Project
Epithelial-Mesenchymal Transition, human (Scheel, 2011)
Navigation
Downloads
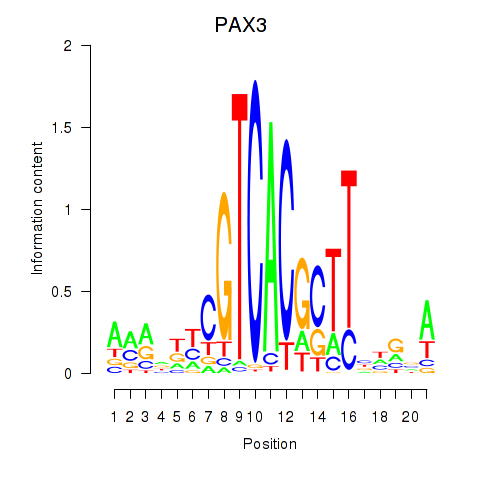
Results for PAX3
Z-value: 0.51
Transcription factors associated with PAX3
| Gene Symbol | Gene ID | Gene Info |
|---|---|---|
|
PAX3
|
ENSG00000135903.14 | PAX3 |
Activity-expression correlation:
| Gene | Promoter | Pearson corr. coef. | P-value | Plot |
|---|---|---|---|---|
| PAX3 | hg19_v2_chr2_-_223163465_223163730 | -0.59 | 1.2e-01 | Click! |
Activity profile of PAX3 motif
Sorted Z-values of PAX3 motif
Network of associatons between targets according to the STRING database.
First level regulatory network of PAX3
| Promoter | Score | Transcript | Gene | Gene Info |
|---|---|---|---|---|
| chr4_+_14113592 | 0.92 |
ENST00000502759.1 ENST00000511200.1 ENST00000512754.1 ENST00000506739.1 |
LINC01085 |
long intergenic non-protein coding RNA 1085 |
| chrX_+_102883620 | 0.54 |
ENST00000372626.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chr2_+_152214098 | 0.52 |
ENST00000243347.3 |
TNFAIP6 |
tumor necrosis factor, alpha-induced protein 6 |
| chr12_-_58131931 | 0.43 |
ENST00000547588.1 |
AGAP2 |
ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| chrX_+_102883887 | 0.26 |
ENST00000372625.3 ENST00000372624.3 |
TCEAL1 |
transcription elongation factor A (SII)-like 1 |
| chr2_+_242811874 | 0.22 |
ENST00000343216.3 |
CXXC11 |
CXXC finger protein 11 |
| chr9_+_33795533 | 0.21 |
ENST00000379405.3 |
PRSS3 |
protease, serine, 3 |
| chr9_-_73029540 | 0.21 |
ENST00000377126.2 |
KLF9 |
Kruppel-like factor 9 |
| chr11_-_73694346 | 0.20 |
ENST00000310473.3 |
UCP2 |
uncoupling protein 2 (mitochondrial, proton carrier) |
| chr19_+_38085731 | 0.19 |
ENST00000589117.1 |
ZNF540 |
zinc finger protein 540 |
| chr5_-_180688105 | 0.19 |
ENST00000327767.4 |
TRIM52 |
tripartite motif containing 52 |
| chr11_+_313503 | 0.19 |
ENST00000528780.1 ENST00000328221.5 |
IFITM1 |
interferon induced transmembrane protein 1 |
| chr19_+_10527449 | 0.19 |
ENST00000592685.1 ENST00000380702.2 |
PDE4A |
phosphodiesterase 4A, cAMP-specific |
| chrX_-_48931648 | 0.19 |
ENST00000376386.3 ENST00000376390.4 |
PRAF2 |
PRA1 domain family, member 2 |
| chr10_+_104178946 | 0.17 |
ENST00000432590.1 |
FBXL15 |
F-box and leucine-rich repeat protein 15 |
| chr10_+_134150835 | 0.17 |
ENST00000432555.2 |
LRRC27 |
leucine rich repeat containing 27 |
| chrX_+_37850026 | 0.17 |
ENST00000341016.3 |
CXorf27 |
chromosome X open reading frame 27 |
| chrX_-_53461305 | 0.16 |
ENST00000168216.6 |
HSD17B10 |
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr3_-_58572760 | 0.16 |
ENST00000447756.2 |
FAM107A |
family with sequence similarity 107, member A |
| chr22_+_42372764 | 0.15 |
ENST00000396426.3 ENST00000406029.1 |
SEPT3 |
septin 3 |
| chr9_+_104161123 | 0.15 |
ENST00000374861.3 ENST00000339664.2 ENST00000259395.4 |
ZNF189 |
zinc finger protein 189 |
| chr12_+_8309630 | 0.15 |
ENST00000396570.3 |
ZNF705A |
zinc finger protein 705A |
| chr9_-_13175823 | 0.15 |
ENST00000545857.1 |
MPDZ |
multiple PDZ domain protein |
| chr1_+_171283331 | 0.15 |
ENST00000367749.3 |
FMO4 |
flavin containing monooxygenase 4 |
| chrX_-_53461288 | 0.15 |
ENST00000375298.4 ENST00000375304.5 |
HSD17B10 |
hydroxysteroid (17-beta) dehydrogenase 10 |
| chr21_-_31744557 | 0.15 |
ENST00000399889.2 |
KRTAP13-2 |
keratin associated protein 13-2 |
| chrX_+_135230712 | 0.15 |
ENST00000535737.1 |
FHL1 |
four and a half LIM domains 1 |
| chr5_+_178977546 | 0.14 |
ENST00000319449.4 ENST00000377001.2 |
RUFY1 |
RUN and FYVE domain containing 1 |
| chr19_-_38085633 | 0.14 |
ENST00000593133.1 ENST00000590751.1 ENST00000358744.3 ENST00000328550.2 ENST00000451802.2 |
ZNF571 |
zinc finger protein 571 |
| chr10_+_91087651 | 0.13 |
ENST00000371818.4 |
IFIT3 |
interferon-induced protein with tetratricopeptide repeats 3 |
| chr6_+_127588020 | 0.13 |
ENST00000309649.3 ENST00000610162.1 ENST00000610153.1 ENST00000608991.1 ENST00000480444.1 |
RNF146 |
ring finger protein 146 |
| chr6_+_127587755 | 0.13 |
ENST00000368314.1 ENST00000476956.1 ENST00000609447.1 ENST00000356799.2 ENST00000477776.1 ENST00000609944.1 |
RNF146 |
ring finger protein 146 |
| chrX_+_43515467 | 0.13 |
ENST00000338702.3 ENST00000542639.1 |
MAOA |
monoamine oxidase A |
| chr11_-_615570 | 0.12 |
ENST00000525445.1 ENST00000348655.6 ENST00000397566.1 |
IRF7 |
interferon regulatory factor 7 |
| chrX_+_153770421 | 0.12 |
ENST00000369609.5 ENST00000369607.1 |
IKBKG |
inhibitor of kappa light polypeptide gene enhancer in B-cells, kinase gamma |
| chr9_+_131549483 | 0.12 |
ENST00000372648.5 ENST00000539497.1 |
TBC1D13 |
TBC1 domain family, member 13 |
| chr17_-_8059638 | 0.12 |
ENST00000584202.1 ENST00000354903.5 ENST00000577253.1 |
PER1 |
period circadian clock 1 |
| chr5_+_49961727 | 0.11 |
ENST00000505697.2 ENST00000503750.2 ENST00000514342.2 |
PARP8 |
poly (ADP-ribose) polymerase family, member 8 |
| chr19_+_4343691 | 0.11 |
ENST00000597036.1 |
MPND |
MPN domain containing |
| chr1_-_24151903 | 0.11 |
ENST00000436439.2 ENST00000374490.3 |
HMGCL |
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr14_+_24584508 | 0.11 |
ENST00000559354.1 ENST00000560459.1 ENST00000559593.1 ENST00000396941.4 ENST00000396936.1 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
| chr16_-_71523179 | 0.11 |
ENST00000564230.1 ENST00000565637.1 |
ZNF19 |
zinc finger protein 19 |
| chr19_+_35168567 | 0.10 |
ENST00000457781.2 ENST00000505163.1 ENST00000505242.1 ENST00000423823.2 ENST00000507959.1 ENST00000446502.2 |
ZNF302 |
zinc finger protein 302 |
| chr3_+_38307293 | 0.10 |
ENST00000311856.4 |
SLC22A13 |
solute carrier family 22 (organic anion/urate transporter), member 13 |
| chr15_+_43622843 | 0.10 |
ENST00000428046.3 ENST00000422466.2 ENST00000389651.4 |
ADAL |
adenosine deaminase-like |
| chr1_+_50513686 | 0.09 |
ENST00000448907.2 |
ELAVL4 |
ELAV like neuron-specific RNA binding protein 4 |
| chr19_-_40596828 | 0.09 |
ENST00000414720.2 ENST00000455521.1 ENST00000340963.5 ENST00000595773.1 |
ZNF780A |
zinc finger protein 780A |
| chr14_+_24583836 | 0.09 |
ENST00000559115.1 ENST00000558215.1 ENST00000557810.1 ENST00000561375.1 ENST00000446197.3 ENST00000559796.1 ENST00000560713.1 ENST00000560901.1 ENST00000559382.1 |
DCAF11 |
DDB1 and CUL4 associated factor 11 |
| chr9_+_131549610 | 0.09 |
ENST00000223865.8 |
TBC1D13 |
TBC1 domain family, member 13 |
| chr19_-_48048518 | 0.09 |
ENST00000595558.1 ENST00000263351.5 |
ZNF541 |
zinc finger protein 541 |
| chr6_+_140175987 | 0.09 |
ENST00000414038.1 ENST00000431609.1 |
RP5-899B16.1 |
RP5-899B16.1 |
| chr19_+_35168633 | 0.09 |
ENST00000505365.2 |
ZNF302 |
zinc finger protein 302 |
| chrX_-_15872914 | 0.09 |
ENST00000380291.1 ENST00000545766.1 ENST00000421527.2 ENST00000329235.2 |
AP1S2 |
adaptor-related protein complex 1, sigma 2 subunit |
| chr19_-_57183114 | 0.09 |
ENST00000537055.2 ENST00000601659.1 |
ZNF835 |
zinc finger protein 835 |
| chr16_-_71523236 | 0.09 |
ENST00000288177.5 ENST00000569072.1 |
ZNF19 |
zinc finger protein 19 |
| chr17_-_7835228 | 0.09 |
ENST00000303731.4 ENST00000571947.1 ENST00000540486.1 ENST00000572656.1 |
TRAPPC1 |
trafficking protein particle complex 1 |
| chr1_-_24151892 | 0.08 |
ENST00000235958.4 |
HMGCL |
3-hydroxymethyl-3-methylglutaryl-CoA lyase |
| chr16_-_18468926 | 0.08 |
ENST00000545114.1 |
RP11-1212A22.4 |
LOC339047 protein; Nuclear pore complex-interacting protein family member A3; Nuclear pore complex-interacting protein family member A5; Protein PKD1P1 |
| chr4_+_128982430 | 0.08 |
ENST00000512292.1 ENST00000508819.1 |
LARP1B |
La ribonucleoprotein domain family, member 1B |
| chr22_-_44258360 | 0.08 |
ENST00000330884.4 ENST00000249130.5 |
SULT4A1 |
sulfotransferase family 4A, member 1 |
| chr7_+_142457315 | 0.08 |
ENST00000486171.1 ENST00000311737.7 |
PRSS1 |
protease, serine, 1 (trypsin 1) |
| chr12_+_10460549 | 0.08 |
ENST00000543420.1 ENST00000543777.1 |
KLRD1 |
killer cell lectin-like receptor subfamily D, member 1 |
| chrX_+_103217207 | 0.08 |
ENST00000563257.1 ENST00000540220.1 ENST00000436583.1 ENST00000567181.1 ENST00000569577.1 |
TMSB15B |
thymosin beta 15B |
| chr2_-_44588893 | 0.07 |
ENST00000409272.1 ENST00000410081.1 ENST00000541738.1 |
PREPL |
prolyl endopeptidase-like |
| chr10_+_124739911 | 0.07 |
ENST00000405485.1 |
PSTK |
phosphoseryl-tRNA kinase |
| chr19_+_38085768 | 0.07 |
ENST00000316433.4 ENST00000590588.1 ENST00000586134.1 ENST00000586792.1 |
ZNF540 |
zinc finger protein 540 |
| chr2_+_44589036 | 0.07 |
ENST00000402247.1 ENST00000407131.1 ENST00000403853.3 ENST00000378494.3 |
CAMKMT |
calmodulin-lysine N-methyltransferase |
| chr3_+_183892635 | 0.07 |
ENST00000427072.1 ENST00000411763.2 ENST00000292807.5 ENST00000448139.1 ENST00000455925.1 |
AP2M1 |
adaptor-related protein complex 2, mu 1 subunit |
| chr1_+_99729813 | 0.07 |
ENST00000457765.1 |
LPPR4 |
Lipid phosphate phosphatase-related protein type 4 |
| chr7_+_99816859 | 0.07 |
ENST00000317271.2 |
PVRIG |
poliovirus receptor related immunoglobulin domain containing |
| chr12_+_10460417 | 0.07 |
ENST00000381908.3 ENST00000336164.4 ENST00000350274.5 |
KLRD1 |
killer cell lectin-like receptor subfamily D, member 1 |
| chr18_-_59274139 | 0.07 |
ENST00000586949.1 |
RP11-879F14.2 |
RP11-879F14.2 |
| chr14_+_22964877 | 0.06 |
ENST00000390494.1 |
TRAJ43 |
T cell receptor alpha joining 43 |
| chr6_-_4135693 | 0.06 |
ENST00000495548.1 ENST00000380125.2 ENST00000465828.1 |
ECI2 |
enoyl-CoA delta isomerase 2 |
| chr19_+_37407212 | 0.06 |
ENST00000427117.1 ENST00000587130.1 ENST00000333987.7 ENST00000415168.1 ENST00000444991.1 |
ZNF568 |
zinc finger protein 568 |
| chr9_-_91793675 | 0.06 |
ENST00000375835.4 ENST00000375830.1 |
SHC3 |
SHC (Src homology 2 domain containing) transforming protein 3 |
| chr19_+_4343584 | 0.06 |
ENST00000596722.1 |
MPND |
MPN domain containing |
| chr19_+_35168547 | 0.06 |
ENST00000502743.1 ENST00000509528.1 ENST00000506901.1 |
ZNF302 |
zinc finger protein 302 |
| chr10_+_96162242 | 0.06 |
ENST00000225235.4 |
TBC1D12 |
TBC1 domain family, member 12 |
| chr2_-_44588679 | 0.06 |
ENST00000409411.1 |
PREPL |
prolyl endopeptidase-like |
| chr6_+_31939608 | 0.06 |
ENST00000375331.2 ENST00000375333.2 |
STK19 |
serine/threonine kinase 19 |
| chr2_-_44588624 | 0.06 |
ENST00000438314.1 ENST00000409936.1 |
PREPL |
prolyl endopeptidase-like |
| chr17_-_3461092 | 0.06 |
ENST00000301365.4 ENST00000572519.1 |
TRPV3 |
transient receptor potential cation channel, subfamily V, member 3 |
| chr12_-_56123444 | 0.06 |
ENST00000546457.1 ENST00000549117.1 |
CD63 |
CD63 molecule |
| chr8_+_107670064 | 0.06 |
ENST00000312046.6 |
OXR1 |
oxidation resistance 1 |
| chr19_+_55105085 | 0.06 |
ENST00000251372.3 ENST00000453777.1 |
LILRA1 |
leukocyte immunoglobulin-like receptor, subfamily A (with TM domain), member 1 |
| chr12_-_89920030 | 0.05 |
ENST00000413530.1 ENST00000547474.1 |
GALNT4 POC1B-GALNT4 |
UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 4 (GalNAc-T4) POC1B-GALNT4 readthrough |
| chr20_-_23807358 | 0.05 |
ENST00000304725.2 |
CST2 |
cystatin SA |
| chr2_-_44588694 | 0.05 |
ENST00000409957.1 |
PREPL |
prolyl endopeptidase-like |
| chr7_+_142031986 | 0.05 |
ENST00000547918.2 |
TRBV7-1 |
T cell receptor beta variable 7-1 (non-functional) |
| chr1_+_154244987 | 0.05 |
ENST00000328703.7 ENST00000457918.2 ENST00000483970.2 ENST00000435087.1 ENST00000532105.1 |
HAX1 |
HCLS1 associated protein X-1 |
| chr6_+_35227449 | 0.05 |
ENST00000373953.3 ENST00000440666.2 ENST00000339411.5 |
ZNF76 |
zinc finger protein 76 |
| chr11_-_615942 | 0.05 |
ENST00000397562.3 ENST00000330243.5 ENST00000397570.1 ENST00000397574.2 |
IRF7 |
interferon regulatory factor 7 |
| chr1_+_84630053 | 0.05 |
ENST00000394838.2 ENST00000370682.3 ENST00000432111.1 |
PRKACB |
protein kinase, cAMP-dependent, catalytic, beta |
| chr22_+_20105259 | 0.05 |
ENST00000416427.1 ENST00000421656.1 ENST00000423859.1 ENST00000418705.2 |
RANBP1 |
RAN binding protein 1 |
| chr19_+_2841433 | 0.05 |
ENST00000334241.4 ENST00000585966.1 ENST00000591539.1 |
ZNF555 |
zinc finger protein 555 |
| chr7_+_128399002 | 0.05 |
ENST00000493278.1 |
CALU |
calumenin |
| chr1_+_203444887 | 0.05 |
ENST00000343110.2 |
PRELP |
proline/arginine-rich end leucine-rich repeat protein |
| chr1_-_205180664 | 0.05 |
ENST00000367161.3 ENST00000367162.3 ENST00000367160.4 |
DSTYK |
dual serine/threonine and tyrosine protein kinase |
| chr4_+_8582287 | 0.05 |
ENST00000382487.4 |
GPR78 |
G protein-coupled receptor 78 |
| chr12_-_75603643 | 0.05 |
ENST00000549446.1 |
KCNC2 |
potassium voltage-gated channel, Shaw-related subfamily, member 2 |
| chr10_-_104178857 | 0.04 |
ENST00000020673.5 |
PSD |
pleckstrin and Sec7 domain containing |
| chr13_-_81801115 | 0.04 |
ENST00000567258.1 |
LINC00564 |
long intergenic non-protein coding RNA 564 |
| chr11_+_73358594 | 0.04 |
ENST00000227214.6 ENST00000398494.4 ENST00000543085.1 |
PLEKHB1 |
pleckstrin homology domain containing, family B (evectins) member 1 |
| chr12_-_57352103 | 0.04 |
ENST00000398138.3 |
RDH16 |
retinol dehydrogenase 16 (all-trans) |
| chr1_-_155214475 | 0.04 |
ENST00000428024.3 |
GBA |
glucosidase, beta, acid |
| chr2_-_218706877 | 0.04 |
ENST00000446688.1 |
TNS1 |
tensin 1 |
| chr17_+_35732955 | 0.04 |
ENST00000300618.4 |
C17orf78 |
chromosome 17 open reading frame 78 |
| chr13_+_53191605 | 0.04 |
ENST00000342657.3 ENST00000398039.1 |
HNRNPA1L2 |
heterogeneous nuclear ribonucleoprotein A1-like 2 |
| chr15_+_99791716 | 0.04 |
ENST00000558172.1 ENST00000561276.1 ENST00000331450.5 |
LRRC28 |
leucine rich repeat containing 28 |
| chr7_-_148580563 | 0.04 |
ENST00000476773.1 |
EZH2 |
enhancer of zeste homolog 2 (Drosophila) |
| chrX_-_13835461 | 0.04 |
ENST00000316715.4 ENST00000356942.5 |
GPM6B |
glycoprotein M6B |
| chr1_-_145715565 | 0.04 |
ENST00000369288.2 ENST00000369290.1 ENST00000401557.3 |
CD160 |
CD160 molecule |
| chr1_+_247670415 | 0.04 |
ENST00000366491.2 ENST00000366489.1 ENST00000526896.1 |
GCSAML |
germinal center-associated, signaling and motility-like |
| chr17_+_35732916 | 0.04 |
ENST00000586700.1 |
C17orf78 |
chromosome 17 open reading frame 78 |
| chr6_-_24721054 | 0.04 |
ENST00000378119.4 |
C6orf62 |
chromosome 6 open reading frame 62 |
| chr1_-_7913089 | 0.04 |
ENST00000361696.5 |
UTS2 |
urotensin 2 |
| chr14_+_22217447 | 0.04 |
ENST00000390427.3 |
TRAV5 |
T cell receptor alpha variable 5 |
| chr12_-_53594227 | 0.04 |
ENST00000550743.2 |
ITGB7 |
integrin, beta 7 |
| chr5_+_140480083 | 0.03 |
ENST00000231130.2 |
PCDHB3 |
protocadherin beta 3 |
| chr1_-_155214436 | 0.03 |
ENST00000327247.5 |
GBA |
glucosidase, beta, acid |
| chr12_-_10562356 | 0.03 |
ENST00000309384.1 |
KLRC4 |
killer cell lectin-like receptor subfamily C, member 4 |
| chr7_+_116502605 | 0.03 |
ENST00000458284.2 ENST00000490693.1 |
CAPZA2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr3_+_179040767 | 0.03 |
ENST00000496856.1 ENST00000491818.1 |
ZNF639 |
zinc finger protein 639 |
| chr15_+_99791567 | 0.03 |
ENST00000558879.1 ENST00000301981.3 ENST00000422500.2 ENST00000447360.2 ENST00000442993.2 |
LRRC28 |
leucine rich repeat containing 28 |
| chr15_+_90808919 | 0.03 |
ENST00000379095.3 |
NGRN |
neugrin, neurite outgrowth associated |
| chr19_-_42463418 | 0.03 |
ENST00000600292.1 ENST00000601078.1 ENST00000601891.1 ENST00000222008.6 |
RABAC1 |
Rab acceptor 1 (prenylated) |
| chr15_-_52861157 | 0.03 |
ENST00000564163.1 |
ARPP19 |
cAMP-regulated phosphoprotein, 19kDa |
| chr2_-_25194963 | 0.03 |
ENST00000264711.2 |
DNAJC27 |
DnaJ (Hsp40) homolog, subfamily C, member 27 |
| chr7_+_116502527 | 0.02 |
ENST00000361183.3 |
CAPZA2 |
capping protein (actin filament) muscle Z-line, alpha 2 |
| chr13_+_76445187 | 0.02 |
ENST00000318245.4 |
C13orf45 |
chromosome 13 open reading frame 45 |
| chr2_+_240323439 | 0.02 |
ENST00000428471.1 ENST00000413029.1 |
AC062017.1 |
Uncharacterized protein |
| chr17_-_59668550 | 0.02 |
ENST00000521764.1 |
NACA2 |
nascent polypeptide-associated complex alpha subunit 2 |
| chr15_+_51973680 | 0.02 |
ENST00000542355.2 |
SCG3 |
secretogranin III |
| chr9_+_100395891 | 0.02 |
ENST00000375147.3 |
NCBP1 |
nuclear cap binding protein subunit 1, 80kDa |
| chr1_+_101185290 | 0.02 |
ENST00000370119.4 ENST00000347652.2 ENST00000294728.2 ENST00000370115.1 |
VCAM1 |
vascular cell adhesion molecule 1 |
| chr3_+_14716606 | 0.02 |
ENST00000253697.3 ENST00000435614.1 ENST00000412910.1 |
C3orf20 |
chromosome 3 open reading frame 20 |
| chrX_+_48398053 | 0.02 |
ENST00000537536.1 ENST00000418627.1 |
TBC1D25 |
TBC1 domain family, member 25 |
| chr16_-_21416640 | 0.02 |
ENST00000542817.1 |
NPIPB3 |
nuclear pore complex interacting protein family, member B3 |
| chr17_+_4855053 | 0.02 |
ENST00000518175.1 |
ENO3 |
enolase 3 (beta, muscle) |
| chr15_+_51973550 | 0.02 |
ENST00000220478.3 |
SCG3 |
secretogranin III |
| chr3_+_112709755 | 0.02 |
ENST00000383678.2 |
GTPBP8 |
GTP-binding protein 8 (putative) |
| chr9_-_35650900 | 0.02 |
ENST00000259608.3 |
SIT1 |
signaling threshold regulating transmembrane adaptor 1 |
| chr16_-_21849091 | 0.01 |
ENST00000537951.1 |
NPIPB4 |
nuclear pore complex interacting protein family, member B4 |
| chr19_-_35719609 | 0.01 |
ENST00000324675.3 |
FAM187B |
family with sequence similarity 187, member B |
| chr6_-_169654139 | 0.01 |
ENST00000366787.3 |
THBS2 |
thrombospondin 2 |
| chr12_-_69080590 | 0.01 |
ENST00000433116.2 ENST00000500695.2 |
RP11-637A17.2 |
RP11-637A17.2 |
| chr11_-_104972158 | 0.01 |
ENST00000598974.1 ENST00000593315.1 ENST00000594519.1 ENST00000415981.2 ENST00000525374.1 ENST00000375707.1 |
CASP1 CARD16 CARD17 |
caspase 1, apoptosis-related cysteine peptidase caspase recruitment domain family, member 16 caspase recruitment domain family, member 17 |
| chr8_+_125463048 | 0.01 |
ENST00000328599.3 |
TRMT12 |
tRNA methyltransferase 12 homolog (S. cerevisiae) |
| chr10_+_118305435 | 0.01 |
ENST00000369221.2 |
PNLIP |
pancreatic lipase |
| chr2_+_171034646 | 0.01 |
ENST00000409044.3 ENST00000408978.4 |
MYO3B |
myosin IIIB |
| chr4_-_168155417 | 0.01 |
ENST00000511269.1 ENST00000506697.1 ENST00000512042.1 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr2_-_68479614 | 0.01 |
ENST00000234310.3 |
PPP3R1 |
protein phosphatase 3, regulatory subunit B, alpha |
| chr7_-_19813192 | 0.01 |
ENST00000422233.1 ENST00000433641.1 |
TMEM196 |
transmembrane protein 196 |
| chr20_-_17511962 | 0.01 |
ENST00000377873.3 |
BFSP1 |
beaded filament structural protein 1, filensin |
| chr6_-_30815936 | 0.01 |
ENST00000442852.1 |
XXbac-BPG27H4.8 |
XXbac-BPG27H4.8 |
| chr21_+_34775181 | 0.01 |
ENST00000290219.6 |
IFNGR2 |
interferon gamma receptor 2 (interferon gamma transducer 1) |
| chr2_-_217724767 | 0.01 |
ENST00000236979.2 |
TNP1 |
transition protein 1 (during histone to protamine replacement) |
| chr22_-_21581926 | 0.01 |
ENST00000401924.1 |
GGT2 |
gamma-glutamyltransferase 2 |
| chr4_-_168155730 | 0.01 |
ENST00000502330.1 ENST00000357154.3 |
SPOCK3 |
sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 3 |
| chr14_-_24711806 | 0.00 |
ENST00000540705.1 ENST00000538777.1 ENST00000558566.1 ENST00000559019.1 |
TINF2 |
TERF1 (TRF1)-interacting nuclear factor 2 |
| chr3_+_127770455 | 0.00 |
ENST00000464451.1 |
SEC61A1 |
Sec61 alpha 1 subunit (S. cerevisiae) |
| chr14_-_64804814 | 0.00 |
ENST00000554572.1 ENST00000358599.5 |
ESR2 |
estrogen receptor 2 (ER beta) |
| chr22_+_18593507 | 0.00 |
ENST00000330423.3 |
TUBA8 |
tubulin, alpha 8 |
| chr6_+_28493753 | 0.00 |
ENST00000469384.1 |
GPX5 |
glutathione peroxidase 5 (epididymal androgen-related protein) |
| chr12_-_127544894 | 0.00 |
ENST00000546062.1 ENST00000512624.2 ENST00000540244.1 |
RP11-575F12.1 |
RP11-575F12.1 |
| chr10_-_75226166 | 0.00 |
ENST00000544628.1 |
PPP3CB |
protein phosphatase 3, catalytic subunit, beta isozyme |
| chr5_+_177457525 | 0.00 |
ENST00000511856.1 ENST00000511189.1 |
FAM153C |
family with sequence similarity 153, member C |
| chr15_-_52861323 | 0.00 |
ENST00000569723.1 ENST00000249822.4 ENST00000567669.1 ENST00000569281.2 ENST00000563566.1 ENST00000567830.1 |
ARPP19 |
cAMP-regulated phosphoprotein, 19kDa |
| chr17_+_37356586 | 0.00 |
ENST00000579260.1 ENST00000582193.1 |
RPL19 |
ribosomal protein L19 |
Gene Ontology Analysis
Gene overrepresentation in biological_process category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0070901 | mitochondrial tRNA methylation(GO:0070901) |
| 0.0 | 0.2 | GO:2000110 | negative regulation of macrophage apoptotic process(GO:2000110) |
| 0.0 | 0.1 | GO:0046103 | adenosine catabolic process(GO:0006154) inosine biosynthetic process(GO:0046103) |
| 0.0 | 0.2 | GO:0051414 | response to cortisol(GO:0051414) |
| 0.0 | 0.2 | GO:0006552 | leucine catabolic process(GO:0006552) |
| 0.0 | 0.1 | GO:1901805 | beta-glucoside metabolic process(GO:1901804) beta-glucoside catabolic process(GO:1901805) positive regulation of neuronal action potential(GO:1904457) |
| 0.0 | 0.1 | GO:2000323 | negative regulation of glucocorticoid receptor signaling pathway(GO:2000323) |
| 0.0 | 0.5 | GO:0030728 | ovulation(GO:0030728) |
| 0.0 | 0.2 | GO:0061179 | negative regulation of insulin secretion involved in cellular response to glucose stimulus(GO:0061179) |
| 0.0 | 0.0 | GO:1904772 | hepatocyte homeostasis(GO:0036333) response to tetrachloromethane(GO:1904772) |
| 0.0 | 0.0 | GO:0046005 | positive regulation of circadian sleep/wake cycle, REM sleep(GO:0046005) |
| 0.0 | 0.3 | GO:0035455 | response to interferon-alpha(GO:0035455) |
| 0.0 | 0.1 | GO:0097056 | selenocysteinyl-tRNA(Sec) biosynthetic process(GO:0097056) |
| 0.0 | 0.3 | GO:0009235 | cobalamin metabolic process(GO:0009235) |
Gene overrepresentation in cellular_component category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.1 | 0.3 | GO:0030678 | mitochondrial ribonuclease P complex(GO:0030678) |
Gene overrepresentation in molecular_function category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.2 | GO:0004419 | hydroxymethylglutaryl-CoA lyase activity(GO:0004419) |
| 0.0 | 0.2 | GO:0017077 | oxidative phosphorylation uncoupler activity(GO:0017077) |
| 0.0 | 0.1 | GO:0023024 | MHC class I protein complex binding(GO:0023024) |
| 0.0 | 0.1 | GO:0008131 | primary amine oxidase activity(GO:0008131) |
| 0.0 | 0.1 | GO:0004499 | N,N-dimethylaniline monooxygenase activity(GO:0004499) |
| 0.0 | 0.1 | GO:0004165 | dodecenoyl-CoA delta-isomerase activity(GO:0004165) |
| 0.0 | 0.3 | GO:0003857 | 3-hydroxyacyl-CoA dehydrogenase activity(GO:0003857) |
| 0.0 | 0.5 | GO:0005540 | hyaluronic acid binding(GO:0005540) |
| 0.0 | 0.1 | GO:0004348 | glucosylceramidase activity(GO:0004348) |
| 0.0 | 0.8 | GO:0050699 | WW domain binding(GO:0050699) |
Gene overrepresentation in C2:CP:REACTOME category:
| Log-likelihood per target | Total log-likelihood | Term | Description |
|---|---|---|---|
| 0.0 | 0.1 | REACTOME IRAK1 RECRUITS IKK COMPLEX | Genes involved in IRAK1 recruits IKK complex |
| 0.0 | 0.2 | REACTOME TRAF6 MEDIATED IRF7 ACTIVATION IN TLR7 8 OR 9 SIGNALING | Genes involved in TRAF6 mediated IRF7 activation in TLR7/8 or 9 signaling |
| 0.0 | 0.3 | REACTOME BRANCHED CHAIN AMINO ACID CATABOLISM | Genes involved in Branched-chain amino acid catabolism |