | 0.06 |
ENST00000594563.1 |
AC026369.1 |
Uncharacterized protein |
| chr10_+_133753533 | 0.06 |
ENST00000422256.2 |
PPP2R2D |
protein phosphatase 2, regulatory subunit B, delta |
| chr16_-_20702578 | 0.06 |
ENST00000307493.4 ENST00000219151.4 |
ACSM1 |
acyl-CoA synthetase medium-chain family member 1 |
| chrX_-_133792480 | 0.06 |
ENST00000359237.4 |
PLAC1 |
placenta-specific 1 |
| chr16_-_75467318 | 0.06 |
ENST00000283882.3 |
CFDP1 |
craniofacial development protein 1 |
| chr1_-_111150048 | 0.06 |
ENST00000485317.1 |
KCNA2 |
potassium voltage-gated channel, shaker-related subfamily, member 2 |
| chr8_-_135522425 | 0.05 |
ENST00000521673.1 |
ZFAT |
zinc finger and AT hook domain containing |
| chr5_-_156772729 | 0.05 |
ENST00000312349.4 |
FNDC9 |
fibronectin type III domain containing 9 |
| chr1_-_29557383 | 0.05 |
ENST00000373791.3 ENST00000263702.6 |
MECR |
mitochondrial trans-2-enoyl-CoA reductase |
| chr9_-_117420052 | 0.05 |
ENST00000423632.1 |
RP11-402G3.5 |
RP11-402G3.5 |
| chr12_-_54653313 | 0.05 |
ENST00000550411.1 ENST00000439541.2 |
CBX5 |
chromobox homolog 5 |
| chr16_-_8962544 | 0.05 |
ENST00000570125.1 |
CARHSP1 |
calcium regulated heat stable protein 1, 24kDa |
| chr1_-_53686261 | 0.05 |
ENST00000294360.4 |
C1orf123 |
chromosome 1 open reading frame 123 |
| chr3_-_187388173 | 0.05 |
ENST00000287641.3 |
SST |
somatostatin |
| chr14_-_24020858 | 0.05 |
ENST00000419474.3 |
ZFHX2 |
zinc finger homeobox 2 |
| chr4_-_70826725 | 0.05 |
ENST00000353151.3 |
CSN2 |
casein beta |
| chrX_+_15525426 | 0.05 |
ENST00000342014.6 |
BMX |
BMX non-receptor tyrosine kinase |
| chr4_+_174818390 | 0.05 |
ENST00000509968.1 ENST00000512263.1 |
RP11-161D15.1 |
RP11-161D15.1 |
| chr16_+_72459838 | 0.05 |
ENST00000564508.1 |
AC004158.3 |
AC004158.3 |
| chr2_+_102413726 | 0.05 |
ENST00000350878.4 |
MAP4K4 |
mitogen-activated protein kinase kinase kinase kinase 4 |
| chr1_+_207262578 | 0.05 |
ENST00000243611.5 ENST00000367076.3 |
C4BPB |
complement component 4 binding protein, beta |
| chr1_+_207262627 | 0.05 |
ENST00000391923.1 |
C4BPB |
complement component 4 binding protein, beta |
| chr18_+_11752040 | 0.04 |
ENST00000423027.3 |
GNAL |
guanine nucleotide binding protein (G protein), alpha activating activity polypeptide, olfactory type |
| chr9_+_140445651 | 0.04 |
ENST00000371443.5 |
MRPL41 |
mitochondrial ribosomal protein L41 |
| chr19_+_37998031 | 0.04 |
ENST00000586138.1 ENST00000588578.1 ENST00000587986.1 |
ZNF793 |
zinc finger protein 793 |
| chr10_+_70980051 | 0.04 |
ENST00000354624.5 ENST00000395086.2 |
HKDC1 |
hexokinase domain containing 1 |
| chr11_-_35440579 | 0.04 |
ENST00000606205.1 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr10_-_121296045 | 0.04 |
ENST00000392865.1 |
RGS10 |
regulator of G-protein signaling 10 |
| chr7_+_43152191 | 0.04 |
ENST00000395891.2 |
HECW1 |
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 1 |
| chr6_+_29068386 | 0.04 |
ENST00000377171.3 |
OR2J1 |
olfactory receptor, family 2, subfamily J, member 1 (gene/pseudogene) |
| chr1_+_207262540 | 0.04 |
ENST00000452902.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr16_-_66952742 | 0.04 |
ENST00000565235.2 ENST00000568632.1 ENST00000565796.1 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr13_+_103459704 | 0.04 |
ENST00000602836.1 |
BIVM-ERCC5 |
BIVM-ERCC5 readthrough |
| chr16_-_66952779 | 0.04 |
ENST00000570262.1 ENST00000394055.3 ENST00000299752.4 |
CDH16 |
cadherin 16, KSP-cadherin |
| chr2_+_241625749 | 0.04 |
ENST00000407635.2 |
AC011298.2 |
AC011298.2 |
| chr2_+_157330081 | 0.04 |
ENST00000409674.1 |
GPD2 |
glycerol-3-phosphate dehydrogenase 2 (mitochondrial) |
| chr7_-_28220354 | 0.04 |
ENST00000283928.5 |
JAZF1 |
JAZF zinc finger 1 |
| chr11_+_8040739 | 0.04 |
ENST00000534099.1 |
TUB |
tubby bipartite transcription factor |
| chr3_-_93747425 | 0.04 |
ENST00000315099.2 |
STX19 |
syntaxin 19 |
| chr19_-_52674896 | 0.04 |
ENST00000322146.8 ENST00000597065.1 |
ZNF836 |
zinc finger protein 836 |
| chr1_-_243326612 | 0.04 |
ENST00000492145.1 ENST00000490813.1 ENST00000464936.1 |
CEP170 |
centrosomal protein 170kDa |
| chr17_+_7461781 | 0.04 |
ENST00000349228.4 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr10_+_71562180 | 0.04 |
ENST00000517713.1 ENST00000522165.1 ENST00000520133.1 |
COL13A1 |
collagen, type XIII, alpha 1 |
| chr17_+_7461849 | 0.04 |
ENST00000338784.4 |
TNFSF13 |
tumor necrosis factor (ligand) superfamily, member 13 |
| chr6_-_154751629 | 0.04 |
ENST00000424998.1 |
CNKSR3 |
CNKSR family member 3 |
| chr4_+_71588372 | 0.03 |
ENST00000536664.1 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr12_-_76817036 | 0.03 |
ENST00000546946.1 |
OSBPL8 |
oxysterol binding protein-like 8 |
| chr7_+_128784712 | 0.03 |
ENST00000289407.4 |
TSPAN33 |
tetraspanin 33 |
| chr2_-_197457335 | 0.03 |
ENST00000260983.3 |
HECW2 |
HECT, C2 and WW domain containing E3 ubiquitin protein ligase 2 |
| chr5_-_76935513 | 0.03 |
ENST00000306422.3 |
OTP |
orthopedia homeobox |
| chr4_-_103747011 | 0.03 |
ENST00000350435.7 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr3_+_137490748 | 0.03 |
ENST00000478772.1 |
RP11-2A4.3 |
RP11-2A4.3 |
| chr19_+_9361606 | 0.03 |
ENST00000456448.1 |
OR7E24 |
olfactory receptor, family 7, subfamily E, member 24 |
| chr3_+_46283916 | 0.03 |
ENST00000395940.2 |
CCR3 |
chemokine (C-C motif) receptor 3 |
| chr22_+_46449674 | 0.03 |
ENST00000381051.2 |
FLJ27365 |
hsa-mir-4763 |
| chr5_+_60933634 | 0.03 |
ENST00000505642.1 |
C5orf64 |
chromosome 5 open reading frame 64 |
| chr4_+_169418255 | 0.03 |
ENST00000505667.1 ENST00000511948.1 |
PALLD |
palladin, cytoskeletal associated protein |
| chr19_+_45596218 | 0.03 |
ENST00000421905.1 ENST00000221462.4 |
PPP1R37 |
protein phosphatase 1, regulatory subunit 37 |
| chr4_-_103746683 | 0.03 |
ENST00000504211.1 ENST00000508476.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr1_+_248201474 | 0.03 |
ENST00000366479.2 |
OR2L2 |
olfactory receptor, family 2, subfamily L, member 2 |
| chr2_+_190541153 | 0.03 |
ENST00000313581.4 ENST00000438402.2 ENST00000431575.2 ENST00000281412.6 |
ANKAR |
ankyrin and armadillo repeat containing |
| chr10_+_114710516 | 0.03 |
ENST00000542695.1 ENST00000346198.4 |
TCF7L2 |
transcription factor 7-like 2 (T-cell specific, HMG-box) |
| chr16_-_69788816 | 0.03 |
ENST00000268802.5 |
NOB1 |
NIN1/RPN12 binding protein 1 homolog (S. cerevisiae) |
| chr1_-_115259337 | 0.03 |
ENST00000369535.4 |
NRAS |
neuroblastoma RAS viral (v-ras) oncogene homolog |
| chr4_-_103746924 | 0.03 |
ENST00000505207.1 ENST00000502404.1 ENST00000507845.1 |
UBE2D3 |
ubiquitin-conjugating enzyme E2D 3 |
| chr11_-_116658695 | 0.02 |
ENST00000429220.1 ENST00000444935.1 |
ZNF259 |
zinc finger protein 259 |
| chr14_+_93357749 | 0.02 |
ENST00000557689.1 |
RP11-862G15.1 |
RP11-862G15.1 |
| chr3_+_63428982 | 0.02 |
ENST00000479198.1 ENST00000460711.1 ENST00000465156.1 |
SYNPR |
synaptoporin |
| chr11_-_116658758 | 0.02 |
ENST00000227322.3 |
ZNF259 |
zinc finger protein 259 |
| chr11_-_35440796 | 0.02 |
ENST00000278379.3 |
SLC1A2 |
solute carrier family 1 (glial high affinity glutamate transporter), member 2 |
| chr3_+_188889737 | 0.02 |
ENST00000345063.3 |
TPRG1 |
tumor protein p63 regulated 1 |
| chr22_-_36220420 | 0.02 |
ENST00000473487.2 |
RBFOX2 |
RNA binding protein, fox-1 homolog (C. elegans) 2 |
| chr19_+_35940486 | 0.02 |
ENST00000246549.2 |
FFAR2 |
free fatty acid receptor 2 |
| chr20_+_36405665 | 0.02 |
ENST00000373469.1 |
CTNNBL1 |
catenin, beta like 1 |
| chr4_+_169418195 | 0.02 |
ENST00000261509.6 ENST00000335742.7 |
PALLD |
palladin, cytoskeletal associated protein |
| chr16_-_2013101 | 0.02 |
ENST00000526586.2 |
RPS2 |
ribosomal protein S2 |
| chr2_-_50201327 | 0.02 |
ENST00000412315.1 |
NRXN1 |
neurexin 1 |
| chr5_+_65440032 | 0.02 |
ENST00000334121.6 |
SREK1 |
splicing regulatory glutamine/lysine-rich protein 1 |
| chr8_+_68864330 | 0.02 |
ENST00000288368.4 |
PREX2 |
phosphatidylinositol-3,4,5-trisphosphate-dependent Rac exchange factor 2 |
| chrX_+_46771848 | 0.02 |
ENST00000218343.4 |
PHF16 |
jade family PHD finger 3 |
| chr3_+_52811596 | 0.02 |
ENST00000542827.1 ENST00000273283.2 |
ITIH1 |
inter-alpha-trypsin inhibitor heavy chain 1 |
| chr15_+_38746307 | 0.02 |
ENST00000397609.2 ENST00000491535.1 |
FAM98B |
family with sequence similarity 98, member B |
| chr3_-_64211112 | 0.02 |
ENST00000295902.6 |
PRICKLE2 |
prickle homolog 2 (Drosophila) |
| chr18_+_29671812 | 0.02 |
ENST00000261593.3 ENST00000578914.1 |
RNF138 |
ring finger protein 138, E3 ubiquitin protein ligase |
| chr10_-_46167722 | 0.02 |
ENST00000374366.3 ENST00000344646.5 |
ZFAND4 |
zinc finger, AN1-type domain 4 |
| chr4_+_187187098 | 0.02 |
ENST00000403665.2 ENST00000264692.4 |
F11 |
coagulation factor XI |
| chr20_+_58571419 | 0.02 |
ENST00000244049.3 ENST00000350849.6 ENST00000456106.1 |
CDH26 |
cadherin 26 |
| chrX_+_46771711 | 0.02 |
ENST00000424392.1 ENST00000397189.1 |
PHF16 |
jade family PHD finger 3 |
| chr2_-_208030647 | 0.02 |
ENST00000309446.6 |
KLF7 |
Kruppel-like factor 7 (ubiquitous) |
| chr2_-_39348137 | 0.02 |
ENST00000426016.1 |
SOS1 |
son of sevenless homolog 1 (Drosophila) |
| chr5_+_138940742 | 0.02 |
ENST00000398733.3 ENST00000253815.2 ENST00000505007.1 |
UBE2D2 |
ubiquitin-conjugating enzyme E2D 2 |
| chr2_-_220174166 | 0.02 |
ENST00000409251.3 ENST00000451506.1 ENST00000295718.2 ENST00000446182.1 |
PTPRN |
protein tyrosine phosphatase, receptor type, N |
| chr9_+_77230499 | 0.02 |
ENST00000396204.2 |
RORB |
RAR-related orphan receptor B |
| chr8_-_86290333 | 0.02 |
ENST00000521846.1 ENST00000523022.1 ENST00000524324.1 ENST00000519991.1 ENST00000520663.1 ENST00000517590.1 ENST00000522579.1 ENST00000522814.1 ENST00000522662.1 ENST00000523858.1 ENST00000519129.1 |
CA1 |
carbonic anhydrase I |
| chr10_-_73975657 | 0.02 |
ENST00000394919.1 ENST00000526751.1 |
ASCC1 |
activating signal cointegrator 1 complex subunit 1 |
| chr17_+_31318886 | 0.02 |
ENST00000269053.3 ENST00000394638.1 |
SPACA3 |
sperm acrosome associated 3 |
| chr1_+_99127225 | 0.02 |
ENST00000370189.5 ENST00000529992.1 |
SNX7 |
sorting nexin 7 |
| chr9_-_70490107 | 0.01 |
ENST00000377395.4 ENST00000429800.2 ENST00000430059.2 ENST00000377384.1 ENST00000382405.3 |
CBWD5 |
COBW domain containing 5 |
| chr1_+_153190053 | 0.01 |
ENST00000368744.3 |
PRR9 |
proline rich 9 |
| chr9_-_28670283 | 0.01 |
ENST00000379992.2 |
LINGO2 |
leucine rich repeat and Ig domain containing 2 |
| chr14_+_22265444 | 0.01 |
ENST00000390430.2 |
TRAV8-1 |
T cell receptor alpha variable 8-1 |
| chr4_+_156824840 | 0.01 |
ENST00000536354.2 |
TDO2 |
tryptophan 2,3-dioxygenase |
| chr7_-_23510086 | 0.01 |
ENST00000258729.3 |
IGF2BP3 |
insulin-like growth factor 2 mRNA binding protein 3 |
| chr2_-_206950781 | 0.01 |
ENST00000403263.1 |
INO80D |
INO80 complex subunit D |
| chr14_+_37131058 | 0.01 |
ENST00000361487.6 |
PAX9 |
paired box 9 |
| chr6_+_158733692 | 0.01 |
ENST00000367094.2 ENST00000367097.3 |
TULP4 |
tubby like protein 4 |
| chrX_-_15332665 | 0.01 |
ENST00000537676.1 ENST00000344384.4 |
ASB11 |
ankyrin repeat and SOCS box containing 11 |
| chr1_+_207262881 | 0.01 |
ENST00000451804.2 |
C4BPB |
complement component 4 binding protein, beta |
| chr13_+_28366780 | 0.01 |
ENST00000302945.2 |
GSX1 |
GS homeobox 1 |
| chr19_-_44952635 | 0.01 |
ENST00000592308.1 ENST00000588931.1 ENST00000291187.4 |
ZNF229 |
zinc finger protein 229 |
| chr4_+_71587669 | 0.01 |
ENST00000381006.3 ENST00000226328.4 |
RUFY3 |
RUN and FYVE domain containing 3 |
| chr7_+_142829162 | 0.01 |
ENST00000291009.3 |
PIP |
prolactin-induced protein |
| chr5_-_156390230 | 0.01 |
ENST00000407087.3 ENST00000274532.2 |
TIMD4 |
T-cell immunoglobulin and mucin domain containing 4 |
| chr8_+_70850403 | 0.01 |
ENST00000602248.1 |
AC090574.1 |
Uncharacterized protein |
| chr3_-_11685345 | 0.01 |
ENST00000430365.2 |
VGLL4 |
vestigial like 4 (Drosophila) |
| chr4_-_69536346 | 0.01 |
ENST00000338206.5 |
UGT2B15 |
UDP glucuronosyltransferase 2 family, polypeptide B15 |
| chr10_+_63661053 | 0.01 |
ENST00000279873.7 |
ARID5B |
AT rich interactive domain 5B (MRF1-like) |
| chr7_-_14026123 | 0.01 |
ENST00000420159.2 ENST00000399357.3 ENST00000403527.1 |
ETV1 |
ets variant 1 |
| chr6_+_56954808 | 0.01 |
ENST00000510483.1 ENST00000370706.4 ENST00000357489.3 |
ZNF451 |
zinc finger protein 451 |
| chr2_+_171034646 | 0.01 |
ENST00000409044.3 ENST00000408978.4 |
MYO3B |
myosin IIIB |
| chr1_-_217262969 | 0.01 |
ENST00000361525.3 |
ESRRG |
estrogen-related receptor gamma |
| chr15_+_76352178 | 0.01 |
ENST00000388942.3 |
C15orf27 |
chromosome 15 open reading frame 27 |
| chr7_-_100239132 | 0.01 |
ENST00000223051.3 ENST00000431692.1 |
TFR2 |
transferrin receptor 2 |
| chr12_+_9144626 | 0.00 |
ENST00000543895.1 |
KLRG1 |
killer cell lectin-like receptor subfamily G, member 1 |
| chr14_+_55034599 | 0.00 |
ENST00000392067.3 ENST00000357634.3 |
SAMD4A |
sterile alpha motif domain containing 4A |
| chrX_+_100805496 | 0.00 |
ENST00000372829.3 |
ARMCX1 |
armadillo repeat containing, X-linked 1 |
| chrX_+_123480194 | 0.00 |
ENST00000371139.4 |
SH2D1A |
SH2 domain containing 1A |
| chr6_+_130339710 | 0.00 |
ENST00000526087.1 ENST00000533560.1 ENST00000361794.2 |
L3MBTL3 |
l(3)mbt-like 3 (Drosophila) |
| chr20_+_43849941 | 0.00 |
ENST00000372769.3 |
SEMG2 |
semenogelin II |
| chr4_-_20985632 | 0.00 |
ENST00000359001.5 |
KCNIP4 |
Kv channel interacting protein 4 |
| chr4_-_102268628 | 0.00 |
ENST00000323055.6 ENST00000512215.1 ENST00000394854.3 |
PPP3CA |
protein phosphatase 3, catalytic subunit, alpha isozyme |
| chr8_+_92261516 | 0.00 |
ENST00000276609.3 ENST00000309536.2 |
SLC26A7 |
solute carrier family 26 (anion exchanger), member 7 |
| chr7_-_121944491 | 0.00 |
ENST00000331178.4 ENST00000427185.2 ENST00000442488.2 |
FEZF1 |
FEZ family zinc finger 1 |
| chr16_-_30621663 | 0.00 |
ENST00000287461.3 |
ZNF689 |
zinc finger protein 689 |
| chr5_-_33297946 | 0.00 |
ENST00000510327.1 |
CTD-2066L21.3 |
CTD-2066L21.3 |
| chr11_+_85359062 | 0.00 |
ENST00000532180.1 |
TMEM126A |
transmembrane protein 126A |
| chr12_+_100594557 | 0.00 |
ENST00000546902.1 ENST00000552376.1 ENST00000551617.1 |
ACTR6 |
ARP6 actin-related protein 6 homolog (yeast) |
| chr2_+_7017796 | 0.00 |
ENST00000382040.3 |
RSAD2 |
radical S-adenosyl methionine domain containing 2 |
| chr4_+_187148556 | 0.00 |
ENST00000264690.6 ENST00000446598.2 ENST00000414291.1 ENST00000513864.1 |
KLKB1 |
kallikrein B, plasma (Fletcher factor) 1 |
| chr11_+_85359002 | 0.00 |
ENST00000528105.1 ENST00000304511.2 |
TMEM126A |
transmembrane protein 126A |
| chr15_+_54901540 | 0.00 |
ENST00000539562.2 |
UNC13C |
unc-13 homolog C (C. elegans) |
| chr5_-_135701164 | 0.00 |
ENST00000355180.3 ENST00000426057.2 ENST00000513104.1 |
TRPC7 |
transient receptor potential cation channel, subfamily C, member 7 |
| chr14_+_69865401 | 0.00 |
ENST00000556605.1 ENST00000336643.5 ENST00000031146.4 |
SLC39A9 |
solute carrier family 39, member 9 |
| chr2_+_166095898 | 0.00 |
ENST00000424833.1 ENST00000375437.2 ENST00000357398.3 |
SCN2A |
sodium channel, voltage-gated, type II, alpha subunit |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0008050 | female courtship behavior(GO:0008050) |
| 0.0 | 0.1 | GO:0045013 | carbon catabolite repression of transcription(GO:0045013) negative regulation of transcription by glucose(GO:0045014) |
| 0.0 | 0.1 | GO:0071418 | cellular response to amine stimulus(GO:0071418) |
| 0.0 | 0.1 | GO:0052199 | negative regulation of catalytic activity in other organism involved in symbiotic interaction(GO:0052199) |
| 0.0 | 0.1 | GO:0035063 | nuclear speck organization(GO:0035063) |
| 0.0 | 0.2 | GO:0048298 | positive regulation of isotype switching to IgA isotypes(GO:0048298) |
| 0.0 | 0.1 | GO:0018874 | benzoate metabolic process(GO:0018874) |
| 0.0 | 0.4 | GO:0071481 | cellular response to X-ray(GO:0071481) |
| 0.0 | 0.0 | GO:1903487 | regulation of lactation(GO:1903487) |
| 0.0 | 0.1 | GO:0030450 | regulation of complement activation, classical pathway(GO:0030450) negative regulation of complement activation, classical pathway(GO:0045959) regulation of opsonization(GO:1903027) |
| 0.0 | 0.5 | GO:0044705 | mating behavior(GO:0007617) multi-organism reproductive behavior(GO:0044705) |
| 0.0 | 0.1 | GO:0090214 | spongiotrophoblast layer developmental growth(GO:0090214) |
| 0.0 | 0.2 | GO:0015747 | urate transport(GO:0015747) |
| 0.0 | 0.1 | GO:0051987 | positive regulation of attachment of spindle microtubules to kinetochore(GO:0051987) |
| 0.0 | 0.1 | GO:0021842 | directional guidance of interneurons involved in migration from the subpallium to the cortex(GO:0021840) chemorepulsion involved in interneuron migration from the subpallium to the cortex(GO:0021842) |
| 0.0 | 0.1 | GO:0021633 | optic nerve structural organization(GO:0021633) |
| 0.0 | 0.2 | GO:0040015 | negative regulation of multicellular organism growth(GO:0040015) |
| 0.0 | 0.1 | GO:0008628 | hormone-mediated apoptotic signaling pathway(GO:0008628) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.4 | GO:0097129 | cyclin D2-CDK4 complex(GO:0097129) |
| 0.0 | 0.2 | GO:0005600 | collagen type XIII trimer(GO:0005600) transmembrane collagen trimer(GO:0030936) |
| 0.0 | 0.3 | GO:0070652 | HAUS complex(GO:0070652) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0004852 | uroporphyrinogen-III synthase activity(GO:0004852) |
| 0.0 | 0.1 | GO:0019166 | trans-2-enoyl-CoA reductase (NADPH) activity(GO:0019166) |
| 0.0 | 0.1 | GO:0001849 | complement component C1q binding(GO:0001849) |
| 0.0 | 0.2 | GO:1901702 | urate transmembrane transporter activity(GO:0015143) salt transmembrane transporter activity(GO:1901702) |
| 0.0 | 0.1 | GO:0070324 | thyroid hormone binding(GO:0070324) |
| 0.0 | 0.1 | GO:0001515 | opioid peptide activity(GO:0001515) |
| 0.0 | 0.1 | GO:0016802 | adenosylhomocysteinase activity(GO:0004013) trialkylsulfonium hydrolase activity(GO:0016802) |
| 0.0 | 0.2 | GO:0005068 | transmembrane receptor protein tyrosine kinase adaptor activity(GO:0005068) |
| 0.0 | 0.0 | GO:0052590 | sn-glycerol-3-phosphate:ubiquinone oxidoreductase activity(GO:0052590) sn-glycerol-3-phosphate:ubiquinone-8 oxidoreductase activity(GO:0052591) |