Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
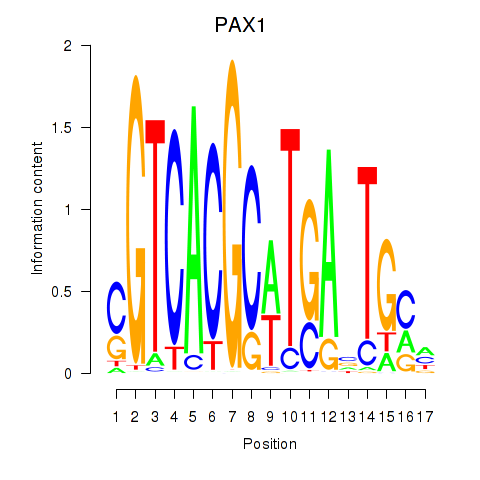
Results for PAX1_PAX9
Z-value: 0.31
Transcription factors associated with PAX1_PAX9
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX1
|
ENSG00000125813.9 | PAX1 |
|
PAX9
|
ENSG00000198807.8 | PAX9 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX9 | hg19_v2_chr14_+_37126765_37126799 | 0.44 | 2.8e-01 | Click! |
| PAX1 | hg19_v2_chr20_+_21686290_21686311 | 0.30 | 4.7e-01 | Click! |
Activity profile of PAX1_PAX9 motif
Sorted Z-values of PAX1_PAX9 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX1_PAX9
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_123747979 | 0.41 |
ENST00000608478.1 |
FGF2 |
fibroblast growth factor 2 (basic) |
| chr4_+_123747834 | 0.35 |
ENST00000264498.3 |
FGF2 |
fibroblast growth factor 2 (basic) |
| chr5_-_133747589 | 0.22 |
ENST00000458198.2 |
CDKN2AIPNL |
CDKN2A interacting protein N-terminal like |
| chr8_+_84824920 | 0.21 |
ENST00000523678.1 |
RP11-120I21.2 |
RP11-120I21.2 |
| chr5_-_133747551 | 0.17 |
ENST00000395009.3 |
CDKN2AIPNL |
CDKN2A interacting protein N-terminal like |
| chr2_-_207629997 | 0.15 |
ENST00000454776.2 |
MDH1B |
malate dehydrogenase 1B, NAD (soluble) |
| chrX_-_153707545 | 0.14 |
ENST00000357360.4 |
LAGE3 |
L antigen family, member 3 |
| chr7_-_19157248 | 0.14 |
ENST00000242261.5 |
TWIST1 |
twist family bHLH transcription factor 1 |
| chrX_-_153707246 | 0.14 |
ENST00000407062.1 |
LAGE3 |
L antigen family, member 3 |
| chr1_-_162838551 | 0.14 |
ENST00000367910.1 ENST00000367912.2 ENST00000367911.2 |
C1orf110 |
chromosome 1 open reading frame 110 |
| chr19_-_40791211 | 0.12 |
ENST00000579047.1 |
AKT2 |
v-akt murine thymoma viral oncogene homolog 2 |
| chr8_-_121824374 | 0.11 |
ENST00000517992.1 |
SNTB1 |
syntrophin, beta 1 (dystrophin-associated protein A1, 59kDa, basic component 1) |
| chr2_+_119699864 | 0.10 |
ENST00000541757.1 ENST00000412481.1 |
MARCO |
macrophage receptor with collagenous structure |
| chr8_-_125384927 | 0.10 |
ENST00000297632.6 |
TMEM65 |
transmembrane protein 65 |
| chr2_-_207630033 | 0.09 |
ENST00000449792.1 |
MDH1B |
malate dehydrogenase 1B, NAD (soluble) |
| chr5_+_133842243 | 0.08 |
ENST00000515627.2 |
AC005355.2 |
AC005355.2 |
| chr19_-_18995029 | 0.08 |
ENST00000596048.1 |
CERS1 |
ceramide synthase 1 |
| chr7_-_144533074 | 0.08 |
ENST00000360057.3 ENST00000378099.3 |
TPK1 |
thiamin pyrophosphokinase 1 |
| chr2_-_175712270 | 0.08 |
ENST00000295497.7 ENST00000444394.1 |
CHN1 |
chimerin 1 |
| chr9_+_33795533 | 0.07 |
ENST00000379405.3 |
PRSS3 |
protease, serine, 3 |
| chr9_+_33750667 | 0.07 |
ENST00000457896.1 ENST00000342836.4 ENST00000429677.3 |
PRSS3 |
protease, serine, 3 |
| chr1_+_26759295 | 0.07 |
ENST00000430232.1 |
DHDDS |
dehydrodolichyl diphosphate synthase |
| chr10_-_99393208 | 0.07 |
ENST00000307450.6 |
MORN4 |
MORN repeat containing 4 |
| chr9_+_33750515 | 0.07 |
ENST00000361005.5 |
PRSS3 |
protease, serine, 3 |
| chr13_+_98086445 | 0.06 |
ENST00000245304.4 |
RAP2A |
RAP2A, member of RAS oncogene family |
| chr5_+_72251857 | 0.06 |
ENST00000507345.2 ENST00000512348.1 ENST00000287761.6 |
FCHO2 |
FCH domain only 2 |
| chr20_-_17641097 | 0.06 |
ENST00000246043.4 |
RRBP1 |
ribosome binding protein 1 |
| chr17_+_39969183 | 0.05 |
ENST00000321562.4 |
FKBP10 |
FK506 binding protein 10, 65 kDa |
| chr12_-_101801505 | 0.05 |
ENST00000539055.1 ENST00000551688.1 ENST00000551671.1 ENST00000261636.8 |
ARL1 |
ADP-ribosylation factor-like 1 |
| chr17_-_7137582 | 0.05 |
ENST00000575756.1 ENST00000575458.1 |
DVL2 |
dishevelled segment polarity protein 2 |
| chr1_+_26758790 | 0.05 |
ENST00000427245.2 ENST00000525682.2 ENST00000236342.7 ENST00000526219.1 ENST00000374185.3 ENST00000360009.2 |
DHDDS |
dehydrodolichyl diphosphate synthase |
| chr17_-_3461092 | 0.04 |
ENST00000301365.4 ENST00000572519.1 |
TRPV3 |
transient receptor potential cation channel, subfamily V, member 3 |
| chr6_-_137366163 | 0.04 |
ENST00000367748.1 |
IL20RA |
interleukin 20 receptor, alpha |
| chr19_+_14184370 | 0.04 |
ENST00000590772.1 |
hsa-mir-1199 |
hsa-mir-1199 |
| chr10_-_99393242 | 0.04 |
ENST00000370635.3 ENST00000478953.1 ENST00000335628.3 |
MORN4 |
MORN repeat containing 4 |
| chr7_+_142457315 | 0.04 |
ENST00000486171.1 ENST00000311737.7 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
| chr14_-_69261310 | 0.04 |
ENST00000336440.3 |
ZFP36L1 |
ZFP36 ring finger protein-like 1 |
| chr9_-_131418944 | 0.04 |
ENST00000419989.1 ENST00000451652.1 ENST00000372715.2 |
WDR34 |
WD repeat domain 34 |
| chr9_+_96793076 | 0.04 |
ENST00000375360.3 |
PTPDC1 |
protein tyrosine phosphatase domain containing 1 |
| chr6_+_3118926 | 0.04 |
ENST00000380379.5 |
BPHL |
biphenyl hydrolase-like (serine hydrolase) |
| chr18_-_33077556 | 0.03 |
ENST00000589273.1 ENST00000586489.1 |
INO80C |
INO80 complex subunit C |
| chr16_+_67197288 | 0.03 |
ENST00000264009.8 ENST00000421453.1 |
HSF4 |
heat shock transcription factor 4 |
| chr15_+_63569731 | 0.03 |
ENST00000261879.5 |
APH1B |
APH1B gamma secretase subunit |
| chr1_-_213031418 | 0.03 |
ENST00000356684.3 ENST00000426161.1 ENST00000424044.1 |
FLVCR1-AS1 |
FLVCR1 antisense RNA 1 (head to head) |
| chr4_+_619386 | 0.03 |
ENST00000496514.1 |
PDE6B |
phosphodiesterase 6B, cGMP-specific, rod, beta |
| chr8_-_101965559 | 0.02 |
ENST00000353245.3 |
YWHAZ |
tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta |
| chr10_-_43904608 | 0.02 |
ENST00000337970.3 |
HNRNPF |
heterogeneous nuclear ribonucleoprotein F |
| chr3_+_15247686 | 0.02 |
ENST00000253693.2 |
CAPN7 |
calpain 7 |
| chr13_+_21141208 | 0.02 |
ENST00000351808.5 |
IFT88 |
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr18_-_47017956 | 0.02 |
ENST00000584895.1 ENST00000332968.6 ENST00000580210.1 ENST00000579408.1 |
RPL17-C18orf32 RPL17 |
RPL17-C18orf32 readthrough ribosomal protein L17 |
| chr6_+_159291090 | 0.02 |
ENST00000367073.4 ENST00000608817.1 |
C6orf99 |
chromosome 6 open reading frame 99 |
| chr13_+_21141270 | 0.02 |
ENST00000319980.6 ENST00000537103.1 ENST00000389373.3 |
IFT88 |
intraflagellar transport 88 homolog (Chlamydomonas) |
| chr18_+_39766626 | 0.02 |
ENST00000593234.1 ENST00000585627.1 ENST00000591199.1 ENST00000586990.1 ENST00000593051.1 ENST00000593316.1 ENST00000591381.1 ENST00000585639.1 ENST00000589068.1 |
LINC00907 |
long intergenic non-protein coding RNA 907 |
| chr6_+_159290917 | 0.02 |
ENST00000367072.1 |
C6orf99 |
chromosome 6 open reading frame 99 |
| chr4_-_171011084 | 0.02 |
ENST00000337664.4 |
AADAT |
aminoadipate aminotransferase |
| chr10_+_82297658 | 0.02 |
ENST00000339284.2 |
SH2D4B |
SH2 domain containing 4B |
| chr11_+_61522844 | 0.01 |
ENST00000265460.5 |
MYRF |
myelin regulatory factor |
| chr19_-_52148798 | 0.01 |
ENST00000534261.2 |
SIGLEC5 |
sialic acid binding Ig-like lectin 5 |
| chr8_+_39792474 | 0.01 |
ENST00000502986.2 |
IDO2 |
indoleamine 2,3-dioxygenase 2 |
| chr4_-_171011323 | 0.01 |
ENST00000509167.1 ENST00000353187.2 ENST00000507375.1 ENST00000515480.1 |
AADAT |
aminoadipate aminotransferase |
| chr2_-_162931052 | 0.01 |
ENST00000360534.3 |
DPP4 |
dipeptidyl-peptidase 4 |
| chr6_-_137494775 | 0.01 |
ENST00000349184.4 ENST00000296980.2 ENST00000339602.3 |
IL22RA2 |
interleukin 22 receptor, alpha 2 |
| chr9_-_21142144 | 0.01 |
ENST00000380229.2 |
IFNW1 |
interferon, omega 1 |
| chr1_+_160121356 | 0.01 |
ENST00000368081.4 |
ATP1A4 |
ATPase, Na+/K+ transporting, alpha 4 polypeptide |
| chrX_+_148622513 | 0.01 |
ENST00000393985.3 ENST00000423421.1 ENST00000423540.2 ENST00000434353.2 ENST00000514208.1 |
CXorf40A |
chromosome X open reading frame 40A |
| chr11_+_6947720 | 0.01 |
ENST00000414517.2 |
ZNF215 |
zinc finger protein 215 |
| chr14_+_39703112 | 0.01 |
ENST00000555143.1 ENST00000280082.3 |
MIA2 |
melanoma inhibitory activity 2 |
| chr19_-_40791302 | 0.01 |
ENST00000392038.2 ENST00000578123.1 |
AKT2 |
v-akt murine thymoma viral oncogene homolog 2 |
| chr3_-_25824925 | 0.00 |
ENST00000396649.3 ENST00000428257.1 ENST00000280700.5 |
NGLY1 |
N-glycanase 1 |
| chr3_-_25824872 | 0.00 |
ENST00000308710.5 |
NGLY1 |
N-glycanase 1 |
| chr15_+_63569785 | 0.00 |
ENST00000380343.4 ENST00000560353.1 |
APH1B |
APH1B gamma secretase subunit |
| chr18_-_47018769 | 0.00 |
ENST00000583637.1 ENST00000578528.1 ENST00000578532.1 ENST00000580387.1 ENST00000579248.1 ENST00000581373.1 |
RPL17 |
ribosomal protein L17 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | GO:2000546 | positive regulation of cell chemotaxis to fibroblast growth factor(GO:1904849) positive regulation of endothelial cell chemotaxis to fibroblast growth factor(GO:2000546) |
| 0.0 | 0.1 | GO:2000276 | negative regulation of peroxisome proliferator activated receptor signaling pathway(GO:0035359) negative regulation of oxidative phosphorylation uncoupler activity(GO:2000276) |
| 0.0 | 0.1 | GO:0016094 | polyprenol biosynthetic process(GO:0016094) |
| 0.0 | 0.1 | GO:0009644 | response to high light intensity(GO:0009644) cellular response to light intensity(GO:0071484) |
| 0.0 | 0.1 | GO:0072720 | cellular response to mycotoxin(GO:0036146) response to dithiothreitol(GO:0072720) |
| 0.0 | 0.1 | GO:0042357 | thiamine diphosphate metabolic process(GO:0042357) |
| 0.0 | 0.1 | GO:0000412 | histone peptidyl-prolyl isomerization(GO:0000412) |
| 0.0 | 0.2 | GO:0006108 | malate metabolic process(GO:0006108) |
| 0.0 | 0.0 | GO:1904582 | regulation of intracellular mRNA localization(GO:1904580) positive regulation of intracellular mRNA localization(GO:1904582) |
| 0.0 | 0.0 | GO:0022007 | neural plate elongation(GO:0014022) convergent extension involved in neural plate elongation(GO:0022007) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.3 | GO:0000408 | EKC/KEOPS complex(GO:0000408) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | GO:0002094 | polyprenyltransferase activity(GO:0002094) |
| 0.0 | 0.2 | GO:0030060 | L-malate dehydrogenase activity(GO:0030060) |
| 0.0 | 0.8 | GO:0005104 | fibroblast growth factor receptor binding(GO:0005104) |
Gene overrepresentation in C2:CP category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.8 | PID GLYPICAN 1PATHWAY | Glypican 1 network |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.8 | REACTOME SIGNALING BY FGFR3 MUTANTS | Genes involved in Signaling by FGFR3 mutants |
| 0.0 | 0.1 | REACTOME NEGATIVE REGULATION OF THE PI3K AKT NETWORK | Genes involved in Negative regulation of the PI3K/AKT network |