Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
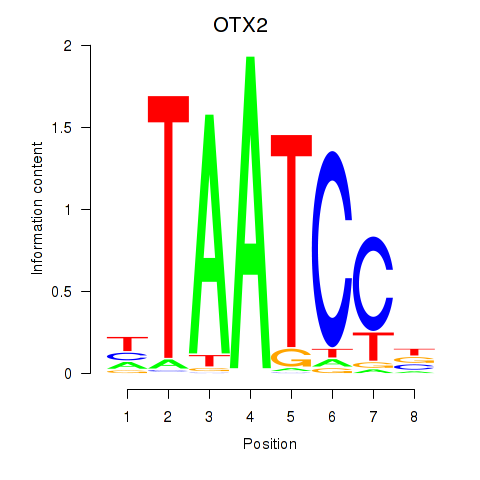
Results for OTX2_CRX
Z-value: 0.67
Transcription factors associated with OTX2_CRX
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
OTX2
|
ENSG00000165588.12 | OTX2 |
|
CRX
|
ENSG00000105392.11 | CRX |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| OTX2 | hg19_v2_chr14_-_57277163_57277168 | -0.77 | 2.5e-02 | Click! |
| CRX | hg19_v2_chr19_+_48325097_48325211 | -0.29 | 4.8e-01 | Click! |
Activity profile of OTX2_CRX motif
Sorted Z-values of OTX2_CRX motif
Network of associatons between targets according to the STRING database.
First level regulatory network of OTX2_CRX
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr8_+_70378852 | 0.52 |
ENST00000525061.1 ENST00000458141.2 ENST00000260128.4 |
SULF1 |
sulfatase 1 |
| chr12_+_6949964 | 0.49 |
ENST00000541978.1 ENST00000435982.2 |
GNB3 |
guanine nucleotide binding protein (G protein), beta polypeptide 3 |
| chr5_-_41510725 | 0.42 |
ENST00000328457.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr5_-_41510656 | 0.42 |
ENST00000377801.3 |
PLCXD3 |
phosphatidylinositol-specific phospholipase C, X domain containing 3 |
| chr12_-_2027639 | 0.32 |
ENST00000586184.1 ENST00000587995.1 ENST00000585732.1 |
CACNA2D4 |
calcium channel, voltage-dependent, alpha 2/delta subunit 4 |
| chr1_+_114522049 | 0.29 |
ENST00000369551.1 ENST00000320334.4 |
OLFML3 |
olfactomedin-like 3 |
| chr12_-_91576561 | 0.29 |
ENST00000547568.2 ENST00000552962.1 |
DCN |
decorin |
| chr12_-_91576429 | 0.29 |
ENST00000552145.1 ENST00000546745.1 |
DCN |
decorin |
| chr12_-_91576750 | 0.28 |
ENST00000228329.5 ENST00000303320.3 ENST00000052754.5 |
DCN |
decorin |
| chr1_+_159141397 | 0.25 |
ENST00000368124.4 ENST00000368125.4 ENST00000416746.1 |
CADM3 |
cell adhesion molecule 3 |
| chrX_+_69488174 | 0.25 |
ENST00000480877.2 ENST00000307959.8 |
ARR3 |
arrestin 3, retinal (X-arrestin) |
| chr9_-_95244781 | 0.24 |
ENST00000375544.3 ENST00000375543.1 ENST00000395538.3 ENST00000450139.2 |
ASPN |
asporin |
| chr2_+_128175997 | 0.23 |
ENST00000234071.3 ENST00000429925.1 ENST00000442644.1 ENST00000453608.2 |
PROC |
protein C (inactivator of coagulation factors Va and VIIIa) |
| chr2_+_120770581 | 0.23 |
ENST00000263713.5 |
EPB41L5 |
erythrocyte membrane protein band 4.1 like 5 |
| chr2_-_175711133 | 0.23 |
ENST00000409597.1 ENST00000413882.1 |
CHN1 |
chimerin 1 |
| chr1_-_165414414 | 0.22 |
ENST00000359842.5 |
RXRG |
retinoid X receptor, gamma |
| chrX_-_101771645 | 0.21 |
ENST00000289373.4 |
TMSB15A |
thymosin beta 15a |
| chr18_-_70532906 | 0.20 |
ENST00000299430.2 ENST00000397929.1 |
NETO1 |
neuropilin (NRP) and tolloid (TLL)-like 1 |
| chr12_+_7941989 | 0.20 |
ENST00000229307.4 |
NANOG |
Nanog homeobox |
| chr7_-_128415844 | 0.19 |
ENST00000249389.2 |
OPN1SW |
opsin 1 (cone pigments), short-wave-sensitive |
| chr6_-_42690312 | 0.19 |
ENST00000230381.5 |
PRPH2 |
peripherin 2 (retinal degeneration, slow) |
| chr5_+_145718587 | 0.19 |
ENST00000230732.4 |
POU4F3 |
POU class 4 homeobox 3 |
| chr5_-_88178964 | 0.18 |
ENST00000513252.1 ENST00000508569.1 ENST00000510942.1 ENST00000506554.1 |
MEF2C |
myocyte enhancer factor 2C |
| chr5_-_88179017 | 0.18 |
ENST00000514028.1 ENST00000514015.1 ENST00000503075.1 ENST00000437473.2 |
MEF2C |
myocyte enhancer factor 2C |
| chr19_+_2476116 | 0.16 |
ENST00000215631.4 ENST00000587345.1 |
GADD45B |
growth arrest and DNA-damage-inducible, beta |
| chr2_+_120770645 | 0.16 |
ENST00000443902.2 |
EPB41L5 |
erythrocyte membrane protein band 4.1 like 5 |
| chr2_-_175499294 | 0.16 |
ENST00000392547.2 |
WIPF1 |
WAS/WASL interacting protein family, member 1 |
| chr11_-_8680383 | 0.16 |
ENST00000299550.6 |
TRIM66 |
tripartite motif containing 66 |
| chr5_-_149324306 | 0.15 |
ENST00000255266.5 |
PDE6A |
phosphodiesterase 6A, cGMP-specific, rod, alpha |
| chr17_-_46690839 | 0.15 |
ENST00000498634.2 |
HOXB8 |
homeobox B8 |
| chr11_-_111794446 | 0.15 |
ENST00000527950.1 |
CRYAB |
crystallin, alpha B |
| chr3_+_141105705 | 0.15 |
ENST00000513258.1 |
ZBTB38 |
zinc finger and BTB domain containing 38 |
| chr19_-_51538148 | 0.14 |
ENST00000319590.4 ENST00000250351.4 |
KLK12 |
kallikrein-related peptidase 12 |
| chr19_-_51538118 | 0.14 |
ENST00000529888.1 |
KLK12 |
kallikrein-related peptidase 12 |
| chr11_-_111783595 | 0.14 |
ENST00000528628.1 |
CRYAB |
crystallin, alpha B |
| chr1_+_159409512 | 0.14 |
ENST00000423932.3 |
OR10J1 |
olfactory receptor, family 10, subfamily J, member 1 |
| chr3_-_54962100 | 0.13 |
ENST00000273286.5 |
LRTM1 |
leucine-rich repeats and transmembrane domains 1 |
| chr17_-_9808887 | 0.13 |
ENST00000226193.5 |
RCVRN |
recoverin |
| chr8_-_25281747 | 0.13 |
ENST00000421054.2 |
GNRH1 |
gonadotropin-releasing hormone 1 (luteinizing-releasing hormone) |
| chr16_-_31076332 | 0.12 |
ENST00000539836.3 ENST00000535577.1 ENST00000442862.2 |
ZNF668 |
zinc finger protein 668 |
| chrX_+_69488155 | 0.12 |
ENST00000374495.3 |
ARR3 |
arrestin 3, retinal (X-arrestin) |
| chr16_+_58010339 | 0.12 |
ENST00000290871.5 ENST00000441824.2 |
TEPP |
testis, prostate and placenta expressed |
| chr1_+_144989309 | 0.12 |
ENST00000596396.1 |
AL590452.1 |
Uncharacterized protein |
| chr6_-_31697563 | 0.11 |
ENST00000375789.2 ENST00000416410.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr14_-_101034407 | 0.11 |
ENST00000443071.2 ENST00000557378.1 |
BEGAIN |
brain-enriched guanylate kinase-associated |
| chr9_-_20622478 | 0.11 |
ENST00000355930.6 ENST00000380338.4 |
MLLT3 |
myeloid/lymphoid or mixed-lineage leukemia (trithorax homolog, Drosophila); translocated to, 3 |
| chr3_+_69915385 | 0.11 |
ENST00000314589.5 |
MITF |
microphthalmia-associated transcription factor |
| chr13_-_33780133 | 0.11 |
ENST00000399365.3 |
STARD13 |
StAR-related lipid transfer (START) domain containing 13 |
| chr9_+_18474098 | 0.11 |
ENST00000327883.7 ENST00000431052.2 ENST00000380570.4 |
ADAMTSL1 |
ADAMTS-like 1 |
| chr1_+_164528866 | 0.11 |
ENST00000420696.2 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr16_-_31076273 | 0.11 |
ENST00000426488.2 |
ZNF668 |
zinc finger protein 668 |
| chr1_+_18957500 | 0.10 |
ENST00000375375.3 |
PAX7 |
paired box 7 |
| chr2_-_119605253 | 0.10 |
ENST00000295206.6 |
EN1 |
engrailed homeobox 1 |
| chr1_-_241799232 | 0.10 |
ENST00000366553.1 |
CHML |
choroideremia-like (Rab escort protein 2) |
| chr6_-_31697255 | 0.10 |
ENST00000436437.1 |
DDAH2 |
dimethylarginine dimethylaminohydrolase 2 |
| chr7_-_97881429 | 0.09 |
ENST00000420697.1 ENST00000379795.3 ENST00000415086.1 ENST00000542604.1 ENST00000447648.2 |
TECPR1 |
tectonin beta-propeller repeat containing 1 |
| chr10_+_95848824 | 0.09 |
ENST00000371385.3 ENST00000371375.1 |
PLCE1 |
phospholipase C, epsilon 1 |
| chr3_-_179692042 | 0.09 |
ENST00000468741.1 |
PEX5L |
peroxisomal biogenesis factor 5-like |
| chr17_-_48207157 | 0.09 |
ENST00000330175.4 ENST00000503131.1 |
SAMD14 |
sterile alpha motif domain containing 14 |
| chr1_+_11751748 | 0.09 |
ENST00000294485.5 |
DRAXIN |
dorsal inhibitory axon guidance protein |
| chr3_-_190580404 | 0.09 |
ENST00000442080.1 |
GMNC |
geminin coiled-coil domain containing |
| chr9_-_13279563 | 0.09 |
ENST00000541718.1 |
MPDZ |
multiple PDZ domain protein |
| chr5_+_5140436 | 0.08 |
ENST00000511368.1 ENST00000274181.7 |
ADAMTS16 |
ADAM metallopeptidase with thrombospondin type 1 motif, 16 |
| chr7_+_90338712 | 0.08 |
ENST00000265741.3 ENST00000406263.1 |
CDK14 |
cyclin-dependent kinase 14 |
| chrX_+_79675965 | 0.08 |
ENST00000308293.5 |
FAM46D |
family with sequence similarity 46, member D |
| chr1_+_207277590 | 0.08 |
ENST00000367070.3 |
C4BPA |
complement component 4 binding protein, alpha |
| chr15_+_33603147 | 0.08 |
ENST00000415757.3 ENST00000389232.4 |
RYR3 |
ryanodine receptor 3 |
| chr3_+_63953415 | 0.08 |
ENST00000484332.1 |
ATXN7 |
ataxin 7 |
| chr5_-_149669192 | 0.08 |
ENST00000398376.3 |
CAMK2A |
calcium/calmodulin-dependent protein kinase II alpha |
| chr17_-_4689649 | 0.08 |
ENST00000441199.2 ENST00000416307.2 |
VMO1 |
vitelline membrane outer layer 1 homolog (chicken) |
| chr10_-_62761188 | 0.08 |
ENST00000357917.4 |
RHOBTB1 |
Rho-related BTB domain containing 1 |
| chr14_-_67981870 | 0.07 |
ENST00000555994.1 |
TMEM229B |
transmembrane protein 229B |
| chr1_+_164529004 | 0.07 |
ENST00000559240.1 ENST00000367897.1 ENST00000540236.1 ENST00000401534.1 |
PBX1 |
pre-B-cell leukemia homeobox 1 |
| chr6_-_111804905 | 0.07 |
ENST00000358835.3 ENST00000435970.1 |
REV3L |
REV3-like, polymerase (DNA directed), zeta, catalytic subunit |
| chr7_-_128171123 | 0.07 |
ENST00000608477.1 |
RP11-212P7.2 |
RP11-212P7.2 |
| chr12_-_13248705 | 0.07 |
ENST00000396310.2 |
GSG1 |
germ cell associated 1 |
| chr10_+_103986085 | 0.07 |
ENST00000370005.3 |
ELOVL3 |
ELOVL fatty acid elongase 3 |
| chrX_+_15767971 | 0.07 |
ENST00000479740.1 ENST00000454127.2 |
CA5B |
carbonic anhydrase VB, mitochondrial |
| chr9_-_13279589 | 0.07 |
ENST00000319217.7 |
MPDZ |
multiple PDZ domain protein |
| chr14_-_22005018 | 0.07 |
ENST00000546363.1 |
SALL2 |
spalt-like transcription factor 2 |
| chr10_+_24528108 | 0.07 |
ENST00000438429.1 |
KIAA1217 |
KIAA1217 |
| chr12_-_13248732 | 0.07 |
ENST00000396302.3 |
GSG1 |
germ cell associated 1 |
| chr10_-_104178857 | 0.07 |
ENST00000020673.5 |
PSD |
pleckstrin and Sec7 domain containing |
| chr11_+_61522844 | 0.06 |
ENST00000265460.5 |
MYRF |
myelin regulatory factor |
| chr6_+_155537771 | 0.06 |
ENST00000275246.7 |
TIAM2 |
T-cell lymphoma invasion and metastasis 2 |
| chr12_-_13248562 | 0.06 |
ENST00000457134.2 ENST00000537302.1 |
GSG1 |
germ cell associated 1 |
| chr7_-_100171270 | 0.06 |
ENST00000538735.1 |
SAP25 |
Sin3A-associated protein, 25kDa |
| chr19_-_44124019 | 0.06 |
ENST00000300811.3 |
ZNF428 |
zinc finger protein 428 |
| chr4_+_70894130 | 0.06 |
ENST00000526767.1 ENST00000530128.1 ENST00000381057.3 |
HTN3 |
histatin 3 |
| chr12_+_133757995 | 0.06 |
ENST00000536435.2 ENST00000228289.5 ENST00000541211.2 ENST00000500625.3 ENST00000539248.2 ENST00000542711.2 ENST00000536899.2 ENST00000542986.2 ENST00000537565.1 ENST00000541975.2 |
ZNF268 |
zinc finger protein 268 |
| chr5_-_95768973 | 0.06 |
ENST00000311106.3 |
PCSK1 |
proprotein convertase subtilisin/kexin type 1 |
| chr11_+_20044096 | 0.06 |
ENST00000533917.1 |
NAV2 |
neuron navigator 2 |
| chr1_+_169079823 | 0.06 |
ENST00000367813.3 |
ATP1B1 |
ATPase, Na+/K+ transporting, beta 1 polypeptide |
| chr20_-_31124186 | 0.06 |
ENST00000375678.3 |
C20orf112 |
chromosome 20 open reading frame 112 |
| chr3_+_129247479 | 0.06 |
ENST00000296271.3 |
RHO |
rhodopsin |
| chr2_+_201170703 | 0.06 |
ENST00000358677.5 |
SPATS2L |
spermatogenesis associated, serine-rich 2-like |
| chr19_+_11457175 | 0.06 |
ENST00000458408.1 ENST00000586451.1 ENST00000588592.1 |
CCDC159 |
coiled-coil domain containing 159 |
| chr12_-_90049828 | 0.06 |
ENST00000261173.2 ENST00000348959.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr2_+_219472488 | 0.05 |
ENST00000450993.2 |
PLCD4 |
phospholipase C, delta 4 |
| chr9_+_75229616 | 0.05 |
ENST00000340019.3 |
TMC1 |
transmembrane channel-like 1 |
| chr19_-_59066452 | 0.05 |
ENST00000312547.2 |
CHMP2A |
charged multivesicular body protein 2A |
| chr5_-_150138061 | 0.05 |
ENST00000521533.1 ENST00000424236.1 |
DCTN4 |
dynactin 4 (p62) |
| chr12_-_90049878 | 0.05 |
ENST00000359142.3 |
ATP2B1 |
ATPase, Ca++ transporting, plasma membrane 1 |
| chr9_+_18474204 | 0.05 |
ENST00000276935.6 |
ADAMTSL1 |
ADAMTS-like 1 |
| chr11_-_128712362 | 0.05 |
ENST00000392664.2 |
KCNJ1 |
potassium inwardly-rectifying channel, subfamily J, member 1 |
| chr19_-_10121144 | 0.05 |
ENST00000264828.3 |
COL5A3 |
collagen, type V, alpha 3 |
| chr12_+_40787194 | 0.05 |
ENST00000425730.2 ENST00000454784.4 |
MUC19 |
mucin 19, oligomeric |
| chr10_-_49482907 | 0.05 |
ENST00000374201.3 ENST00000407470.4 |
FRMPD2 |
FERM and PDZ domain containing 2 |
| chr14_+_88471468 | 0.05 |
ENST00000267549.3 |
GPR65 |
G protein-coupled receptor 65 |
| chr3_-_50605077 | 0.05 |
ENST00000426034.1 ENST00000441239.1 |
C3orf18 |
chromosome 3 open reading frame 18 |
| chr5_+_133451254 | 0.05 |
ENST00000517851.1 ENST00000521639.1 ENST00000522375.1 ENST00000378560.4 ENST00000432532.2 ENST00000520958.1 ENST00000518915.1 ENST00000395023.1 |
TCF7 |
transcription factor 7 (T-cell specific, HMG-box) |
| chr13_-_49975632 | 0.05 |
ENST00000457041.1 ENST00000355854.4 |
CAB39L |
calcium binding protein 39-like |
| chr10_+_18240834 | 0.05 |
ENST00000377371.3 ENST00000539911.1 |
SLC39A12 |
solute carrier family 39 (zinc transporter), member 12 |
| chr17_-_14683517 | 0.05 |
ENST00000379640.1 |
AC005863.1 |
AC005863.1 |
| chr12_-_53171128 | 0.05 |
ENST00000332411.2 |
KRT76 |
keratin 76 |
| chr3_-_149095652 | 0.05 |
ENST00000305366.3 |
TM4SF1 |
transmembrane 4 L six family member 1 |
| chr15_-_31453359 | 0.05 |
ENST00000542188.1 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
| chr4_+_129730947 | 0.05 |
ENST00000452328.2 ENST00000504089.1 |
PHF17 |
jade family PHD finger 1 |
| chr6_-_9933500 | 0.05 |
ENST00000492169.1 |
OFCC1 |
orofacial cleft 1 candidate 1 |
| chr1_-_117664317 | 0.05 |
ENST00000256649.4 ENST00000369464.3 ENST00000485032.1 |
TRIM45 |
tripartite motif containing 45 |
| chr4_+_154125565 | 0.05 |
ENST00000338700.5 |
TRIM2 |
tripartite motif containing 2 |
| chr11_+_72975559 | 0.05 |
ENST00000349767.2 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr3_-_50605150 | 0.04 |
ENST00000357203.3 |
C3orf18 |
chromosome 3 open reading frame 18 |
| chr5_+_173316341 | 0.04 |
ENST00000520867.1 ENST00000334035.5 |
CPEB4 |
cytoplasmic polyadenylation element binding protein 4 |
| chr14_+_23846328 | 0.04 |
ENST00000382809.2 |
CMTM5 |
CKLF-like MARVEL transmembrane domain containing 5 |
| chr22_-_38380543 | 0.04 |
ENST00000396884.2 |
SOX10 |
SRY (sex determining region Y)-box 10 |
| chr1_+_20396649 | 0.04 |
ENST00000375108.3 |
PLA2G5 |
phospholipase A2, group V |
| chr20_+_4702548 | 0.04 |
ENST00000305817.2 |
PRND |
prion protein 2 (dublet) |
| chr2_-_121223697 | 0.04 |
ENST00000593290.1 |
FLJ14816 |
long intergenic non-protein coding RNA 1101 |
| chr11_+_72975524 | 0.04 |
ENST00000540342.1 ENST00000542092.1 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr15_+_58430368 | 0.04 |
ENST00000558772.1 ENST00000219919.4 |
AQP9 |
aquaporin 9 |
| chr11_+_72975578 | 0.04 |
ENST00000393592.2 |
P2RY6 |
pyrimidinergic receptor P2Y, G-protein coupled, 6 |
| chr9_+_82188077 | 0.04 |
ENST00000425506.1 |
TLE4 |
transducin-like enhancer of split 4 (E(sp1) homolog, Drosophila) |
| chr1_-_146082633 | 0.04 |
ENST00000605317.1 ENST00000604938.1 ENST00000339388.5 |
NBPF11 |
neuroblastoma breakpoint family, member 11 |
| chr19_-_44123734 | 0.04 |
ENST00000598676.1 |
ZNF428 |
zinc finger protein 428 |
| chr9_+_17134980 | 0.04 |
ENST00000380647.3 |
CNTLN |
centlein, centrosomal protein |
| chr11_-_67290867 | 0.04 |
ENST00000353903.5 ENST00000294288.4 |
CABP2 |
calcium binding protein 2 |
| chr8_-_10512569 | 0.04 |
ENST00000382483.3 |
RP1L1 |
retinitis pigmentosa 1-like 1 |
| chr3_-_186262166 | 0.04 |
ENST00000307944.5 |
CRYGS |
crystallin, gamma S |
| chr19_+_46001697 | 0.04 |
ENST00000451287.2 ENST00000324688.4 |
PPM1N |
protein phosphatase, Mg2+/Mn2+ dependent, 1N (putative) |
| chr10_+_18240753 | 0.04 |
ENST00000377369.2 |
SLC39A12 |
solute carrier family 39 (zinc transporter), member 12 |
| chr11_+_67222818 | 0.04 |
ENST00000325656.5 |
CABP4 |
calcium binding protein 4 |
| chr3_+_191046810 | 0.04 |
ENST00000392455.3 ENST00000392456.3 |
CCDC50 |
coiled-coil domain containing 50 |
| chr10_+_18240814 | 0.04 |
ENST00000377374.4 |
SLC39A12 |
solute carrier family 39 (zinc transporter), member 12 |
| chr7_+_28452130 | 0.04 |
ENST00000357727.2 |
CREB5 |
cAMP responsive element binding protein 5 |
| chr4_+_129730779 | 0.04 |
ENST00000226319.6 |
PHF17 |
jade family PHD finger 1 |
| chr1_+_151682909 | 0.04 |
ENST00000326413.3 ENST00000442233.2 |
RIIAD1 AL589765.1 |
regulatory subunit of type II PKA R-subunit (RIIa) domain containing 1 Uncharacterized protein; cDNA FLJ36032 fis, clone TESTI2017069 |
| chr10_+_134150835 | 0.04 |
ENST00000432555.2 |
LRRC27 |
leucine rich repeat containing 27 |
| chr1_-_173174681 | 0.04 |
ENST00000367718.1 |
TNFSF4 |
tumor necrosis factor (ligand) superfamily, member 4 |
| chr2_+_170366203 | 0.04 |
ENST00000284669.1 |
KLHL41 |
kelch-like family member 41 |
| chr11_-_30608413 | 0.04 |
ENST00000528686.1 |
MPPED2 |
metallophosphoesterase domain containing 2 |
| chr12_-_7848364 | 0.04 |
ENST00000329913.3 |
GDF3 |
growth differentiation factor 3 |
| chr5_+_161494521 | 0.04 |
ENST00000356592.3 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr6_+_54173227 | 0.04 |
ENST00000259782.4 ENST00000370864.3 |
TINAG |
tubulointerstitial nephritis antigen |
| chr18_+_29769978 | 0.03 |
ENST00000269202.6 ENST00000581447.1 |
MEP1B |
meprin A, beta |
| chr10_-_65028817 | 0.03 |
ENST00000542921.1 |
JMJD1C |
jumonji domain containing 1C |
| chr6_+_111580508 | 0.03 |
ENST00000368847.4 |
KIAA1919 |
KIAA1919 |
| chr15_+_58430567 | 0.03 |
ENST00000536493.1 |
AQP9 |
aquaporin 9 |
| chr3_-_137851220 | 0.03 |
ENST00000236709.3 |
A4GNT |
alpha-1,4-N-acetylglucosaminyltransferase |
| chr8_-_114449112 | 0.03 |
ENST00000455883.2 ENST00000352409.3 ENST00000297405.5 |
CSMD3 |
CUB and Sushi multiple domains 3 |
| chr15_-_74495188 | 0.03 |
ENST00000563965.1 ENST00000395105.4 |
STRA6 |
stimulated by retinoic acid 6 |
| chr10_-_65028938 | 0.03 |
ENST00000402544.1 |
JMJD1C |
jumonji domain containing 1C |
| chr9_+_34179003 | 0.03 |
ENST00000545103.1 ENST00000543944.1 ENST00000536252.1 ENST00000540348.1 ENST00000297661.4 ENST00000379186.4 |
UBAP1 |
ubiquitin associated protein 1 |
| chrX_+_153409678 | 0.03 |
ENST00000369951.4 |
OPN1LW |
opsin 1 (cone pigments), long-wave-sensitive |
| chr15_+_54901540 | 0.03 |
ENST00000539562.2 |
UNC13C |
unc-13 homolog C (C. elegans) |
| chr1_-_53608249 | 0.03 |
ENST00000371494.4 |
SLC1A7 |
solute carrier family 1 (glutamate transporter), member 7 |
| chr1_-_102312517 | 0.03 |
ENST00000338858.5 |
OLFM3 |
olfactomedin 3 |
| chr17_-_39661849 | 0.03 |
ENST00000246635.3 ENST00000336861.3 ENST00000587544.1 ENST00000587435.1 |
KRT13 |
keratin 13 |
| chr3_-_9291063 | 0.03 |
ENST00000383836.3 |
SRGAP3 |
SLIT-ROBO Rho GTPase activating protein 3 |
| chrM_+_9207 | 0.03 |
ENST00000362079.2 |
MT-CO3 |
mitochondrially encoded cytochrome c oxidase III |
| chr21_-_35016231 | 0.03 |
ENST00000438788.1 |
CRYZL1 |
crystallin, zeta (quinone reductase)-like 1 |
| chrX_+_100645812 | 0.03 |
ENST00000427805.2 ENST00000553110.3 ENST00000392994.3 ENST00000409338.1 ENST00000409170.3 |
RPL36A RPL36A-HNRNPH2 |
ribosomal protein L36a RPL36A-HNRNPH2 readthrough |
| chr13_+_36050881 | 0.03 |
ENST00000537702.1 |
NBEA |
neurobeachin |
| chr13_+_31191920 | 0.03 |
ENST00000255304.4 |
USPL1 |
ubiquitin specific peptidase like 1 |
| chr19_-_51537982 | 0.03 |
ENST00000525263.1 |
KLK12 |
kallikrein-related peptidase 12 |
| chr17_-_55822653 | 0.03 |
ENST00000299415.2 |
AC007431.1 |
coiled-coil domain containing 182 |
| chr15_+_96869165 | 0.03 |
ENST00000421109.2 |
NR2F2 |
nuclear receptor subfamily 2, group F, member 2 |
| chr3_-_170626418 | 0.03 |
ENST00000474096.1 ENST00000295822.2 |
EIF5A2 |
eukaryotic translation initiation factor 5A2 |
| chr1_+_26644441 | 0.03 |
ENST00000374213.2 |
CD52 |
CD52 molecule |
| chr5_+_63461642 | 0.03 |
ENST00000296615.6 ENST00000381081.2 ENST00000389100.4 |
RNF180 |
ring finger protein 180 |
| chr1_-_151965048 | 0.02 |
ENST00000368809.1 |
S100A10 |
S100 calcium binding protein A10 |
| chr10_-_48390973 | 0.02 |
ENST00000224600.4 |
RBP3 |
retinol binding protein 3, interstitial |
| chr16_-_58004992 | 0.02 |
ENST00000564448.1 ENST00000251102.8 ENST00000311183.4 |
CNGB1 |
cyclic nucleotide gated channel beta 1 |
| chr5_+_161494770 | 0.02 |
ENST00000414552.2 ENST00000361925.4 |
GABRG2 |
gamma-aminobutyric acid (GABA) A receptor, gamma 2 |
| chr19_+_18208603 | 0.02 |
ENST00000262811.6 |
MAST3 |
microtubule associated serine/threonine kinase 3 |
| chr15_-_31453157 | 0.02 |
ENST00000559177.1 ENST00000558445.1 |
TRPM1 |
transient receptor potential cation channel, subfamily M, member 1 |
| chr19_-_48547294 | 0.02 |
ENST00000293255.2 |
CABP5 |
calcium binding protein 5 |
| chrX_+_100646190 | 0.02 |
ENST00000471855.1 |
RPL36A |
ribosomal protein L36a |
| chr15_+_79603404 | 0.02 |
ENST00000299705.5 |
TMED3 |
transmembrane emp24 protein transport domain containing 3 |
| chr4_+_129730839 | 0.02 |
ENST00000511647.1 |
PHF17 |
jade family PHD finger 1 |
| chr13_-_62001982 | 0.02 |
ENST00000409186.1 |
PCDH20 |
protocadherin 20 |
| chr2_+_183943464 | 0.02 |
ENST00000354221.4 |
DUSP19 |
dual specificity phosphatase 19 |
| chr1_+_202431859 | 0.02 |
ENST00000391959.3 ENST00000367270.4 |
PPP1R12B |
protein phosphatase 1, regulatory subunit 12B |
| chr10_-_94003003 | 0.02 |
ENST00000412050.4 |
CPEB3 |
cytoplasmic polyadenylation element binding protein 3 |
| chr20_-_34330129 | 0.02 |
ENST00000397370.3 ENST00000528062.3 ENST00000407261.4 ENST00000374038.3 ENST00000361162.6 |
RBM39 |
RNA binding motif protein 39 |
| chr3_+_130650738 | 0.02 |
ENST00000504612.1 |
ATP2C1 |
ATPase, Ca++ transporting, type 2C, member 1 |
| chr22_-_41258074 | 0.02 |
ENST00000307221.4 |
DNAJB7 |
DnaJ (Hsp40) homolog, subfamily B, member 7 |
| chr1_+_87380299 | 0.02 |
ENST00000370551.4 ENST00000370550.5 |
HS2ST1 |
heparan sulfate 2-O-sulfotransferase 1 |
| chr2_-_30144432 | 0.02 |
ENST00000389048.3 |
ALK |
anaplastic lymphoma receptor tyrosine kinase |
| chr6_-_139613269 | 0.02 |
ENST00000358430.3 |
TXLNB |
taxilin beta |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0014846 | esophagus smooth muscle contraction(GO:0014846) |
| 0.1 | 0.2 | GO:0044537 | regulation of circulating fibrinogen levels(GO:0044537) |
| 0.1 | 0.2 | GO:0003172 | primary heart field specification(GO:0003138) sinoatrial valve development(GO:0003172) sinoatrial valve morphogenesis(GO:0003185) |
| 0.1 | 0.2 | GO:0070171 | negative regulation of tooth mineralization(GO:0070171) |
| 0.1 | 0.9 | GO:1900747 | negative regulation of vascular endothelial growth factor signaling pathway(GO:1900747) |
| 0.0 | 0.2 | GO:0007509 | mesoderm migration involved in gastrulation(GO:0007509) |
| 0.0 | 0.2 | GO:2000312 | regulation of kainate selective glutamate receptor activity(GO:2000312) |
| 0.0 | 0.1 | GO:1990637 | response to prolactin(GO:1990637) |
| 0.0 | 0.2 | GO:0001714 | endodermal cell fate specification(GO:0001714) |
| 0.0 | 0.1 | GO:0061743 | motor learning(GO:0061743) |
| 0.0 | 0.1 | GO:0045897 | positive regulation of transcription during mitosis(GO:0045897) |
| 0.0 | 0.2 | GO:0060041 | retina development in camera-type eye(GO:0060041) |
| 0.0 | 0.1 | GO:0097498 | endothelial tube lumen extension(GO:0097498) |
| 0.0 | 0.1 | GO:1903722 | regulation of centriole elongation(GO:1903722) |
| 0.0 | 0.2 | GO:0021562 | vestibulocochlear nerve development(GO:0021562) |
| 0.0 | 0.3 | GO:0007021 | tubulin complex assembly(GO:0007021) |
| 0.0 | 0.1 | GO:0006651 | diacylglycerol biosynthetic process(GO:0006651) |
| 0.0 | 0.1 | GO:0015722 | canalicular bile acid transport(GO:0015722) |
| 0.0 | 0.3 | GO:0018298 | protein-chromophore linkage(GO:0018298) |
| 0.0 | 0.1 | GO:1990034 | cellular response to corticosterone stimulus(GO:0071386) calcium ion export from cell(GO:1990034) |
| 0.0 | 0.3 | GO:0021516 | dorsal spinal cord development(GO:0021516) |
| 0.0 | 0.0 | GO:2000523 | regulation of T cell costimulation(GO:2000523) positive regulation of T cell costimulation(GO:2000525) |
| 0.0 | 0.1 | GO:1903281 | protein transport into plasma membrane raft(GO:0044861) positive regulation of calcium:sodium antiporter activity(GO:1903281) positive regulation of potassium ion import(GO:1903288) |
| 0.0 | 0.2 | GO:0006527 | arginine catabolic process(GO:0006527) |
| 0.0 | 0.2 | GO:0036066 | protein O-linked fucosylation(GO:0036066) |
| 0.0 | 0.1 | GO:0060005 | vestibular reflex(GO:0060005) |
| 0.0 | 0.1 | GO:0018344 | protein geranylgeranylation(GO:0018344) |
| 0.0 | 0.0 | GO:0071393 | cellular response to progesterone stimulus(GO:0071393) |
| 0.0 | 0.0 | GO:0006663 | platelet activating factor biosynthetic process(GO:0006663) |
| 0.0 | 0.1 | GO:0016560 | protein import into peroxisome matrix, docking(GO:0016560) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.9 | GO:0005589 | collagen type VI trimer(GO:0005589) collagen beaded filament(GO:0098647) |
| 0.0 | 0.1 | GO:0005968 | Rab-protein geranylgeranyltransferase complex(GO:0005968) |
| 0.0 | 0.1 | GO:0035841 | new growing cell tip(GO:0035841) |
| 0.0 | 0.1 | GO:0005588 | collagen type V trimer(GO:0005588) |
| 0.0 | 0.2 | GO:0098839 | postsynaptic density membrane(GO:0098839) |
| 0.0 | 0.1 | GO:0000308 | cytoplasmic cyclin-dependent protein kinase holoenzyme complex(GO:0000308) |
| 0.0 | 0.3 | GO:0097512 | cardiac myofibril(GO:0097512) |
| 0.0 | 0.1 | GO:0098554 | cytoplasmic side of endoplasmic reticulum membrane(GO:0098554) |
| 0.0 | 0.1 | GO:0016035 | zeta DNA polymerase complex(GO:0016035) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.5 | GO:0008449 | N-acetylglucosamine-6-sulfatase activity(GO:0008449) |
| 0.1 | 0.4 | GO:0002046 | opsin binding(GO:0002046) |
| 0.1 | 0.2 | GO:0004886 | 9-cis retinoic acid receptor activity(GO:0004886) |
| 0.0 | 0.2 | GO:0016403 | dimethylargininase activity(GO:0016403) |
| 0.0 | 0.1 | GO:0045029 | UDP-activated nucleotide receptor activity(GO:0045029) |
| 0.0 | 0.1 | GO:0048763 | calcium-induced calcium release activity(GO:0048763) |
| 0.0 | 0.3 | GO:0008020 | G-protein coupled photoreceptor activity(GO:0008020) |
| 0.0 | 0.1 | GO:0005275 | amine transmembrane transporter activity(GO:0005275) |
| 0.0 | 0.2 | GO:0003680 | AT DNA binding(GO:0003680) |
| 0.0 | 0.1 | GO:0005052 | peroxisome matrix targeting signal-1 binding(GO:0005052) |
| 0.0 | 0.9 | GO:0050840 | extracellular matrix binding(GO:0050840) |
| 0.0 | 0.3 | GO:0005212 | structural constituent of eye lens(GO:0005212) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.7 | PID CONE PATHWAY | Visual signal transduction: Cones |
| 0.0 | 1.0 | NABA PROTEOGLYCANS | Genes encoding proteoglycans |
| 0.0 | 0.2 | PID RHODOPSIN PATHWAY | Visual signal transduction: Rods |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.9 | REACTOME CS DS DEGRADATION | Genes involved in CS/DS degradation |
| 0.0 | 0.6 | REACTOME SYNTHESIS SECRETION AND INACTIVATION OF GLP1 | Genes involved in Synthesis, Secretion, and Inactivation of Glucagon-like Peptide-1 (GLP-1) |
| 0.0 | 0.3 | REACTOME OPSINS | Genes involved in Opsins |
| 0.0 | 0.2 | REACTOME GAMMA CARBOXYLATION TRANSPORT AND AMINO TERMINAL CLEAVAGE OF PROTEINS | Genes involved in Gamma-carboxylation, transport, and amino-terminal cleavage of proteins |